Abstract
To assess the source and public health significance of Cryptosporidium oocyst contamination in storm runoff, a PCR-restriction fragment length polymorphism technique based on the small-subunit rRNA gene was used in the analysis of 94 storm water samples collected from the Malcolm Brook and N5 stream basins in New York over a 3-year period. The distribution of Cryptosporidium in this study was compared with the data obtained from 27 storm water samples from the Ashokan Brook in a previous study. These three watersheds represented different levels of human activity. Among the total of 121 samples analyzed from the three watersheds, 107 were PCR positive, 101 of which (94.4%) were linked to animal sources. In addition, C. hominis (W14) was detected in six samples collected from the Malcolm Brook over a 2-week period. Altogether, 22 Cryptosporidium species or genotypes were found in storm water samples from these three watersheds, only 11 of which could be attributed to known species/groups of animals. Several Cryptosporidium spp. were commonly found in these three watersheds, including the W1 genotype from an unknown animal source, the W4 genotype from deer, and the W7 genotype from muskrats. Some genotypes were found only in a particular watershed. Aliquots of 113 samples were also analyzed by the Environmental Protection Agency (EPA) Method 1623; 63 samples (55.7%) were positive for Cryptosporidium by microscopy, and 39 (78%) of the 50 microscopy-negative samples were positive by PCR. Results of this study demonstrate that molecular techniques can complement traditional detection methods by providing information on the source of contamination and the human-infective potential of Cryptosporidium oocysts found in water.
Waterborne cryptosporidiosis has been reported worldwide and remains one of the prominent public health concerns (22). Cryptosporidium spp. are a threat to water supplies because they are resistant to chlorine disinfections, have a small infectious dose, and are harbored by many animal species (4). Farm animals and humans have been considered major sources of contamination of Cryptosporidium oocysts in surface water (15, 20, 36). Thus, controlling agricultural and human sewage discharge is important in watershed protection. However, wildlife are also commonly infected (1, 18, 21, 26) and can be a source of water contamination with Cryptosporidium oocysts (25). Controlling wildlife contamination remains largely beyond the reach of water management efforts.
With the exception of C. hominis (previously known as the C. parvum human genotype or genotype I) (17), which almost exclusively infects humans, C. parvum (previously known as the C. parvum bovine genotype or genotype II) can infect not only humans but also ruminants and perhaps a few other animals (27). Some researchers believe the natural ecology of C. parvum probably involves at least two cycles; one is a zoonotic cycle in agricultural settings involving humans and farm animals, particularly dairy cattle and sheep (2, 29, 30), and the other is a cycle with transmission among wild mammals (21, 26). Although wildlife can contribute to Cryptosporidium contamination in water through aquatic activities, runoff, or snowmelt, recent molecular characterizations of Cryptosporidium from wildlife indicate that most wild mammals are infected with host-adapted species or genotypes (37). Thus, most wild mammals are probably not infected with C. parvum.
Limited studies have examined Cryptosporidium loading in rivers during stormy weather (12, 31). After rainfall or snowmelt, there are often massive increases in the turbidity of creeks in mountain ranges, which is frequently used as an indication of microbial contamination. The number of Giardia cysts and Cryptosporidium oocysts in water is known to increase due to runoff after storms (7, 9). Rainfall can transport wildlife stools deposited in high areas into the watershed, which in turn increases the contamination in source water. This sudden loading of pathogens contributes to the occurrence of spikes in the level of Cryptosporidium contamination in a watershed, most of which is probably of wildlife origin. Thus, it is important to monitor and characterize Cryptosporidium spp. in storm water as part of the scientific management and protection of watersheds.
This study compares the distribution of Cryptosporidium species in the storm water of three watersheds located in the state of New York. Results of the study indicate that ecologic settings of the watershed affect the distribution of various Cryptosporidium spp. in water and that most Cryptosporidium spp. found in storm runoff are from wildlife and are not known human pathogens.
MATERIALS AND METHODS
Sample collection.
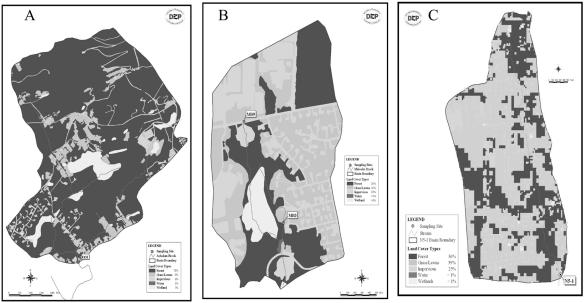
All storm samples for this study were collected from stream subbasins within the New York City water supply in New York. Initial samples were collected from the Ashokan Brook between May 1999 and March 2000, in the Catskill region, and results were reported previously (31). More recent samples were collected from the Malcolm Brook between March 2002 and December 2003 and the N5 basin between November 2002 and April 2004. Malcolm Brook and the N5 basin are located in the Kensico Reservoir basin in Valhalla, N.Y. The storm water samples were taken from two sampling sites in Malcolm Brook, MB9 and MB3. Site MB9 is located upstream at the headwaters of the brook, the drainage of which flows through only grass/lawn areas. Site MB3 is located at the lower section of the brook (Fig. 1). Although the Malcolm Brook watershed has many large wooded areas (39% of the total area), it also has corporate parks (22%) and relatively high-density suburban residential lots (34%) with public sewer systems. The N5 stream basin is very close to the Malcolm Brook basin and is almost entirely residential lots (91%). The area of the N5 stream basin is 298 acres compared to only 95 acres for the Malcolm Brook. In contrast, the Ashokan Brook watershed (878 acres) is mostly an undeveloped and forested area (88%). These differences in land use provided a good foundation for a comparison of contamination sources (Table 1; Fig. 1).
FIG. 1.
Differences in environmental settings among the Ashokan Brook (A), Malcolm Brook (B),and N5 basin (C) examined in this study. For each watershed, dark areas on the maps are wooded areas, white areas are wetlands, and gray areas are residential lots and office parks. Small rectangular boxes on the map indicate the four sampling sites.
TABLE 1.
