Abstract
Culturability and coexistence of bacterioplankton exhibiting different life strategies were investigated in the Baltic Sea and Skagerrak Sea. Bacterial numbers were estimated using a dilution-to-extinction culturing assay (DCA) and calculated as the most probable number, based on six different methods to detect bacterial growth in the DCA. Irrespective of the method used to detect growth, the fraction of multiplying cells never exceeded 10%, using the total count of 4′,6′-diamidino-2-phenylindole (DAPI)-stainable cells as a reference. Furthermore, the data also showed that non-colony-forming bacteria made up the majority of the viable cells, confirming molecular results showing dominance of non-colony-forming bacteria in clone libraries. The results obtained are in agreement with previous observations, indicating that bacterial assemblages in seawater are dominated by small, active subpopulations coexisting with a large group of inactive cells. The ratio of colony-forming to non-colony-forming bacteria was approximately 10 to 20 times higher in the brackish Baltic Sea than in the Skagerrak Sea. These two sea areas differ in (for example) their levels of bacterial production, dissolved organic carbon, and salinity. We suggest that the relative importance of colony-forming versus non-colony-forming bacterioplankton may be linked to environmental characteristics.
Marine bacterioplankton face a heterogeneous environment with hot spots of particulate organic carbon surrounded by oligotrophic water: these diverse microniches select for bacteria adapted to different conditions (1). It has been speculated that to allow exploitation of nutrients in hotspots, bacteria must be motile, grow fast, and be able to colonize surfaces (17, 23). These features are common among eutrophic bacteria able to grow under rich conditions (45). Organisms that are adapted to consume nutrients in oligotrophic water surrounding a hot spot likely require a completely different, more-energy-saving life strategy. These bacteria need to be small, free living, and slow growing (38). Hence, the bacterioplankton assemblage can be divided into two widely separated life strategies, the eutrophic and the oligotrophic. The eutrophic bacteria represent opportunistic ecotypes that can be expected to show substantial fluctuations in abundance, due to large variations in growth and death rates, while oligotrophic ecotypes are likely to show small variations in growth rates and abundance (37). Intermediate life strategies between these two extremes are likely to persist in a continuum of behavioral adaptations competing for resources.
During the last 2 decades, knowledge about marine bacteria has primarily been gained by culture-independent approaches. These approaches have been motivated by “the great plate count anomaly,” signifying the fact that <1% of the bacteria seen in the microscope form colonies on solid media (20, 40). The most widely used culture-independent approaches to study bacterioplankton diversity have been cloning of the 16S rRNA gene and fluorescence in situ hybridization (FISH). In 16S rRNA gene libraries from surface water, collected all over the world, a major proportion of the clones belong to the alpha subclass of the Proteobacteria. A large fraction of these clones are affiliated with the SAR11 clade, a group of slow-growing, minuscule bacteria that do not form colonies on an agar surfaces (33). In turn, these properties have been found to be associated with the life strategy of oligotrophic organisms, showing dispersed and slow growth to optimize their access to nutrients (38). For these bacteria, colony formation has been suggested to be nonadaptive, which would explain their absence on solid media.
Several other explanations have been given for the low numbers of CFU and the selective isolation of easily cultured eutrophic bacteria on agar plates. For instance, that the fundamental needs (chemical, nutritional, and physical) of marine bacteria are unclear makes it difficult to provide a culturing medium that is suitable for all microorganisms sampled from a heterogeneous environment. The use of conventional cultivation techniques have consequently resulted in a biased isolation of bacterioplankton in relation to the diversity derived from cultivation-independent techniques (42). An explanation for the relatively unsuccessful attempts to culture marine bacteria has been that rich media such as marine broth exclude oligotrophic or facultative oligtrophic cells adapted to nutrient-poor environments because of nutrient stress (6). Also, constant exposure to other stress factors such as substrate limitation, UV irradiation, and oxidative stress could lead to reduced viability of the bacterial cell (27). Alternatively, the low plating efficiency could be due to virus infection (34). Approximately 10 to 50% of the bacterial mortality in surface waters is caused by viral infections, and up to 7% of the bacterial community carries mature phages at any given time (31, 32). Infection by lytic phages is obviously lethal, but lysogenic phages may also increase bacterial mortality, since it is likely that a shift to rich media will trigger virus induction and cell lysis (46). In summary, there are several possibilities for bacteria to become moribund, which makes it difficult to evaluate the dominance of bacteria with different life strategies in situ, due to the presence of nonmultiplying cells.
