Abstract
The Escherichia coli O45 O-antigen gene cluster of strain O45:H2 96-3285 was sequenced, and conventional (singleplex), multiplex, and real-time PCR assays were designed to amplify regions in the wzx (O-antigen flippase) and wzy (O-antigen polymerase) genes. In addition, PCR assays targeting the E. coli O55 wzx and wzy genes were designed based on previously published sequences. PCR assays targeting E. coli O45 showed 100% specificity for this serogroup, whereas by PCR assays specific for E. coli O55, 97/102 strains serotyped as E. coli O55 were positive for wzx and 98/102 for wzy. Multiplex PCR assays targeting the E. coli O45 and the E. coli O55 wzx and wzy genes were used to detect the organisms in fecal samples spiked at levels of 106 and 108 CFU/0.2 g feces. Thus, the PCR assays can be used to detect and identify E. coli serogroups O45 and O55.
Strains of Escherichia coli belonging to serogroup O45 have been isolated from animals and humans and classified as both enterotoxigenic E. coli and Shiga toxin-producing enterohemorrhagic E. coli (4, 16, 25). In addition, E. coli O45 strains isolated from diarrheic dairy calves produced cytotoxic necrotizing factor and were designated as strains of necrotoxigenic E. coli (10). Many E. coli O45 strains isolated from swine have been associated with postweaning diarrhea and demonstrated the attaching and effacing (A/E) phenotype, allowing them to adhere to intestinal epithelial cells (3). Six E. coli serogroups, including O45, were associated with feral pigeons and produced a variant of Shiga toxin 2 called Stx2f (9).
E. coli strains belonging to serogroup O55 have been classified as human enteropathogenic strains and are believed to be genetically related to enterohemorrhagic E. coli O157:H7, shown to have been derived from an O55:H7 ancestral clone (15, 23). Because of the potential pathogenicity of E. coli isolates belonging to serogroups O45 and O55, rapid and reliable assays for detecting and identifying these serogroups in food, environmental, and clinical samples are needed.
Conventionally, antigenic analysis of the ca. 179 different O serogroups in E. coli is performed by agglutination reactions using antisera raised in rabbits against the O standard reference strains. O serotyping is laborious, and cross-reactions between different serogroups often occur, giving equivocal results. Furthermore, E. coli strains occasionally undergo transition from smooth to rough forms as a result of mutations in one or more of the multiple genes controlling synthesis and polymerization of the O antigen. Rough isolates do not produce an O antigen and therefore cannot be typed by using antisera. In view of these facts, there is a need for the development of alternative methods for identifying and typing E. coli serogroups. Lipopolysaccharides (LPS) are essential components of the gram-negative bacterial outer membrane. They are composed of three parts: lipid A, which is composed of sugars and fatty acids and anchors the LPS in the outer membrane through covalent linkages; an oligosaccharide core made of sugars and sugar derivatives; and a lateral polysaccharide chain (O antigen) responsible for the antigenic specificity of the smooth form of each of the E. coli serogroups. The O antigen contains many repeats (often 10 to 30) of an oligosaccharide unit (O unit) generally composed of three to six sugars. Different combinations of sugars in the O unit, as well as diversity of the chemical linkages between the sugars, determine the diversity of the O antigens. Further levels of variation are conferred by the addition of nonsugar moieties (such as O-acetyl residues or amino acids) and by differences in the modal length of the polysaccharide chains (13).
In E. coli, genes encoding the enzymes involved in O-antigen synthesis are clustered in a chromosomal region referred to as the O-antigen gene cluster (formerly known as the rfb cluster), which is generally found between the galF gene and the gnd gene that encodes the housekeeping enzyme, 6-phosphogluconate dehydrogenase, involved in the pentose phosphate pathway (1, 2, 13). Upstream of the O-antigen gene cluster, there is a highly conserved 39-bp element called the JUMPstart sequence (8). The number of genes in the clusters varies depending on the complexity of the polysaccharide, and strains of different serogroups can show completely different gene sets. PCR-based tests amplifying certain genes in the E. coli O-antigen gene clusters have been found to be serogroup specific (5, 6, 19-22). The objective of this study was to develop rapid and specific PCR assays for detecting and typing E. coli serogroups O45 and O55 by targeting the wzx and wzy genes of the respective O-antigen gene clusters.