Differences in land usage among the three studied watersheds in New York
| Ashokan Brook | Malcolm Brook | N5 basin | |
|---|---|---|---|
| Area (acres) | 878 | 95 | 298 |
| Residential | 12% | 34% | 91% |
| Office park | <1% | 22% | 4% |
| Recreation | <1% | 5% | <1% |
| Wooded area | 88% | 39% | 4% |
Water samples were collected with preset autosamplers (model 6700; ISCO, Inc. Lincoln, NE) that were designed to capture significant precipitation events. These events were characterized by periods of rainfall or rapid snowmelt. In general, autosamplers were set to trigger when either a predetermined flow rate was reached or the stream reached two times the base flow in the fall, winter, and spring, and three times the base flow in the summer. Once the trigger value was reached, subsequent 1,000-ml samples were collected at volume-weighted intervals based on the predicted intensity of the event. These intervals of collection were predicted with the main objective of collecting several aliquots over the period of the rising limb of the hydrograph to optimize the capture of Cryptosporidium oocysts. Each 1,000-ml aliquot was combined into a single 20-liter carboy, which was the total target volume for each sample. After collection, the 20-liter carboys were put in coolers on ice, not allowed to freeze, and delivered to the Pathogen Laboratory of the New York City Department of Environmental Protection (NYC DEP) for initial processing.
Sample processing.
Samples were filtered through an Envirochek HV filter (Pall Gelman Laboratory, Ann Arbor, MI) in the laboratory using procedures described in U.S. Environmental Protection Agency (EPA) Method 1623 (28) whereby the 20-liter carboy contents from each storm were captured on the filter. Material on the filter was eluted, and the eluate was centrifuged to combine the oocysts to a concentrated pellet. Once the total pellet size was measured, 0.5 ml was saved at the NYC DEP laboratory for oocyst enumeration following procedures described in Method 1623 whereby Cryptosporidium oocysts were ultimately identified by apple-green fluorescence, size, and shape and confirmed by differential interference contrast microscopy. The remaining sample concentrate was sealed and transported to the laboratory at the Centers for Disease Control and Prevention for molecular analysis. Genotyping for each sample was done without the knowledge of microscopy results.
DNA extraction.
For DNA extraction, Cryptosporidium oocysts were isolated from 0.5-ml water pellets by immunomagnetic separation using magnetic beads coated with an anti-Cryptosporidium monoclonal antibody (Dynal, Lake Success, N.Y.) and manufacturer-recommended procedures. Magnetic beads without dissociation of oocysts were directly used in DNA extraction with the QIAamp DNA mini kit (QIAGEN, Valencia, Calif.). Briefly, 180 μl ATL buffer from the kit was added into 1.5-ml tubes containing the beads and subjected to five freeze-thaw cycles at −70°C and 56°C for at least 1 h to break the oocyst wall. The manufacturer-recommended protocol was followed for the reminder of the DNA extraction.
PCR-restriction fragment length polymorphism (RFLP).
Each sample was analyzed by a small-subunit (SSU) rRNA-based nested-PCR previously described (32, 33, 35, 36) with six different volumes of DNA template (0.5, 1.0, 1.5, 2.0, 2.5, and 3.0 μl), with the exception of the use of 400 ng/μl of nonacetylated bovine serum albumin (Sigma, St. Louis, MO) in primary PCR and the use of a modified reverse primer (5′ CTC ATA AGG TGC TGA AGG AGT A 3′) in secondary PCR. The secondary PCR products were visualized by 1.5% agarose gel electrophoresis. For the restriction fragment analysis, 5 μl of the secondary PCR products was digested at 37°C for 1 h in a total of 40 μl of reaction mixture, which contained 20 U of SspI (New England BioLabs, Beverly, Mass.) or VspI (Promega, Madison, Wis.). The digested products were fractionated on 2.0% agarose gel and visualized by ethidium bromide staining.
Sequence analysis.
After purification by Microcon PCR (Amicon Inc., Beverly, Mass.), the secondary PCR products were sequenced directly with secondary PCR primers using an ABI 3100 autosequencer (PerkinElmer, Foster City, Calif.). Sequence accuracy was confirmed by two-directional sequencing, and sequencing of another PCR product of the same genotype from the same sample if an unusual sequence for the genotype was obtained. For a few samples, mixed genotypes (judged by RFLP profiles) were concurrently present in some PCR products. For these samples, only PCR products with a single RFLP profile were sequenced to avoid the generation of erroneous sequences. Nucleotide sequences obtained were aligned with reference Cryptosporidium sequences using the ClustalX 1.81 package (ftp://ftp-igbmc.u-strasbg.fr/pub/ClustalX/) and the default setting. Phylogenetic analysis was carried out to assess the relationship between parasites in storm water samples and known Cryptosporidium spp. Neighbor-joining trees were constructed using the TreeCon package (http://www.psb.rug.ac.be/bioinformatics/psb/Userman/treeconw.html), based on the evolutionary distances between sequences calculated by the Kimura two-parameter model. An SSU rRNA sequence of Eimeria tenella (GenBank accession no. AF026388) was used as the outgroup. The reliability of various clusters was evaluated by the bootstrap method with 1,000 pseudoreplicates.
Nucleotide sequence accession numbers.
The GenBank accession numbers of the SSU rRNA reference sequences used in this study for alignments and phylogenetic tree construction were as follows: AF026388 for Eimeria tenella, AY007254 for Cryptosporidium genotype W12, AF262329 for Cryptosporidium genotype W2, AF262331 for Cryptosporidium genotype W6, AF093498 for C. muris, AB089285 for C. andersoni, AY120912 for Cryptosporidium goose genotype I, AY504515 for Cryptosporidium goose genotype II, AY504514 for Cryptosporidium duck genotype, AY168847 for C. galli, AF093502 for C. serpentis, AY524773 for C. molnari, AY120914 for Cryptosporidium tortoise genotype, AY120913 for Cryptosporidium snake genotype, AF093495 for C. baileyi, AY120910 for Cryptosporidium deer genotype, AY120911 for Cryptosporidium bovine genotype B, AF093489 for C. hominis, AF164102 for C. parvum, AY120901 for Cryptosporidium rabbit genotype, AF112571 for Cryptosporidium mouse genotype, AF112572 for Cryptosporidium ferret genotype, AF329187 for C. meleagridis, AF115378 for C. wrairi, AY120903 for Cryptosporidium skunk genotype, AF112570 for Cryptosporidium marsupial genotype I, AY120902 for Cryptosporidium opossum genotype I, AY120906 for Cryptosporidium opossum genotype II, AF247535 for Cryptosporidium bear genotype, AJ493209 for C. canis, AY120904 for Cryptosporidium muskrat genotype I, AY545546, AY545547, and AY545548 for Cryptosporidium muskrat genotype II, AF108862 for C. felis, and AF115377 for Cryptosporidium pig genotype I. Unique partial SSU rRNA sequences obtained from storm water during the study were deposited in the GenBank database under accession numbers AY737556-AY737603.