Microenvironments favoring bacteria expressing either eutrophic or oligotrophic life strategies cannot be separated during sampling. However, their relative importance may depend on environmental conditions that can be observed in different sea areas. Results from the Baltic Sea, obtained by DNA-DNA hybridization, demonstrated that bacterial species able to form colonies on solid media could be dominant in a bacterioplankton community. Results from the North Sea, based on FISH, have indicated that marine colony-forming bacteria (CFB) constitute an insignificant fraction of the bacterioplankton community (10, 29). Could these seemingly opposing views somehow be reconciled? There are important differences between these sampling sites; the Baltic Sea proper is a brackish sea area largely influenced by the drainage area, resulting in a high dissolved organic carbon (DOC) level while the North Sea (Skagerrak) is characterized by “normal marine” conditions such as high salinity and a low-DOC level. In this paper, we examined the culturability and coexistence of bacterioplankton with different life strategies in these two sea areas and found variation in the relative abundance of colony-forming versus non-colony-forming bacteria.
MATERIALS AND METHODS
Sample collection.
Seawater was collected at a 2-m depth in the Baltic Sea (57°27′N, 17°05′E) on 8 October 2003 and from the Skagerrak Sea (58°15′N, 11°22′E) on 15 May 2003 using a 10 l Niskin bottle. The samples were collected in 5 l polycarbonate bottles and processing occurred within 1.5 h. The sampled area in the Baltic has a salinity of 7 ppt and a DOC concentration of 300 to 325 μM C, whereas the Skagerrak salinity and DOC concentrations typically vary around 28 ppt and 150 μM C, respectively (44).
Dilution-to-extinction culturing assay (DCA).
The dilution-to-extinction method for culturing marine bacterioplankton in 96-well culture plates (Falcon) or 96-culture glass tubes was modified after Button et al. (6). Seawater was collected and prepared for use as a medium the day prior to sampling. The water used as a medium was filtered (47-mm-diameter, 0.2-μm-pore-size Supor-200 filter; Pall Corporation) and autoclaved. Samples for total bacterial counts were fixed using formaldehyde (2% [wt/vol]) and, when possible, counted prior to the dilution assay. Fixed seawater was filtered through 0.2-μm-pore-size black polycarbonate filters and stained with 4′,6′-diamidino-2-phenylindole (DAPI) (final concentration, 4 μg/ml; Sigma), and cells were counted with an epifluorescence microscope (Zeiss Axioplan) (30). The inocula were initially diluted based on the total count in the seawater sample to reach a cell concentration of one or no cells in the last dilution vials (in the Baltic, the inoculum was diluted 1:100 based on prior experience, since microscopic counts could not be done at sea). Twofold dilutions were made in 12 successive rows of vials. In the Baltic DCA, 96-well plates each with a culture volume of 200 μl were used, while the Skagerrak DCA were made in 96 glass or plastic tubes each with a culture volume of 10 ml. When compared, the assays generated identical most-probable-number (MPN) values irrespective of the culturing volumes (data not shown). Culture tubes were incubated for 40 days at 15°C in a cycle consisting of 12 h of light and 12 h of darkness.
CFU.
On the day of sampling, 100 μl (each) from undiluted, 10×, and 100× diluted seawater was spread on ZoBell agar plates on five replicate plates from each dilution. Plates were incubated at 15°C for 2 weeks and counted three times to detect slow-growing colonies.
MPN.
MPN was calculated according to the U.S. Food and Drug Administration online manual Appendix 2: Most Probable Number from Serial Dilutions (http://www.cfsan.fda.gov/∼ebam/bam-a2.html) (3).
MPN-DAPI.
Growth in 96-well plate or 96-tube DCA was detected by microscopy after DAPI staining as described previously (7). This MPN estimate corresponds to the culturability of both non-colony-forming and colony-forming bacteria.
MPN-CFB (colony-forming bacteria).