The bacterial strains used in the study were from the culture collection of the Gastroenteric Disease Center at The Pennsylvania State University. E. coli O45:H2 strain 96-3285 (Centers for Disease Control and Prevention, Atlanta, GA), which was used for DNA sequencing of the O-antigen gene cluster, and E. coli O55:H6 strain Su 3972-41 (World Health Organization) (12) were used for the development of the PCR assays. The other reference standard strains used belonged to serogroups O1 through O175 with the exceptions of serogroups O31, O47, O72, O93, O94, and O122, which are not designated (12). Fifty-seven strains belonging to E. coli O45, 117 strains belonging to E. coli O55, and 47 non-O45 and non-O55 strains isolated from animals, humans, and the environment from the reference collection at the Gastroenteric Disease Center were used for determining the specificity of the assays. Other bacteria (n = 21) tested included Bacillus cereus, Citrobacter freundii, Enterobacter cloacae, Enterobacter erogenes, Enterococcus faecalis, Hafnia alvei, Klebsiella pneumoniae, Listeria monocytogenes, Pseudomonas aeruginosa, Proteus vulgaris, Salmonella enterica serovars Enterica, Arizonae, Choleraesuis, Enteritidis, and Typhimurium, Serratia marcescens, Staphylococcus aureus, Shigella boydii, Vibrio cholerae, and Yersinia enterocolitica. All bacteria were grown in Luria-Bertani (LB) broth or on LB agar plates.
Sequencing of the O-antigen gene cluster of E. coli O45:H2 was performed as previously described following amplification of the cluster by long PCR using primers targeting the JUMPstart and gnd regions, DNase I digestion, and cloning (6). The assembled sequences were imported into Artemis (14), the open reading frames (ORFs) were located (a cutoff of 150 was used to determine ORFs), and the putative coding regions were ascertained by analyzing the similarity with other published sequences in GenBank. Analysis of the DNA sequence of the 14,483-bp region containing the O45 O-antigen gene cluster showed that it contained 13 complete ORFs, with all having the same transcriptional direction. The genes within the cluster, identified with various degrees of precision and named in accordance with the system proposed by Reeves et al. (13), are shown in Table 1. The E. coli O45 cluster consisted of genes involved in sugar biosynthesis pathways, sugar transferase genes, and O-antigen-processing genes, including the O-antigen flippase gene (wzx) that transports the repeat sugar units across the cytoplasmic membrane and the O-antigen polymerase gene (wzy) that polymerizes the repeat units. The transmembrane regions of the proteins were analyzed as described previously (17, 18). The wzx and wzy genes, located between nucleotides 7144 and 8403 and nucleotides 9366 and 10514, respectively, were selected as targets for PCR assay development. In addition, the E. coli O55 wzx and wzy genes (GenBank accession number AF461121) (21) were also targeted for PCR assay development. Due to the relatively low similarity in Wzx and Wzy among different E. coli serogroups, the genes coding for these enzymes are suitable targets for serogroup-specific PCR assay development (5, 6, 19, 20, 21)
TABLE 1.