RESULTS
Comparison of efficiency of microscopy and PCR methods in detection of Cryptosporidium spp. in storm water samples.
A total of 50 storm water samples were collected from the Malcolm Brook between March 2002 and December 2003. With the exception of four samples from the MB3 sampling site, the number of oocysts was determined by EPA Method 1623 before PCR identification. All six samples from site MB9 were negative by both microscopy and the SSU rRNA-based nested PCR methods. Among 40 samples from site MB3, 22 (55%) were positive by microscopy, with oocyst numbers ranging from 1 to 61 per sample, whereas 41 of 44 samples (93.2%) were positive by PCR (Table 2). All 22 microscopy-positive samples were positive in PCR detection. Three of the four samples from the MB3 site that were not analyzed by microscopy detection were positive by PCR. Thus, 41 of 44 samples (93.2%) from site MB3 were positive by PCR, with 3 PCR-negative samples also being negative by microscopy.
TABLE 2.
Comparison of the performance between microscopy and PCR in the detection of Cryptosporidium spp. in storm water samples from the Ashokan, Malcolm, and N5 basins
| Watershed | Sampling site | Total no. of samples | No. of samples
|
Total no. of genotypes | No. of genotypes per sample
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Microscopy positive | PCR positive | 0 | 1 | 2 | 3 | 4 | >4 | ||||
| Malcolm Brook | MB3 | 44 | 22/40 | 41/44 | 14/22 | 3 | 15 | 13 | 10 | 2 | 1 |
| Malcolm Brook | MB9 | 6 | 0/6 | 0/6 | 0/22 | 6 | 0 | 0 | 0 | 0 | 0 |
| N5 basin | N51 | 44 | 27/40 | 40/44 | 18/22 | 4 | 9 | 8 | 6 | 8 | 9 |
| Ashokan Brook | E13i | 27 | 14/27 | 26/27 | 12/22 | 1 | 14 | 10 | 2 | 0 | 0 |
Results of 44 storm water samples from the N5 basin collected between November 2002 and April 2004 were similar to those of the Malcolm Brook. Forty of 44 samples (90.9%) were positive by PCR (Table 2). Among 40 samples analyzed by microscopy, 27 samples (67.5%) were positive, with oocyst numbers ranging from 1 to 25 per sample, whereas 4 samples that were not analyzed by microscopy due to limited volume of samples were positive by PCR. Twenty-five of the 27 microscopy-positive samples were positive by PCR.
Similar results were also found in the previous study of the Ashokan Brook (31), in which 26 of 27 samples (96.3%) were positive by the PCR method, including all microscopy-positive samples and 12 of 13 samples that were negative by microscopy (Table 2). Because 0.5 ml of concentrated water pellet per sample was analyzed by both microscopy and PCR, these results suggest that the SSU rRNA-based PCR method is more sensitive than microscopy.
Characterization of Cryptosporidium spp. in storm water samples by RFLP.
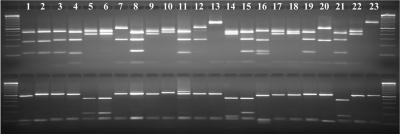
All PCR products were digested with restriction enzymes SspI and VspI to differentiate Cryptosporidium spp. in storm water. Multiple banding patterns were seen for both SspI and VspI. Five obvious SspI banding patterns were seen, depending on the number and size of the bands (Fig. 2). One group had only one large visible SspI band (Fig. 2, lane 13 and 23; C. galli, not shown). Three groups had two visible SspI bands, with each group having different sizes of the two bands (Fig. 2, lanes 7, 12, and 20 versus lanes 5, 6, 9, 14, 17, 18 and 22 or lane 10). The fifth group had three visible SspI bands (Fig. 2, lanes 1, 2, 3, 4, 8, 11, 15, 16, 19, and 21). Minor differences in the size of each band were also seen between members of the sample group, but it was difficult to differentiate them without sequencing. Likewise, four VspI banding patterns were seen among the samples. One group had only two visible VspI bands (Fig. 2, lanes 2, 3, 4, 7 through 13, 17 through 20, 22, and 23; C. galli, not shown). Three groups had three visible VspI bands, with each group having different sizes of the three bands (Fig. 2, lanes 5, 6, 15 and 21 versus lanes 1 and 16 or lane 14). Most samples had more than one banding pattern in RFLP analyses of multiple PCR products (each sample was analyzed six times by PCR using different volumes of DNA), suggesting the presence of mixed genotypes.
FIG. 2.
Differentiation of Cryptosporidium in storm water samples by an SSU rRNA-based PCR-RFLP. Secondary PCR products were digested by SspI (upper panel) or VspI (lower panel) and visualized by 2.0% agarose gel electrophoresis. Lanes 1 and 2, C. hominis and C. parvum, respectively; lanes 3 through 23, storm water Cryptosporidium genotypes W1, W2, W3, W4, W5, W6, W7, W8, W9, W10, W11, W12, W13, W14, W15, W16, W17, W18, W19, W20, and W21, respectively. The lower SspI band in lane 16 was an artifact present in the PCR product. The upper VspI band in lane 11 was due to the presence of another minor genotype in the sample. The RFLP pattern for W22 genotype is not shown.
Distribution of Cryptosporidium genotypes in three watersheds.