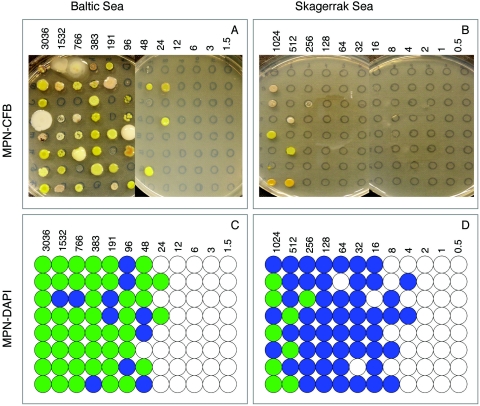
After 40 days of incubation of the DCA, a drop (∼5 μl) was transferred from each well or tube onto a marked circle on a ZoBell agar plate. Each circle represented one well/tube from the DCA (see Fig. 3A and B). The plates were incubated for 1 week at 15°C. Tubes/wells showing growth on the agar plate were recorded as positive for colony-forming bacteria. This MPN estimate corresponded to colony-forming bacteria only.
FIG. 3.
Photographs showing MPN-CFB agar plates inoculated with 5 μl from each dilution well/tube. (A) Baltic Sea DCA; (B) Skagerrak DCA. Illustrations of positive growth detected by epifluorescence microscopy after DAPI staining and MPN-DAPI. Blue circles indicate growth detected only by microscopy, representing non-colony-forming bacteria; green circles indicate tubes/wells with growth detected by both microscopy and spot tests on agar plates. These wells/tubes contain colony-forming bacteria and possibly non-colony-forming bacteria. (C) Baltic Sea DCA; (D) Skagerrak DCA. The numbers of bacteria inoculated are indicated in the respective panels.
MPN estimated by tracer uptake.
In the Skagerrak DCA, 1.5-ml dilution tubes were incubated with [3H]thymidine (5 nM final concentration; 3.11 TBeq/mmol), [14C]leucine (5 nM final concentration; 13 Gbq/mmol) and assayed for growth according to Smith and Azam (39). An MPN estimate was calculated from tubes showing positive radioactive uptake, defined as three times the standard deviation above the background level. Tubes from the three last dilutions (rows) served as negative controls generating the background counts, since these were inoculated from the same dilutions used for the MPN-DAPI assay, where no cells were detected by microscopy.
MPN estimated by PCR amplification.
To confirm growth detected by microscopy with the Skagerrak DCA, DNA from each well/tube was PCR amplified using the universal bacterial 16S rRNA gene primers, 27F (5′-AGAGTTTGATCATGGCTCAG) and 1492R (5′-TACGGYTACCTTGTTACGACTT) (14). PCR amplifications producing visible bands on agarose gels were considered positive for the presence of bacterial growth. Furthermore, to determine the presence of SAR11 16S rRNA genes, real-time quantitative PCR was used (Sequence Detection System, ABI PRISM 7700; Applied Biosystems) (41). A linear plasmid containing a genomic 16S rRNA gene from an SAR11 cluster member, clone KRB2 (closest neighbor, HTCC1062) was used as a standard for the 5′ nuclease PCR assay. An MPN estimate was calculated from wells or tubes showing a positive PCR signal, representing multiplying cells in general and the SAR11 clade specifically.
DNA extraction.
DNA was extracted from 1.5-ml cultures from the Skagerrak DCA, harvested by centrifugation (15 min at 14,000 × g). The pellet was resuspended in 175 μl lysis buffer (400 mM NaCl, 750 mM sucrose, 20 mM EDTA, 50 mM Tris-Hcl; pH 9.0) and incubated with lysozyme (final concentration, 1 mg/ml; Sigma) at 37°C for 30 min. Sodium dodecyl sulfate (final concentration, 1%; Sigma) and proteinase K (final concentration, 100 μg/ml; Roche Molecular Biochemicals) were added, and the samples were incubated at 55°C overnight. Nucleic acid was precipitated with sodium acetate (0.1 volume, 3 M; pH 5.2) and 2.5 volumes of 99.5% ethanol. Samples were resuspended in 1× Tris-EDTA buffer (pH 8.0).
RESULTS
Different estimates of culturability (Skagerrak Sea DCA).
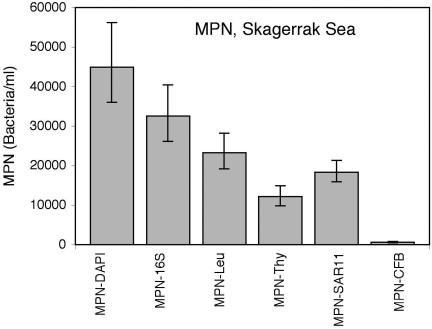
The CFU assay has been the method traditionally used to count multiplying cells and to isolate bacterial strains. For oligotrophic bacterioplankton lacking the ability to form colonies, a different measure of viability is needed. Thus, a DCA, together with an MPN estimate, was used to determine the total number of multiplying cells (3, 6). Six different methods were used to confirm growth in a DCA experiment conducted using seawater collected from the Skagerrak Sea. Based on direct count of bacteria in the original seawater sample, the inoculum was diluted to extinction, and the DCA tubes were incubated for 40 days. The experimental setup is illustrated in Fig. 1. MPN values were calculated for each technique, and the results are shown in Fig. 2 and Table 1. Culturability was calculated as the fraction of MPN/total count and expressed as a percentage.