Genes in the O-antigen gene cluster of E. coli serogroup O45
| ORF no. | Proposed gene name | Location (nucleotides) | No. of amino acids in gene product | Putative function | Most significant homolog(s) (accession no.) | % Identity/% similarity |
|---|---|---|---|---|---|---|
| 1 | rmlB | 59-1198 | 379 | dTDP-d-glucose-4,6-dehydratase | RmlB, dTDP-glucose 4,6-dehydratase, Salmonella enterica (AAG09513) | 83/87 |
| 2 | wbhP | 1246-2196 | 316 | NAD-dependent epimerase/dehydratase | ORF_12; similar to NAD-dependent epimerase/dehydratase family, Pseudomonas aeruginosa (AAM27579) | 42/58 |
| 3 | wbhQ | 2196-3230 | 344 | Glycosyltransferase | Probable N-acetylgalatosaminyl transferase, trsF (S51265), and WbcO, Yersinia enterocolitica (CAA87703) | 50/68 |
| 4 | wbhR | 3231-3791 | 186 | Acetyltransferase | Acetyltransferase, Oceanobacillus iheyensis (NP_693813) (BAC14847) | 37/58 |
| 5 | wbhS | 3830-5716 | 628 | TrsG protein homolog, role in galactose modification | TrsG protein homolog (T44517), ORF9P (BAA85014), and WbgZ (AAG17416), Plesiomonas shigelloides | 61/79 |
| 6 | rmlC | 5797-6336 | 179 | dTDP-4-dehydrorhamnose-3,5-epimerase | Putative dTDP-4-dehydrorhamnose-3,5-epimerase, Aeromonas hydrophila (AAM22546) | 66/80 |
| 7 | wbhT | 6333-7151 | 272 | dTDP-glucose-4,6-dehydratase | Putative dTDP-glucose-4,6-dehydratase, Aeromonas hydrophila (AAM22547) | 55/69 |
| 8 | wzx | 7144-8403 | 419 | O-antigen flippase | Membrane protein involved in the export of O-antigen and teichoic acid, 12 transmembrane domains, Burkholderia fungorum (ZP_00278649) | 25/44 |
| 9 | wbhU | 8387-9352 | 321 | Glycosyltransferase | Putative glycosyltransferase, Pyrococcus furiosus (NP_579088) | 29/49 |
| 10 | wzy | 9366-10514 | 382 | O-antigen polymerase | Transmembrane protein, 9 transmembrane domains, Ralstonia solanacearum (NP_519421.1) | 27/45 |
| 11 | wbhV | 10507-11043 | 178 | Serine acetyltransferase | Putative acetyltransferase, Clostridium thermocellum (ZP_00313397.1) | 37/60 |
| 12 | wbhW | 11045-12055 | 336 | Unknown | Eps11, Streptococcus thermophilus (AAN63793) | 30/49 |
| 13 | rmlA | 12102-12974 | 290 | Glucose-1-phosphate thymidylyltransferase | RmlA, Raoultella terrigena (AAQ82933.1) | 78/89 |
Template DNA for the PCR assays was prepared by mixing a colony from the LB agar in sterile distilled water and heating at 100°C for 20 min in a heating block. The suspension was centrifuged at 13,000 × g for 5 min, and the supernatant containing the DNA was used for the PCR. PCR assays were developed by using the primers listed in Table 2, designed by using the Primer3 software program, for amplifications of regions within the wzx and wzy genes in the O-antigen gene clusters of E. coli O45 and O55 (21). For the singleplex (wzx or wzy) PCR assays, each of the primer sets shown in Table 2 was used separately in the PCRs, and reaction mix contents for each PCR (11-μl total reaction mix volume) consisted of 3 μl of template DNA, 0.5 μM of primers (Integrated DNA Technologies Inc., Coralville, IA), 0.18 mM concentration of each of the four deoxynucleoside triphosphates, 2 mM MgCl2 (for the O55 wzx, O45 wzx, and O45 wzy PCR assays) and 3 mM MgCl2 (for the O55 wzy PCR assays), 0.4 U of Taq DNA polymerase (PGC Scientific, Gaithersburg, MD), 50 mM Tris (pH 8.3), 250 μg/ml bovine serum albumin, 2% sucrose, and 0.1 mM cresol red. The PCR was performed in a RapidCycler (Idaho Technologies Inc., Salt Lake City, UT) by using a rapid-cycle DNA amplification method (24) and consisted of 30 cycles of template denaturation at 94°C, primer annealing and primer extension at temperatures and times indicated in Table 3. The amplification products were subjected to electrophoresis in 1% agarose gels at 200 V for 1 h for all assays except for the O55 wzy PCR that was analyzed using a 2% gel. The gels were stained with ethidium bromide and visualized under UV light. Positive samples were identified based on the presence of bands of the expected sizes compared to results with O45 and O55 control standard reference strains.
TABLE 2.