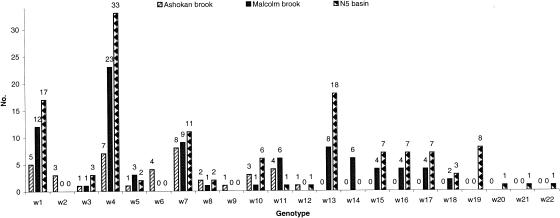
Storm water samples from three different environmental and ecologic settings of watersheds were used in this study to compare the distribution of Cryptosporidium species or genotypes. All PCR-positive products with single RFLP profiles were sequenced to confirm the diagnosis. Altogether, combining these with the 12 Cryptosporidium genotypes identified in the previous study of the Ashokan Brook, a total of 22 Cryptosporidium genotypes were identified in the three watersheds. The dominant Cryptosporidium species or genotypes in three watersheds (Fig. 3) were the W4 (cervine) genotype from deer (63/121), the W1 genotype from an unknown animal (34/121), and the W7 genotype from muskrats (28/121). Two Cryptosporidium genotypes (W2 from opossums and W6 from an unknown animal) were detected only in the Ashokan Brook samples. Likewise, four Cryptosporidium genotypes (W19, W20, and W21 from unknown animals and W22 [C. galli]) were detected in only the N5 basin. Five common Cryptosporidium genotypes were detected in the Malcolm Brook and the N5 basin, W13 from skunks and raccoons (26/94), W15 (11/94), W17 (11/94), and W18 (5/94) from unknown animals and W16 (11/94) from muskrats, and were not detected in the samples from the Ashokan Brook. In addition, the detection of C. hominis (W14 genotype), an anthroponotic parasite, was noted in six samples collected from three storm events at the Malcolm Brook over a 2-week period (Fig. 3), whereas C. hominis was not detected in the Ashokan Brook (31) nor the N5 basin. Twenty-six of the 41 PCR-positive samples from the Malcolm Brook (MB3 sampling site), 31 of the 40 PCR-positive samples from the N5 basin, and 12 of the 26 PCR-positive samples from the Ashokan Brook had multiple Cryptosporidium genotypes in the repeated PCR analysis of samples (Table 2). Nevertheless, most PCR products had only a single dominant Cryptosporidium genotype, which allowed the sequencing of the PCR products.
FIG. 3.
Distribution of Cryptosporidium genotypes in storm water from the Ashokan, Malcolm, and N5 basins, New York. Ashokan data were from a previous study (31) of the E13i sampling site. The genotypes in the Malcolm Brook were detected between March 2002 and December 2003 at the MB3 sampling site since all samples from another sampling site (MB9) were negative. The genotypes in the N5 basin were detected between November 2002 and April 2004. Numbers above the bars represent the numbers of times each genotype was detected.
Phylogenic relationship of Cryptosporidium spp. in storm water.
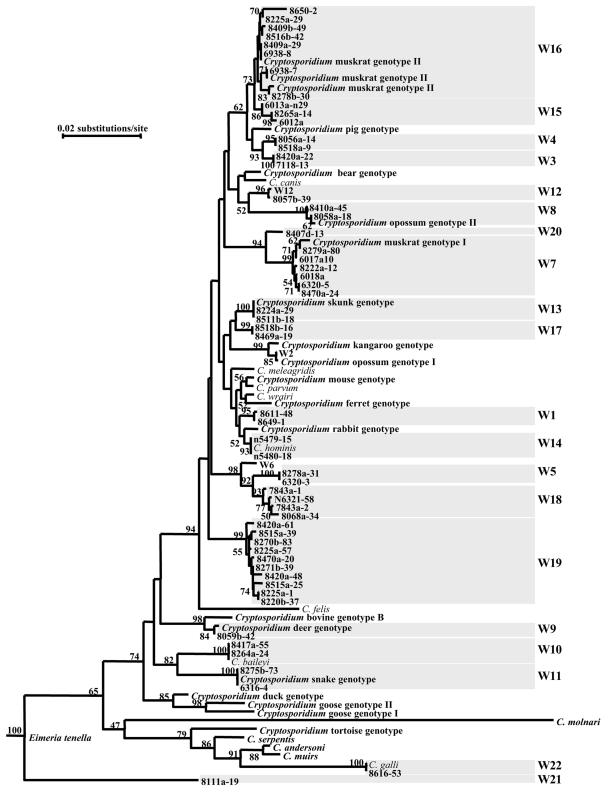
A multiple-sequence alignment in the range of 193 to 1,043 residues of C. hominis (GenBank accession number AF093489), which included the most polymorphic region of the SSU rRNA gene, was used in the confirmation of genotype identification and the assessment of the phylogenetic relationship among Cryptosporidium genotypes found in the study. Most storm water genotypes (W1 to W20) were clustered in the group containing the intestinal Cryptosporidium parasites with high bootstrap value support (74%) (Fig. 4). Only W22 (C. galli), which was found only once in the N5 basin, clustered with the gastric group. However, one genotype (W21) did not group with either intestinal or gastric clusters of Cryptosporidium spp. (36), despite the fact that the results of GenBank BLAST indicated that the W21 genotype was a Cryptosporidium (data not shown). Genetic distances among Cryptosporidium genotypes were in the range of 0.36 to 11.16 nucleotide substitutions per 100 bp, but most genotypes differed from each other by more than 1%, which is more than the differences between C. parvum and C. hominis (0.71%) (Table 3). Therefore, genotypes W1 to W22 were probably different species of Cryptosporidium. Presently, the designation of Cryptosporidium species is based on morphological and biological differences and genetic characterizations (34). Because of the lack of morphological and biological characterizations, many Cryptosporidium genotypes currently have no species designation (34). Even though the genetic distance between W3 and W4 was 0.36 nucleotide substitution per 100 bp (Table 3), phylogenetic analysis showed that these two genotypes formed two separate clades with 95 to 100% bootstrap support (Fig. 4).
FIG. 4.
Phylogenetic relationship among various Cryptosporidium genotypes in storm water samples and known Cryptosporidium species or genotypes, as inferred by a neighbor-joining analysis of the SSU rRNA sequences. The evolutionary distances between sequences were calculated by the Kimura two-parameter model. A sequence of Eimeria tenella (AF026388) was used as the outgroup. Numbers at branches are percent bootstrapping values (>50) using 1,000 replicates.
TABLE 3.