FIG. 1.
Experimental setup of the Skagerrak DCA experiment. For the Baltic Sea DCA, the experimental setup was adopted for use with 96-well plates and analyzed for MPN-DAPI and MPN-CFB.
FIG. 2.
For the Skagerrak DCA, six different detection methods were compared to determine the total abundance of multiplying cells, calculated by MPN. The total count of DAPI-stainable particles in the seawater sample was 7.4 × 105 cells ml−1.
TABLE 1.
Abundance and culturability of the active bacterioplankton fraction in the Skagerrak Sea and Baltic Sea DCAs as estimated by MPN
| Fraction | Skagerrak Sea (15 May 2003)
|
Baltic Sea (8 October 2003)
|
||
|---|---|---|---|---|
| Bacteria (103 ml−1)a | % Culturabilityb | Bacteria (103 ml−1)a | % Culturabilityb | |
| Total count (DAPI) | 741.2 ± 27.2 | 1531.6 ± 25.8 | ||
| MPN-DAPI | 45.0 ± 0.2 | 6.1 | 37.9 ± 9.6 | 2.5 |
| MPN-16S | 32.5 ± 7.2 | 4.4 | ||
| MPN-leucine | 23.3 ± 4.5 | 3.1 | ||
| MPN-thymidine | 12.1 ± 2.6 | 1.6 | ||
| MPN-SAR11 | 18.3 ± 2.7 | |||
| MPN-CFB | 0.6 ± 0.2 | 0.08 | 6.7 ± 1.3 | 0.4 |
| Plate count CFU | 1.0 ± 0.1 | 0.14 | 7.1 ± 0.6 | 0.5 |
Values are expressed as the mean ± standard error.
Values of culturability are calculated as MPN/total count.
The culturability as determined by MPN-DAPI was 6.1%. This value was based on the number of tubes in each dilution of the DCA where positive growth was recorded by microscopy. This number includes growth of both colony-forming and non-colony-forming bacteria (Table 1). The MPN-DAPI value was compared to that of MPN-16S, an alternative measure of total cells based on the presence of 16S rRNA genes in the dilution tubes. Bacterial 16S rRNA genes could be amplified from a majority (73%) of the dilution tubes scored positive by the MPN-DAPI assay (Fig. 2; Table 1).
Since the16S rRNA genes most commonly found in clone libraries belong to the oligotrophic non-colony-forming SAR11 clade, the presence of SAR11 in the dilution tubes was investigated, using a real-time quantitative PCR assay specific for SAR11 16S rRNA genes (15, 41). The MPN value for SAR11 cells in the Skagerrak DCA was 1.8 × 104 cells/ml or 40% of the MPN-DAPI number (Table 1).
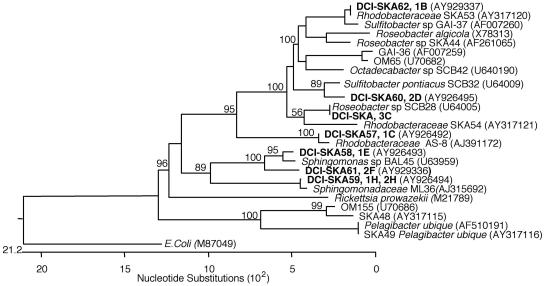
The number of tubes containing CFB in the DCA was used to calculate the MPN-CFB, which was 0.6 ×103± 0.2 ×103 cells/ml. This value was in agreement with the CFU value of 1.0 ×103± 0.1 ×103 cells/ml, determined from the plate count on the day of sampling, resulting in a culturability of colony-forming bacteria of approximately 0.1% of the total DAPI-stained count (Table 1). The colonies on the agar plates used in the spot test for the MPN-CFB showed a number of different colony morphologies (Fig. 3B). These isolates were characterized by their 16S rRNA gene sequences and were all found to be members of the marine α-Proteobacteria. Three isolates clustered within the Roseobacter clade, one clustered with the Rhodobacter clade, four isolates clustered within the Sphingomonas clade, and one spot consisted of a mixed culture (Fig. 4).