Oligonucleotide primers and probes
| Target gene | Sequence (5′ to 3′)a | Location in cluster (nucleotides) | Amplicon size (bp) | Accession no. or source |
|---|---|---|---|---|
| Singleplex and multiplex PCR | ||||
| O45 wzx1 | (F) CCG GGT TTC GAT TTG TGA AGG TTG | 7769-8295 | 527 | This study |
| (R) CAC AAC AGC CAC TAC TAG GCA GAA | ||||
| O45 wzx2 | (F) TAT GAC AGG CAC ATG GAT CTG TGG | 7347-7601 | 255 | |
| (R) TTG AGA CGA GCC TGG CTT TGA TAC | ||||
| O45 wzy1 | (F) GAA ATT ATG CCA TCT TGG CGA GCG | 9570-10066 | 497 | This study |
| (R) CAT GTG AAG CCT GAA GGC AAA CTC | ||||
| O45 wzy2 | (F) CTG ATG TCA GGC CTC GTG GAA ATA | 9858-10308 | 451 | |
| (R) ATG TAA CCA CAA TAA GGG AGC CCG | ||||
| O55 wzx1 | (F) AAT GGA ACA TTG CAA CAG CA | 10746-10895 | 150 | AF461121 |
| (R) TGT GGA TTC CAG AAA AGC AA | ||||
| O55 wzx2 | (F) TCT TGT AAC TAA GTG GCC ACA GGC | 10823-11505 | 683 | |
| (R) ATA ACA CCC AAC CTA TAC CTC CCG | ||||
| O55 wzy1 | (F) GTG GTT TTG ACG ACT CGC TT | 9380-9526 | 147 | AF461121 |
| (R) CCA AAA AGC CCT GCA ACT AA | ||||
| Real-time PCR | ||||
| O45 wzx | (F) CGT TGT GCA TGG TGG CAT | 7472-7543 | 72 | This study |
| (R) TGG CCA AAC CAA CTA TGA ACT G | ||||
| O45 wzy | (F) GGT GCT TTG TGA TAA TTC CTG ATG | 9616-9691 | 76 | This study |
| (R) TTA TAG CCG CCC CTA AAT TGC | ||||
| O45 wzx probe | 6-FAM d(ATT TTT TGC TGC AAG TGG GCT GTC CA)BHQ-1 | 7494-7519 | This study | |
| O45 wzy probe | 6-FAM d(TTG CTG CTG GCG GGA TAC CAA TGA T)BHQ-1 | 9643-9667 | This study | |
| O55 wzx | (F) AAT TAA CGA ACA TAA CAC CCA ACC | 11416-11516 | 101 | AF461121 |
| (R) ATA TCT CTT CGT TAC TGT GTG TAT TTC | ||||
| O55 wzy | (F) AGC TTT CCT GGC GGG TTT | 10118-10204 | 87 | AF461121 |
| (R) GCA CCA CGC TAT CTT TTT TCT TAA T | ||||
| O55 wzx probe | 6-FAM d(ACC TCC CGC TAA AAC CCC AAC TCT AGT AG)BHQ-1 | 11461-11489 | AF461121 | |
| O55 wzy probe | 6-FAM d(CCG CGG CGA TAT TGG GTA CTG C)BHQ-1 | 10142-10163 |
F, forward; R, reverse.
TABLE 3.