Genetic distances among Cryptosporidium genotypes (W1 to W22) in storm watera
| Genotypeb | C. hominis | C. parvum | W1 | W2 | W3 | W4 | W5 | W6 | W7 | W8 | W9 | W10 | W11 | W12 | W13 | W14 | W15 | W16 | W17 | W18 | W19 | W20 | W21 | W22 | E. tenella |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C. hominis | 0.00 | ||||||||||||||||||||||||
| C. parvum | 0.71 | 0.00 | |||||||||||||||||||||||
| W1 | 0.95 | 0.71 | 0.00 | ||||||||||||||||||||||
| W2 | 2.29 | 2.04 | 1.80 | 0.00 | |||||||||||||||||||||
| W3 | 1.81 | 2.18 | 2.42 | 2.67 | 0.00 | ||||||||||||||||||||
| W4 | 1.94 | 2.18 | 2.43 | 2.55 | 0.36 | 0.00 | |||||||||||||||||||
| W5 | 2.81 | 2.69 | 2.81 | 3.06 | 2.81 | 2.82 | 0.00 | ||||||||||||||||||
| W6 | 2.06 | 1.94 | 2.06 | 2.19 | 2.19 | 2.19 | 1.32 | 0.00 | |||||||||||||||||
| W7 | 3.17 | 3.18 | 3.29 | 2.80 | 2.17 | 2.17 | 3.57 | 3.06 | 0.00 | ||||||||||||||||
| W8 | 3.65 | 3.54 | 3.52 | 3.16 | 2.65 | 2.54 | 3.80 | 3.17 | 2.38 | 0.00 | |||||||||||||||
| W9 | 4.22 | 3.84 | 3.96 | 3.71 | 4.23 | 4.23 | 3.95 | 3.19 | 4.89 | 4.35 | 0.00 | ||||||||||||||
| W10 | 4.72 | 4.59 | 4.72 | 4.46 | 4.58 | 4.46 | 4.32 | 3.93 | 5.11 | 4.71 | 3.06 | 0.00 | |||||||||||||
| W11 | 4.98 | 4.86 | 4.98 | 4.47 | 4.72 | 4.73 | 4.32 | 3.82 | 5.25 | 5.24 | 3.57 | 2.68 | 0.00 | ||||||||||||
| W12 | 2.43 | 2.43 | 2.55 | 3.56 | 1.55 | 1.80 | 3.43 | 2.93 | 2.90 | 3.15 | 4.47 | 5.22 | 4.71 | 0.00 | |||||||||||
| W13 | 1.68 | 1.44 | 1.68 | 2.17 | 1.93 | 1.93 | 2.30 | 1.32 | 3.05 | 3.15 | 3.57 | 4.31 | 4.72 | 2.05 | 0.00 | ||||||||||
| W14 | 0.12 | 0.59 | 0.83 | 2.17 | 1.81 | 1.81 | 2.68 | 1.94 | 3.05 | 3.53 | 4.09 | 4.59 | 4.85 | 2.30 | 1.55 | 0.00 | |||||||||
| W15 | 1.94 | 1.81 | 2.06 | 2.45 | 1.45 | 1.45 | 3.19 | 2.44 | 2.43 | 2.56 | 4.75 | 4.98 | 5.25 | 2.30 | 2.44 | 1.82 | 0.00 | ||||||||
| W16 | 1.82 | 1.94 | 2.06 | 2.82 | 0.72 | 0.84 | 2.82 | 2.20 | 2.44 | 2.19 | 4.36 | 4.60 | 5.13 | 1.94 | 1.57 | 1.70 | 0.72 | 0.00 | |||||||
| W17 | 1.19 | 0.96 | 1.32 | 1.68 | 1.94 | 1.69 | 1.94 | 1.32 | 2.43 | 3.16 | 3.46 | 4.20 | 4.21 | 2.18 | 0.83 | 1.07 | 2.32 | 1.82 | 0.00 | ||||||
| W18 | 2.69 | 2.56 | 2.81 | 3.06 | 2.95 | 2.70 | 1.20 | 1.44 | 3.58 | 3.68 | 3.83 | 4.07 | 4.21 | 3.57 | 2.43 | 2.56 | 3.33 | 2.96 | 2.19 | 0.00 | |||||
| W19 | 2.19 | 2.20 | 2.44 | 2.44 | 1.69 | 1.69 | 2.81 | 2.20 | 2.44 | 2.81 | 3.97 | 4.32 | 4.73 | 2.55 | 2.19 | 2.07 | 2.32 | 2.07 | 1.95 | 2.57 | 0.00 | ||||
| W20 | 2.56 | 2.57 | 2.68 | 2.07 | 2.18 | 2.18 | 2.45 | 2.20 | 0.94 | 2.52 | 4.25 | 4.61 | 4.88 | 3.18 | 2.31 | 2.44 | 2.32 | 2.32 | 1.82 | 2.58 | 2.32 | 0.00 | |||
| W21 | 10.05 | 9.41 | 9.68 | 9.13 | 9.44 | 9.05 | 8.71 | 8.57 | 10.15 | 10.49 | 8.83 | 8.66 | 8.96 | 9.85 | 9.61 | 9.92 | 9.06 | 9.20 | 9.11 | 8.20 | 9.31 | 8.94 | 0.00 | ||
| W22 | 9.07 | 8.40 | 8.81 | 8.54 | 8.43 | 8.71 | 8.56 | 8.55 | 8.72 | 8.52 | 8.05 | 6.30 | 6.96 | 8.69 | 8.91 | 8.94 | 8.44 | 8.58 | 8.39 | 8.46 | 8.43 | 8.34 | 11.16 | 0.00 | |
| E. tenella | 23.20 | 23.07 | 24.04 | 22.98 | 23.05 | 22.81 | 21.06 | 21.97 | 22.94 | 24.01 | 22.04 | 21.54 | 20.77 | 22.98 | 23.22 | 23.07 | 21.63 | 22.36 | 23.10 | 21.27 | 21.79 | 22.02 | 21.95 | 20.77 | 0.00 |
Genetic distances (nucleotide substitutions 100 bp) based on Kimura two-parameter analysis were calculated with sequences corresponding to residues 193 to 1043 of C. hominis (accession number AF093489).
Intragenotypic variations in Cryptosporidium genotypes.