FIG. 4.
Phylogenetic tree showing the affiliation of the colony-forming isolates obtained from the Skagerrak Sea DCA experiment. The isolates all belonged to the alpha subclass of Proteobacteria.
Uptake of radioactively labeled thymidine and leucine are routinely used to monitor DNA synthesis and protein synthesis, respectively (12, 24). These tracers should therefore be suitable to detect growth in the dilution tubes. The MPN calculated from the [3H]thymidine-positive tubes was lower than that for [14C]leucine, which in turn resulted in a 45%-lower MPN value than the MPN-DAPI value (Fig. 2; Table 1).
The precision of the MPN-DAPI assay was checked for a Baltic Sea sample using four replicate 96-well plates. The test resulted in a mean value of 4.3 × 104± 0.6 × 104 cells/ml. Furthermore, the use of 10-ml culture tubes versus 200-μl wells was tested using Skagerrak water; the result obtained was within the previously established limit of the method. In all assays, MPN was calculated with reference to direct counts of DAPI-stainable bacteria in the inoculum. Irrespective of the method used, culturability never exceeded 10% (Table 1).
Culturability and coexistence in marine and brackish water.
In this study, the bacterial community from the two sea areas was compared with respect to culturability and relative abundance of colony-forming versus non-colony-forming bacteria (MPN-DAPI and MPN-CFB). For the Baltic Sea DCA, 96-well microtiter plates (culturing volume, 200 μl/well) were used instead of 10-ml tubes. The numbers of multiplying bacteria in the two sea areas were similar when calculated as MPN-DAPI (Table 1). The estimates of culturability based on MPN-DAPI were 6.1% and 2.5% for the Skagerrak Sea and the Baltic Sea, respectively, while based on the CFU count at the sampling occasion the values were 0.1% and 0.5%, respectively (Table 1).
The MPN-CFB, determined from a spot test on agar plates, allowed growth of eutrophic colony-forming bacteria to be distinguished from the oligotrophic non-colony-forming bacteria (Fig. 3A and B). In the Skagerrak DCA, the last row where colonies appeared was in the row with 256 bacteria/well, compared to 24 bacteria/well in the Baltic DCA. This means that the ratio of colony-forming to non-colony-forming bacteria was approximately 10 to 20 times higher in the Baltic. In the respective sea area, the MPN-CFB allowed an estimate showing that non-colony-forming bacteria were 100-fold-more numerous than colony-forming bacteria in the Skagerrak, while only 5-fold-more numerous in the Baltic (Fig. 3C and D; Table 1).
DISCUSSION
In the present paper, we have addressed the issue of culturability of bacterioplankton by expanding the DCAs to include six different methods designed to detect the accumulation of bacterial cells in the dilution tubes. We incubated the DCA for 40 days before analysis to ensure cell concentrations high enough for detection by microscopy and uptake of radioactive tracers, even if the growth rates were as low as 0.3 day−1. Regardless of the method used, the fraction of multiplying cells never exceeded 10% of the total DAPI count (Table 1). Bruns et al. reported that the addition of signal molecules such as cyclic AMP increased culturability in Baltic MPN assays. However, their data from the oxic upper layer only showed a moderate (10%) increase in culturability; although this effect may be of importance for the understanding of bacterioplankton behavior, this treatment was not included in our study (5). Compilations of available data from previous dilution culture experiments also showed a low fraction of multiplying cells (mean, 6.2%± 6.7%; n = 11) (2, 7, 9, 47) (Table 2). In these studies, natural seawater was used as a medium, and the fraction of multiplying cells was estimated with the microscopic total count of cells as a reference.
TABLE 2.