PCR conditions used for the different assays
| PCR gene(s) | Primer or primer pair | Temp (°C) [(time in s)]
|
Amplicon size(s) (bp) | |
|---|---|---|---|---|
| Annealing | Extension | |||
| Singleplex PCR | ||||
| O45 wzx | O45wzx1, O45wzx2 | 59 (0) | 72 (30) | 527, 255 |
| O45 wzy | O45wzy1, O45wzy2 | 59 (0) | 72 (30) | 497, 451 |
| O55 wzx | O55wzx1, O55wzx2 | 50 (0) | 72 (8) | 150, 683 |
| O55 wzy | O55wzy1 | 50 (0) | 72 (8) | 147 |
| Multiplex PCR | ||||
| O45 (wzx and wzy) | O45wzx2, O45wzy2 | 59 (0) | 72 (30) | 255, 451 (Fig. 1A) |
| O55 (wzx and wzy) | O55wzx2, O55wzy1 | 55 (0) | 72 (32) | 683, 147 (Fig. 1B) |
| O45 (wzx) and O55 (wzx) | O45wzx2, O55wzx2 | 55 (0) | 72 (32) | 255, 683 (not shown) |
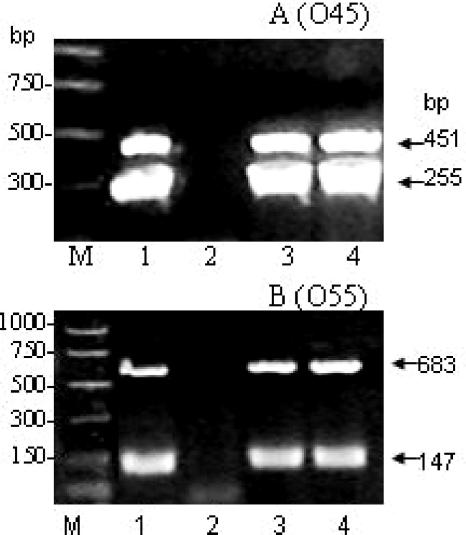
A multiplex PCR assay targeting the E. coli O45 wzx and wzy genes (Fig. 1) was performed by using primers O45wzx2 and O45wzy2 (Table 2) at 20 μM concentrations for amplifying O45 wzx, following the thermocycling conditions presented in Table 3. A multiplex PCR assay targeting the E. coli O55 wzx and wzy genes (Fig. 1) was performed using primers O55wzx2 and O55wzy1 (Table 2) at 50 μM and 20 μM concentrations, respectively, according to the PCR conditions listed in Table 3. To establish that the multiplex PCR assay could be used to detect E. coli O45 or O55 in environmental samples, chicken feces (0.2 g) were spiked with E. coli belonging to either serogroup O45 or O55 at 106 and 108 CFU concentrations. The fecal matter was vortexed until homogeneous, and the DNA was extracted from the samples using the QIAamp DNA stool mini kit (QIAGEN, Inc.). Multiplex PCR assays were performed, and the results are shown in Fig. 1. Multiplex PCRs targeting O45 wzx and O55 wzx genes were also performed (data not shown) for rapid detection of these serogroups by use of primer pairs for O45 wzx2 and O55 wzx2 (Table 2) using the PCR conditions described in Table 3. Multiplex PCRs were as sensitive and specific as singleplex PCR.
FIG. 1.
Multiplex PCRs targeting the E. coli O45 wzx and wzy (A) and O55 wzx and wzy (B) genes. (A) Lane M, molecular weight markers; lane 1, E. coli O45 (positive control) (multiplex PCR showing the amplified wzx [255 bp] and wzy [451 bp] genes); lane 2, E. coli K-12 (negative control); lane 3, results from a chicken fecal sample spiked with E. coli O45 at 106 CFU/0.2 g; lane 4, result from a chicken fecal sample spiked with E. coli O45 at 108 CFU/0.2 g. (B) Lane M, molecular weight markers; lane 1, E. coli O55 (positive control) (multiplex PCR showing the amplified wzx [683 bp] and wzy [147 bp] genes); lane 2, E. coli K-12, (negative control); lane 3, results from a chicken fecal sample spiked with E. coli O55 at 106 CFU/0.2 g; lane 4, results from a chicken fecal sample spiked with E. coli O55 at 108 CFU/0.2 g.