Sequence heterogeneity was observed in genotypes W4, W7, W15, W16, W18, and W19. Most of the sequence variations occurred in the highly polymorphic region (residues 624 to 797 of C. hominis). These variations included nucleotide substitutions, deletions, and insertions (Fig. 5). Intragenotypic variations in the SSU rRNA gene were most common in genotypes W7, W16, and W19. Nevertheless, phylogenetic analysis showed that these intragenetic variable sequences clustered together for each genotype (Fig. 4).
FIG. 5.
Intragenotypic variations in the W4, W7, W15, W16, W18, and W19 genotypes in the highly polymorphic region (nucleotides 624 to 698 and 726 to 797 of C. hominis, accession number AF093489) of the SSU rRNA gene. Dots denote nucleotides identical to the first sequence, and dashes indicate deletions.
DISCUSSION
Currently, the identification of Cryptosporidium oocysts in environmental samples relies largely on immunofluorescent microscopy (IFA) using the EPA ICR method or Method 1622/1623 or the United Kingdom regulatory method, which is time consuming, labor intensive, and expensive (11). Recent studies (3, 6, 8, 10, 13, 14, 24) have shown that the use of molecular methods, such as PCR-RFLP, has some advantages over the traditional IFA detection method. This and the previous study (31) clearly indicate that detection of Cryptosporidium in storm water samples by PCR is more sensitive than the IFA method. Other studies have also shown that PCR has higher sensitivities in the analysis of both clinical and environmental samples (3, 6, 8, 10, 13, 14, 24). It is possible that some oocysts present in storm samples were not stained by immunofluorescence due to the loss of surface antigens as a result of long exposure to the adverse environment. It has been shown that most antibodies used in the immunofluorescence detection of Cryptosporidium oocysts recognize carbohydrate epitopes on the oocyst wall (16, 38) which are labile to chlorine treatment and other environmental conditions without the loss of oocyst viability (16).
The other advantage of PCR over microscopy is the ability of PCR tools to differentiate Cryptosporidium spp. by incorporating RFLP analysis, DNA sequencing, or other molecular procedures, which allows the differentiation of Cryptosporidium species or genotypes. In contrast, because antibodies used in IFA can react with most known Cryptosporidium spp. surface antigens (5, 38), IFA cannot differentiate Cryptosporidium species from each other. The presence of only a few Cryptosporidium species in humans and host-adapted Cryptosporidium species or genotypes in animals makes it possible to assess the human-infective potential and animal sources of Cryptosporidium oocysts in water, as demonstrated in the genetic characterization of Cryptosporidium oocysts in storm water samples in this and a previous study (31).
Results of genetic characterizations of Cryptosporidium in storm water collected from the Malcolm, N5, and Ashokan basins of New York indicate that wildlife is the major source of Cryptosporidium contamination in these areas. Altogether, 22 Cryptosporidium genotypes were found in the three watersheds, most of which have never been found in humans or domestic animals before and some of which are known pathogens of wild animals. Some Cryptosporidium species or genotypes commonly found in animals were seen in these three watersheds, including W4 (cervine genotype) and W9 (deer genotype) from deer, W2 (opossum genotype I) and W8 (opossum genotype II) from opossums, W7 (muskrat genotype I) from muskrats, W13 (skunk genotypes) from skunks and raccoons, W16 (muskrat genotype II) from muskrats, W10 (C. baileyi) from birds, W11 (snake genotype) from snakes, and W22 (C. galli) from birds. All these animals are common in feral and suburban areas. Quite a few of these genotypes (W1, W3, W5, W6, W12, W15, W17, W18, W19, W20, and W21) found in storm water represent new Cryptosporidium spp. Since they have never been seen in humans or domestic animals before, they are likely parasites of wild animals.
With the exception of one Cryptosporidium genotype (W21), other Cryptosporidium genotypes in storm water (W1 to W20 and W22) clustered together with the intestinal and gastric groups of Cryptosporidium spp. Genotype W21 was found only in the presence of W1, W4, and W13 in one N5 basin sample. W21 had big genetic distances to other storm water genotypes, from 8.20 to 10.49 nucleotide substitutions per 100 bp, compared to the greater than 20% nucleotide substitution between Cryptosporidium and Eimeria tenella (Table 3). Nevertheless, the results of GenBank BLAST analysis indicated that the W21 genotype was Cryptosporidium (data not shown). The newly described C. molnari in fish also formed a separate branch in phylogenetic analysis in a recent study (23). Even though C. molnari formed a cluster with gastric Cryptosporidium in this study; the C. molnari branch was very long, indicating that the placement of C. molnari in Fig. 4 might not be accurate. Thus, like C. molnari, W21 may also represent a more ancient Cryptosporidium from fish or amphibians.
The environmental and ecologic settings of watersheds apparently affected the distribution of Cryptosporidium genotypes in water. The N5 basin catchment consists mostly of high-density residential lots (less than 4% wooded area and less than 1% wetland). The Malcolm Brook catchment consists of relatively high-density suburban residential lots (approximately four lots per acre) with public sewer systems, corporate office parks, and substantial grass or forested areas (39% wooded land). In contrast, the Ashokan Brook drainage basin consists of mostly undisturbed grass or forested areas (88%), and differs in topography, fauna, and flora from the Malcolm Brook and N5 basins (Table 1). As expected, even though samples from three watersheds all contained Cryptosporidium spp. from some common wild animals such as deer (W4), muskrats (W7), opossums (W8), birds (W10), and snakes (W11), the distribution of other Cryptosporidium genotypes differed among three watersheds. This was probably a reflection of different animal activities in these three different subbasins. Also, the occurrence of Cryptosporidium spp. varied in different sample locations within the same brook. The MB3 sampling site of the Malcolm Brook is in the lower segment of the brook and flows through areas of human and wildlife activities. In contrast, the MB9 site lies in the upper stream and flows through only grassland. All samples from MB9 were negative by both the IFA and PCR methods, whereas 41 of 44 samples (94.1%) from MB3 were positive for Cryptosporidium by PCR.