Fraction of multiplying cells in dilution culture experiments with unamended autoclaved natural seawater as medium
| Seawater source (date) | Reference | Depth (m) | Total count (cells ml−6) | % Proportion of multiplying (culturability) | Method of growth detection |
|---|---|---|---|---|---|
| Bay of Banyuls-sur-Mer (2 November 1998) | 2 | 2 | 0.68 | 1.6 | FCMb |
| Oregon (May-July)c | 7 | Surface | 1.1-5.6 | 8.8 | DAPIc |
| Oregon (October-April)a | 7 | Surface | 0.9-1.9 | 1.2 | DAPI |
| Continental shelf, Japan (12 June 1998) | 9 | 350 | 0.31 | 7.7 | DAPI |
| Pacific Ocean (4 June 1998) | 9 | 500 | 0.55 | 20 | DAPI |
| Baltic Sea (October 1993) | 47 | 0-30 | 2.5-3.3 | 0.1-0.3 | DAPI |
| Baltic Sea (May 1994) | 47 | 0-100 | 0.7-1.2 | 7-14 | [3H]thymidine |
| Baltic Sea (May 1994) | 47 | 0-200 | 0.6-2.7 | 6-15 | [3H]thymidine |
| North Sea Skagerrak (August 1994) | 47 | 0-25 | 1.1-1.4 | 0.5-0.6 | [3H]thymidine |
| North Sea Skagerrak (August 1994) | 47 | 25-340 | 0.2-0.8 | 0.2-0.8 | [3H]thymidine |
| Mediterranean Sea (October 1994) | 47 | Surface | 0.5 | 16 | DAPI |
Samples were collected 1998, 1999, and 2000.
Flow cytometry.
DAPI staining, fluorescence microscopy.
Metabolic activity detected using radioisotope-labeled precursors of DNA and protein synthesis has been used extensively to monitor bacterial growth (13, 24). In the DCA, we therefore used [3H]thymidine and [14C]leucine to monitor the increase of biomass in the tubes. The MPN of bacteria measured with these two markers was less than half of the MPN obtained from DAPI staining. While we lack information as to why some bacteria failed to incorporate [14C]leucine, several bacterioplankton species are known to be unable to utilize thymidine, which may explain unsuccessful incorporation (8, 21). However, the important observation was that while the radioactive tracer failed to detect growth in some of the tubes scored positive by the microscopic method (MPN-DAPI), the opposite was never recorded; in both cases, the fraction of multiplying bacteria appeared to be <10% of the total count. This is in agreement with previous observations indicating that bacterial assemblages in seawater are dominated by small, active subpopulations coexisting with a large group of dead or temporarily inactive cells (47). Our results are consistent with the image of bacteria trying to multiply in the dilution vials where the success rate is very limited (<10%). Cells were defined as viable when they were shown to be capable of growth and division by techniques that allowed direct demonstration of culturability (4). With these prerequisites, we concluded that the culturability of bacterioplankton in the dilution culture assays was approximately 1 to 2 orders of magnitude higher than when determined from regular CFU counts but still remained low in relation to the total counts. The fate of the bacteria that failed to grow in the dilution vials is, however, unknown to us.
It was not until the dilution-to-extinction culturing technique made it possible to isolate formerly “uncultured” bacteria such as SAR11 that the views of the microbial community on molecular techniques and cultivation techniques became congruous (7). In the past, isolation of bacteria from agar plates inoculated with seawater samples has mostly resulted in the selection of fast-growing eutrophic bacteria. Nevertheless, this standard procedure has resulted in the isolation of bacteria with an astonishing genetic and physiological diversity (28). The biased selection on agar plates appears even more evident with the results of Simu and Hagström, showing that oligotrophic bacterioplankton frequently lack the ability to form colonies on agar plates and thus can escape isolation (38). In the current DCA experiment, we have shown that dominant species found by cloning and hybridization techniques are the same as the dominant species found in the dilution-to-extinction cultures. The members of the SAR11 clade (non-colony-forming oligotrophic bacteria) found in the Skagerrak dilution cultures accounted for 40% of the total counts of multiplying cells (MPN-DAPI) (Fig. 2; Table 1). The abundance of the SAR11 clade was previously estimated by Morris et al. in the northwestern Sargasso Sea and in the coastal waters of Oregon by FISH (26). They found that SAR11 cells made up to 50% of the total bacterioplankton community and concluded that SAR11 is the most successful organism on Earth. Also, the dominant colony-forming bacteria isolated from the Skagerrak dilution assay were members of the marine alpha subclass of the Proteobacteria and the Sphingomonas clade. These bacteria have previously been shown to be important populations in the costal bacterioplankton community. Gonzalez and Moran estimated the marine alpha group to account for up to 28% of the total bacterial community off the coast of the southeastern United States by dot blot DNA hybridization (16). The same high abundance of the marine alpha group was found in the North Sea as determined by FISH (10). Members of the Spingomonas clade have been reported to constitute from 15 to 35% of the bacterioplankton community in the North Sea by Schut et al. and down to <1% of the DAPI stainable cells by Eilers et al. (11, 36). In our DCA experiments, the fraction of colony-forming bacteria never exceeded 1%, and the fraction of multiplying cells (MPN-DAPI, including non-colony-forming bacteria) never exceeded 10% of total DAPI counts (Table 1). In conclusion, available results show that the same dominant bacterioplankton species detected in the clone libraries and by the FISH technique are found in the dilution assays.