TaqMan-based real-time PCR assays were also developed for detection of E. coli O45 and O55 targeting the wzx and wzy genes. Because the assay allows the detection of pathogens in real time during DNA amplification, this method is faster than conventional PCR and is advantageous for diagnostic purposes. Primers and probes were designed based on DNA sequences of the E. coli O45 wzx and wzy genes determined in the current study and on the O55 wzx and wzy genes as described previously (21). The TaqMan probes were labeled with a fluorescent reporter dye, 6-carboxyfluorescein (FAM), at the 5′ end and a quencher dye, BHQ1, at the 3′ end (Biosearch Technologies Inc., CA). The primers and probes used for the assays are depicted in Table 2. The real-time PCR assays were performed using the ABI PRISM 7700 sequence detection system. The reaction mix contained 25 μl of Universal Master Mix (P/N 4304437; Applied Biosciences Inc.), 2 μl of each primer (10 μM), 8 μl of probe (1 μM), and 42 μl of water. DNA from pure cultures was purified by using the QIAquick PCR purification kit (QIAGEN Inc., CA) by following the manufacturer's protocol. DNA (8 μl) was added to the mix, and the tubes were subjected to 50°C for 2 min and 95°C for 10 min, followed by 45 cycles of 95°C for 15 s and 60°C for 1 min. The tubes were held at 25°C for 2 min, and the fluorescence generated by the sequence-specific probes was measured. The ΔRn is the fluorescence signal increase due to template amplification. The amplification plots were generated with the ΔRn mean value on the y axis and the cycle number on the x axis (7). The threshold cycle is the cycle number at which the reporter fluorescence generated by the cleavage of the probe passes a fixed threshold value above baseline. A standard deviation of 10 units above baseline was used to determine the fixed threshold. The real-time PCR assays developed for E. coli O45 reproducibly showed a detectable fluorescence signal above the threshold at 14 cycles for 107 copies of wzx and wzy, and for E. coli O55, fluorescence above threshold was generated at 13 cycles for wzx at 108 copies of DNA and at 15 cycles for 107 copies of wzy.
The specificities of the PCR assays targeting the E. coli O45 and O55 wzx and wzy genes were determined by using 168 O standard reference strains. Only E. coli O45 and O55 standard reference strains were positive for the O45 wzx and wzy and O55 wzx and wzy genes, respectively. Fifty-seven strains belonging to serogroup O45 collected from different sources over the last twenty years exhibited the presence of wzx and wzy by the PCR. Forty-seven randomly selected E. coli isolates belonging to serogroups other than O45 were negative for the presence of the E. coli O45 wzx and wzy genes. On the other hand, when 119 isolates belonging to E. coli serogroup O55 selected from our reference collection isolated during the past thirty-seven years were tested for the presence of the O55 wzx and wzy genes, 17 isolates out of 119 (14.2%) were negative for both genes. The 17 isolates that did not exhibit the presence of O55 wzx and wzy were again serotyped by conventional agglutination reactions using 179 different O antisera raised in rabbits against the standard reference strains as described previously (11). Out of the 17 isolates that were previously designated O55, 9 cross-reacted with antisera raised against E. coli O83, O55, and O22, 4 cross-reacted with O55 and O83, 2 cross-reacted with O55 and O23, and 2 showed a very weak reaction with the O55 antiserum, indicating that these strains may not be O55 but could be related to some other O-antigenic types (Table 4). It is not uncommon to find cultures that cross-react with O22, O83, O55, and O23 antisera. In our large collection of E. coli strains, there were 29 isolates that cross-reacted with O83 and O22, 8 cultures that cross-reacted with O55 and O83, 5 cultures that cross-reacted with O22 and O55, and 3 strains that cross-reacted with O23 and O55 antisera. Therefore, these 17 isolates were probably not O55 and cross-reacted with the O55 antiserum. Long PCR performed with all of these isolates, targeting the regions between gnd and JUMPstart, exhibited product sizes that were different than that of the reference O55 strain. EcoRI was used to digest the amplified gene cluster of all these isolates, and restriction fragment length polymorphism (RFLP) profiles were determined. While the RFLP pattern for O55 standard strain exhibited five fragments of the expected sizes, none of the other isolates showed a profile similar to the standard (data not shown). It was apparent that these 17 isolates did not belong to serogroup O55. Therefore, the PCR assays were more specific for E. coli serogroup O55 strains than conventional serotyping. None of the 21 non-E. coli bacteria exhibited the presence of the O45 and O55 wzx or wzy genes by the PCR exhibiting the specificity of the reactions.
TABLE 4.