Most of the Cryptosporidium genotypes in storm water identified in the Malcolm, N5, and Ashokan basins probably do not have high public health importance. The finding of mostly wildlife Cryptosporidium genotypes in runoff even at sites with extensive residential development (N5 basin and Malcolm Brook) is somewhat surprising. Nevertheless, these results reaffirm the importance of wildlife feces in the contamination of watershed by Cryptosporidium oocysts, especially after storms or rainfall. Of the 22 genotypes identified in three watersheds, only W4 (cervine genotype) and W14 (C. hominis) are two known human pathogens. Humans are infected mostly with five Cryptosporidium spp., including C. hominis, C. parvum, C. meleagridis, C. felis, and C. canis, with the former two most prevalent (32). Even though C. hominis was found in the Malcolm Brook, which is consistent with the existence of residential and office complexes in the surrounding area of the watershed, it was found in only a few samples over a brief 2-week period. The cervine genotype was more prevalent and was found in all three watersheds. This parasite, however, has been found in only two human cases in Canada (19), and thus is unlikely to be a major human pathogen and probably requires higher numbers of oocysts to infect humans than C. hominis and C. parvum.
In summary, results of this study demonstrate that the SSU rRNA-based PCR-RFLP tool can be used effectively in the analysis of environmental samples and can complement the traditional detection methods by providing data with greater sensitivity on the sources and human-infective potential of Cryptosporidium oocysts in water. Results of the study also indicate that Cryptosporidium oocysts in storm runoff from both protected watersheds and watersheds with human activities are likely to be mostly from wildlife and not human infective. Thus, the human health impact could be overestimated if Cryptosporidium oocysts in water are detected only by microscopy or if risk assessment models do not consider the nature of Cryptosporidium in water. Periodic determination of the species of Cryptosporidium oocysts in the watershed or source water may be helpful in the development of strategies for the scientific management and protection of source water.
Acknowledgments
This study was supported in part by funds from the Awwa Research Foundation.
We acknowledge the work of the NYC DEP Pathogen Laboratory staff for microscopy results and the Pathogen and Hydrology field groups for sample collection during storm events.
REFERENCES
- 1.Deng, M. Q., and D. O. Cliver. 1999. Improved immunofluorescence assay for detection of Giardia and Cryptosporidium from asymptomatic adult cervine animals. Parasitol. Res. 85:733-736. [DOI] [PubMed] [Google Scholar]
- 2.Faubert, G. M., and Y. Litvinsky. 2000. Natural transmission of Cryptosporidium parvum between dams and calves on a dairy farm. J. Parasitol. 86:495-500. [DOI] [PubMed] [Google Scholar]
- 3.Fayer, R., J. M. Trout, E. J. Lewis, M. Santin, L. Zhou, A. A. Lal, and L. Xiao. 2003. Contamination of Atlantic coast commercial shellfish with Cryptosporidium. Parasitol. Res. 89:141-145. [DOI] [PubMed] [Google Scholar]
- 4.Fox, K. R., and D. A. Lytle. 1996. Milwaukee's crypto outbreak: investigation and recommendations. J. Am. Water Assoc. 88:87-94. [Google Scholar]
- 5.Graczyk, T. K., M. R. Cranfield, and R. Fayer. 1996. Evaluation of commercial enzyme immunoassay (EIA) and immunofluorescent antibody (FA) test kits for detection of Cryptosporidium oocysts of species other than Cryptosporidium parvum. Am. J. Trop. Med. Hyg. 54:274-279. [DOI] [PubMed] [Google Scholar]
- 6.Hallier-Soulier, S., and E. Guillot. 2000. Detection of Cryptosporidia and Cryptosporidium parvum oocysts in environmental water samples by immunomagnetic separation-polymerase chain reaction. J. Appl. Microbiol. 89:5-10. [DOI] [PubMed] [Google Scholar]
- 7.Hansen, J. S., and J. E. Ongerth. 1991. Effects of time and watershed characteristics on the concentration of Cryptosporidium oocysts in river water. Appl. Environ. Microbiol. 57:2790-2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson, D. W., N. J. Pieniazek, D. W. Griffin, L. Misener, and J. B. Rose. 1995. Development of a PCR protocol for sensitive detection of Cryptosporidium oocysts in water samples. Appl. Environ. Microbiol. 61:3849-3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kistemann, T., T. Classen, C. Koch, F. Dangendorf, R. Fischeder, J. Gebel, V. Vacata, and M. Exner. 2002. Microbial load of drinking water reservoir tributaries during extreme rainfall and runoff. Appl. Environ. Microbiol. 68:2188-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kostrzynska, M., M. Sankey, E. Haack, C. Power, J. E. Aldom, A. H. Chagla, S. Unger, G. Palmateer, H. Lee, J. T. Trevors, and S. A. De Grandis. 1999. Three sample preparation protocols for polymerase chain reaction based detection of Cryptosporidium parvum in environmental samples. J. Microbiol. Methods 35:65-71. [DOI] [PubMed] [Google Scholar]
- 11.LeChevallier, M. W., W. D. Norton, J. E. Siegel, and M. Abbaszadegan. 1995. Evaluation of the immunofluorescence procedure for detection of Giardia cysts and Cryptosporidium oocysts in water. Appl. Environ. Microbiol. 61:690-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loge, F. J., D. E. Thompson, and D. R. Call. 2002. PCR detection of specific pathogens in water: a risk-based analysis. Environ. Sci. Technol. 36:2754-2759. [DOI] [PubMed] [Google Scholar]
- 13.Lowery, C. J., J. E. Moore, B. C. Millar, K. A. McCorry, J. Xu, P. J. Rooney, and J. S. Dooley. 2001. Occurrence and molecular genotyping of Cryptosporidium spp. in surface waters in Northern Ireland. J. Appl. Microbiol. 91:774-779. [DOI] [PubMed] [Google Scholar]