Specificities of 119 putative E. coli O55 isolates and 68 non-O55 E. coli isolates and other bacteria
| Serological reaction | No. of cultures | Sources (no.)b | Conclusions from RFLP analyses of O-antigen gene cluster | No. of positive isolates
|
|
|---|---|---|---|---|---|
| O55 wzx PCR | O55 wzy PCR | ||||
| O55 | 102 | Humans, animals, and environment | Not tested | 97 | 98 |
| O55, O83 | 4 | Cows (3) and unlisted (1) | Not O55 | 0 | 0 |
| O55, O83, O22 | 9 | Humans (7), mouse (1), and unlisted (1) | Not O55 | 0 | 0 |
| O55, O23 | 2 | Chicken (1) and environment (1) | Not O55 | 0 | 0 |
| O55 (weak reaction)a | 2 | Sea gull (2) | Not O55 | 0 | 0 |
| Non-O55 | 47 | Human, animals, and environment | Not tested | 0 | 0 |
| Non-E. coli | 21 | ATCC and unknown | Not tested | 0 | 0 |
These strains may not be O55 but may be an unidentified O type and/or related to O55.
ATCC, American Type Culture Collection.
We have developed conventional (singleplex), multiplex, and real-time PCR assays for detection and typing of E. coli serogroups O45 and O55. The assays were found to be highly specific for the respective serogroups and can potentially replace conventional serotyping assays that are time-consuming and less specific. The PCR assays were used to detect E. coli O45 and O55 serogroup strains in spiked fecal samples and can potentially be used for detecting the presence of these serogroups in food, fecal, and environmental samples.
Nucleotide sequence accession number.
The DNA sequence of the 14,483-bp region containing the O45 O-antigen gene cluster has been assigned GenBank accession no. AY771223.
Acknowledgments
We thank Connie Briggs, Lori Fortis, and Kimberly Lynch (USDA Agricultural Research Service, Eastern Regional Research Center, Wyndmoor, PA) for their assistance in performing the DNA sequencing of the O45 O-antigen gene cluster and James Kundrat (Gastroenteric Disease Center, The Pennsylvania State University, University Park, PA) for his assistance in performing the PCR assays.
REFERENCES
- 1.Bastin, D. A., G. Stevenson, A. H. Brown, and P. R. Reeves. 1993. Repeat unit polysaccharides of bacteria: a model for polymerization resembling that of ribosomes and fatty acid synthetase with novel mechanism for determining chain length. Mol. Microbiol. 7:725-734. [DOI] [PubMed] [Google Scholar]
- 2.Batchelor, R. A., P. Alifano, E. Biffali, S. I. Hull, and R. A. Hull. 1992. Nucleotide sequences of the genes regulating O-polysaccharide antigen chain length (rol) from Escherichia coli and Salmonella typhimurium: protein homology and functional complementation. J. Bacteriol. 174:5228-5236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batisson, I., M. P. Guimond, F. Girard, H. An, C. Zhu, E. Oswald, J. M. Fairbrother, M. Jacques, and J. Harel. 2003. Characterization of the novel factor paa involved in the early steps of the adhesion mechanism of attaching and effacing Escherichia coli. Infect. Immun. 71:4516-4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanco, J., M. Blanco, J. I. Garabal, and E. A. Gonzalez. 1991. Enterotoxins, colonization factors and serotypes of enterotoxigenic Escherichia coli from humans and animals. Microbiologica 7:52-73. [PubMed] [Google Scholar]
- 5.DebRoy, C., E. Roberts, J. Kundrat, M. A. Davis, C. E. Briggs, and P. M. Fratamico. 2004. Detection of Escherichia coli serogroups O26 and O113 by PCR amplification of the wzx and wzy genes. Appl. Environ. Microbiol. 70:1830-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fratamico, P. M., C. E. Briggs, D. Needle, C. Chen, and C. DebRoy. 2003. Sequence of the Escherichia coli O121 O-antigen gene cluster and detection of enterohemorrhagic E. coli O121 by PCR amplification of wzx and wzy genes. J. Clin. Microbiol. 41:3379-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibson, U. E. M., C. A. Heid, and P. M. Williams. 1996. A novel method for real time quantitative RT-PCR. Genome Res. 6:995-1001. [DOI] [PubMed] [Google Scholar]
- 8.Hobbs, M., and P. R. Reeves. 1994. The JUMPstart sequence: 39 bp element common to several polysaccharide gene clusters. Mol. Microbiol. 12:855-856. [DOI] [PubMed] [Google Scholar]
- 9.Morabito, S., G. Dell'Omo, U. Agrimi, H. Schmidt, H. Karch, T. Cheasty, and A. Caprioli. 2001. Detection and characterization of Shiga-toxin producing Escherichia coli in feral pigeons. Vet. Microbiol. 82:275-283. [DOI] [PubMed] [Google Scholar]
- 10.Orden, J. A., J. A. Ruiz-Santa-Quiteria, D. Cid, S. Garcia, and R. de la Fuente. 1999. Prevalence and characteristics of necrotoxigenic Escherichia coli (NTEC) strains isolated from diarrhoeic dairy calves. Vet. Microbiol. 66:265-273. [DOI] [PubMed] [Google Scholar]
- 11.Orskov, F., and I. Orskov. 1975. Escherichia coli O:H serotypes isolated from human blood. Acta Pathol. Microbiol. Scand. Sect. B 83:595-600. [PubMed] [Google Scholar]
- 12.Orskov, I., F. Orskov, and K. Jann. 1977. Serology, chemistry and genetics of O and K antigens of Escherichia coli. Bacteriol. Rev. 41:667-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reeves, P. R., M. Hobbs, M. A. Valvano, M. Skurnik, C. Whitfield, D. Coplin, N. Kido, J. Klena, D. Maskell, C. R. H. Raetz, and P. D. Rick. 1996. Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 4:495-503. [DOI] [PubMed] [Google Scholar]
- 14.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M.-A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 15.Tarr, P. I., L. M. Schoening, Y. Lea, T. R. Ward, S. Jelacic, and T. A. Whittam. 2000. Acquisition of the rfb-gnd cluster in evolution of Escherichia coli O55 and O157. J. Bacteriol. 182:6183-6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toth, I., V. Karcagi, B. Nagi, I. Gado, and H. Milch. 1994. Examination of verocytotoxin producing capacity and determination of the presence of Shiga-like toxin genes in human Escherichia coli isolates. Acta Microbiol. Immunol. Hung. 41:259-264. [PubMed] [Google Scholar]
- 17.Tusnady, G. E., and I. Simon. 1998. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J. Mol. Biol. 283:489-506. [DOI] [PubMed] [Google Scholar]
- 18.Tusnady, G. E., and I. Simon. 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics 17:849-850. [DOI] [PubMed] [Google Scholar]
- 19.Wang, L., C. E. Briggs, D. Rothemund, P. Fratamico, J. B. Luchansky, and P. R. Reeves. 2001. Sequence of the E. coli O104 antigen gene cluster and identification of O104 specific genes. Gene 270:231-236. [DOI] [PubMed] [Google Scholar]
- 20.Wang, L., H. Curd, W. Qu, and P. R. Reeves. 1998. Sequencing of Escherichia coli O111 O-antigen gene cluster and identification of O111-specific genes. J. Clin. Microbiol. 36:3182-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang, L., S. Huskic, A. Cisterne, D. Rothemund, and P. R. Reeves. 2002. The O-antigen gene cluster of Escherichia coli O55:H7 and identification of a new UDP-GlcNAc C4 epimerase. J. Bacteriol. 184:2620-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang, L., and P. R. Reeves. 1998. Organization of Escherichia coli O157 O antigen gene cluster and identification of its specific genes. Infect. Immun. 66:3545-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whittam, T., M. Wolfe, I. Wachsmuth, F. Orskov, I. Orskov, and R. A. Wilson. 1993. Clonal relationship among Escherichia coli strains that cause hemorrhagic colitis and infantile diarrhea. Infect. Immun. 61:1619-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wittwer, C. T., G. B. Reed, and K. M. Ririe. 1994. Rapid cycle DNA amplification, p. 174-181. In K. B. Mullis, F. Ferré, and R. A. Gibbs (ed.), The polymerase chain reaction. Birkauser, Boston, Mass.
- 25.Zhao, S., D. G. White, B. Ge, S. Ayers, S. Friedman, L. English, D. Wagner, S. Gaines, and J. Meng. 2001. Identification and characterization of integron mediated antibiotic resistance among Shiga toxin-producing Escherichia coli isolates. Appl. Environ. Microbiol. 67:1558-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]