- 14.Lowery, C. J., P. Nugent, J. E. Moore, B. C. Millar, X. Xiru, and J. S. Dooley. 2001. PCR-IMS detection and molecular typing of Cryptosporidium parvum recovered from a recreational river source and an associated mussel (Mytilus edulis) bed in Northern Ireland. Epidemiol. Infect. 127:545-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meinhardt, P. L., D. P. Casemore, and K. B. Miller. 1996. Epidemiologic aspects of human cryptosporidiosis and the role of waterborne transmission. Epidemiol. Rev. 18:118-136. [DOI] [PubMed] [Google Scholar]
- 16.Moore, A. G., G. Vesey, A. Champion, P. Scandizzo, D. Deere, D. Veal, and K. L. Williams. 1998. Viable Cryptosporidium parvum oocysts exposed to chlorine or other oxidising conditions may lack identifying epitopes. Int. J. Parasitol. 28:1205-1212. [DOI] [PubMed] [Google Scholar]
- 17.Morgan-Ryan, U. M., A. Fall, L. A. Ward, N. Hijjawi, I. Sulaiman, R. Fayer, R. C. Thompson, M. Olson, A. Lal, and L. Xiao. 2002. Cryptosporidium hominis n. sp. (Apicomplexa: Cryptosporidiidae) from Homo sapiens. J. Eukaryot. Microbiol. 49:433-440. [DOI] [PubMed] [Google Scholar]
- 18.O'Donoghue, P. J. 1995. Cryptosporidium and cryptosporidiosis in man and animals. Int. J. Parasitol. 25:139-195. [DOI] [PubMed] [Google Scholar]
- 19.Ong, C. S., D. L. Eisler, A. Alikhani, V. W. Fung, J. Tomblin, W. R. Bowie, and J. L. Isaac-Renton. 2002. Novel Cryptosporidium genotypes in sporadic cryptosporidiosis cases: first report of human infections with a cervine genotype. Emerg. Infect. Dis. 8:263-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ono, K., H. Tsuji, S. K. Rai, A. Yamamoto, K. Masuda, T. Endo, H. Hotta, T. Kawamura, and S. Uga. 2001. Contamination of river water by Cryptosporidium parvum oocysts in western Japan. Appl. Environ. Microbiol. 67:3832-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perz, J. F., and S. M. Le Blancq. 2001. Cryptosporidium parvum infection involving novel genotypes in wildlife from lower New York State. Appl. Environ. Microbiol. 67:1154-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rose, J. B., D. E. Huffman, and A. Gennaccaro. 2002. Risk and control of waterborne cryptosporidiosis. FEMS Microbiol. Rev. 26:113-123. [DOI] [PubMed] [Google Scholar]
- 23.Ryan, U., A. O'Hara, and L. Xiao. 2004. Molecular and biological characterization of a Cryptosporidium molnari-like isolate from a guppy (Poecilia reticulata). Appl. Environ. Microbiol 70:3761-3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scorza, A. V., M. M. Brewer, and M. R. Lappin. 2003. Polymerase chain reaction for the detection of Cryptosporidium spp. in cat feces. J. Parasitol. 89:423-426. [DOI] [PubMed] [Google Scholar]
- 25.Smith, H. V., and J. B. Rose. 1998. Waterborne cryptosporidiosis: current status. Parasitol. Today 14:14-22. [DOI] [PubMed] [Google Scholar]
- 26.Sturdee, A. P., R. M. Chalmers, and S. A. Bull. 1999. Detection of Cryptosporidium oocysts in wild mammals of mainland Britain. Vet. Parasitol. 80:273-280. [DOI] [PubMed] [Google Scholar]
- 27.Sulaiman, I. M., L. Xiao, C. Yang, L. Escalante, A. Moore, C. B. Beard, M. J. Arrowood, and A. A. Lal. 1998. Differentiating human from animal isolates of Cryptosporidium parvum. Emerg. Infect. Dis. 4:681-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.U.S. Environmental Protection Agency. 2001. Method 1623: Cryptosporidium and Giardia in water by IFA/IMS/FA. EPA 821-R-01-025. Office of Water, U.S. Environmental Protection Agency, Washington D.C.
- 29.Wade, S. E., H. O. Mohammed, and S. L. Schaaf. 2000. Prevalence of Giardia sp. Cryptosporidium parvum and Cryptosporidium andersoni (syn. C. muris) [correction of Cryptosporidium parvum and Cryptosporidium muris (C. andersoni)] in 109 dairy herds in five counties of southeastern New York. Vet. Parasitol. 93:1-11. [DOI] [PubMed] [Google Scholar]
- 30.Webster, J. P., and D. W. Macdonald. 1995. Parasites of wild brown rats (Rattus norvegicus) on UK farms. Parasitology 111:247-255. [DOI] [PubMed] [Google Scholar]
- 31.Xiao, L., K. Alderisio, J. Limor, M. Royer, and A. A. Lal. 2000. Identification of species and sources of Cryptosporidium oocysts in storm waters with a small-subunit rRNA-based diagnostic and genotyping tool. Appl. Environ. Microbiol. 66:5492-5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao, L., C. Bern, J. Limor, I. Sulaiman, J. Roberts, W. Checkley, L. Cabrera, R. H. Gilman, and A. A. Lal. 2001. Identification of 5 types of Cryptosporidium parasites in children in Lima, Peru. J. Infect. Dis. 183:492-497. [DOI] [PubMed] [Google Scholar]
- 33.Xiao, L., L. Escalante, C. Yang, I. Sulaiman, A. A. Escalante, R. J. Montali, R. Fayer, and A. A. Lal. 1999. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl. Environ. Microbiol. 65:1578-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiao, L., R. Fayer, U. M. Ryan, and S. J. Upton. 2004. Cryptosporidium taxonomy: recent advances and implications for public health. Clin. Microbiol. Rev. 17:72-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiao, L., U. M. Morgan, J. Limor, A. Escalante, M. Arrowood, W. Shulaw, R. C. Thompson, R. Fayer, and A. A. Lal. 1999. Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Appl. Environ. Microbiol. 65:3386-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiao, L., A. Singh, J. Limor, T. K. Graczyk, S. Gradus, and A. Lal. 2001. Molecular characterization of Cryptosporidium oocysts in samples of raw surface water and wastewater. Appl. Environ. Microbiol. 67:1097-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiao, L., I. M. Sulaiman, U. M. Ryan, L. Zhou, E. R. Atwill, M. L. Tischler, X. Zhang, R. Fayer, and A. A. Lal. 2002. Host adaptation and host-parasite co-evolution in Cryptosporidium: implications for taxonomy and public health. Int. J. Parasitol. 32:1773-1785. [DOI] [PubMed] [Google Scholar]
- 38.Yu, J. R., S. P. O'Hara, J. L. Lin, M. E. Dailey, and G. Cain. 2002. A common oocyst surface antigen of Cryptosporidium recognized by monoclonal antibodies. Parasitol. Res. 88:412-420. [DOI] [PubMed] [Google Scholar]