Pinhassi et al. showed that bacteria able to form colonies on agar plates occupied a substantially higher proportion of the bactierioplankton community than indicated by their number in CFU counts and expressed the view that this situation could be a general feature (29). This was opposed by Eilers et al., claiming that non-colony-forming bacteria are in fact much more numerous and that colony-forming bacteria make up an almost insignificant fraction of the bacterioplankton community (10). These opposing views have been difficult to reconcile. In the present study, a 10- to 20-fold-higher number of colony-forming bacteria were estimated in dilution assays from the Baltic Sea than for the Skagerrak Sea (Fig. 3). The reason for this difference is unclear, but it can be speculated that the high DOC in the Baltic may supply a suitable substrate for bacteria with eutrophic growth. This substrate would allow a higher baseline level of colony-forming bacteria in the Baltic, whereas the similar numbers of non-colony-forming bacteria in both the Baltic Sea (high DOC) and the Skagerrak Sea (low DOC) indicate that they are able to utilize roughly the same level of substrate (Fig. 3). The samples tested in this study were collected at different seasons; thus, a temporal effect could be suspected. However, previous data from the Skagerak show a high ratio of non-colony-forming versus colony-forming bacteria during autumn, similar to our values, obtained during spring. Also, the low ratio between non-colony-forming and colony-forming bacteria in the Baltic appears to lack seasonal variation (unpublished data).
From the present study, it seems that colony-forming bacteria in certain environments like the Baltic Sea can be as numerous as non-colony-forming bacteria. This was also the case when Pinhassi et al. demonstrated that 69% of the Baltic community consisted of colony-forming bacteria, using whole DNA probes from colony-forming bacteria to hybridize against extracted environmental DNA (29). In the study by Pinhassi et al., as well as in the present study, the estimates of culturability of the colony-forming bacteria were low (CFU, ≈1%). At the same time, our data showed that in other, more marine environments, such as the Skagerrak Sea, the fraction of non-colony-forming bacteria may be dominant. Thus, the opposing statements by Pinhassi et al. and Eilers et al. may both be correct.
In most studies, the degree of culturability has been normalized against the total count of bacteria measured by fluorescent stains such as DAPI that bind to DNA. The discrepancy between the total number of cells/particles and the estimated number of active cells has been difficult to explain. Zweifel and Hagström suggested the nonspecific-stained “ghost cell” concept to explain the presence of inactive cells (47). These authors showed that excess stain could be washed away from some supposedly damaged cells with isopropanol, whereas the intact cells remained stained. Vosjan and van Noort observed that storage of formaldehyde-fixed seawater samples reduced the number of DAPI-stainable bacteria, suggesting that damaged cells might be susceptible to degradation during storage (43). Furthermore, Heissenberger et al. demonstrated electron microscopic evidence showing that a large fraction of the bacterioplankton cells were damaged (18). These results have been taken as evidence that a majority of the cells are damaged and thus given as an explanation to low culturability. The results from FISH assays show that this view may not be correct, since a large fraction of presumably intact cells can be hybridized with universal bacterial 16S rRNA probes. Karner and Furhman compared universal 16S rRNA probes, autoradiography, and nucleoid staining (total counts using DAPI-stained cells after rinsing with isopropanol) for the determination of active bacterioplankton cells (22). In their comparison, all of the above methods yielded lower cell counts than DAPI total count. Universal probe counts averaged about half of the corresponding DAPI counts and were highly correlated to autoradiography counts. Nucleoid-containing cell counts could be lower than DAPI by as much as 1 order of magnitude but sometimes matched autoradiography or probe counts. The different results suggest the existence of factors that still are largely unresolved. In the present paper, we thus decided to use the conventional DAPI-staining protocol as a reference for the estimate of culturability when comparing the outcome of the MPN assay by different screening methods (30).
In addition to the suggestion that cells are dead, other possible explanations for low culturability and the large fraction of inactive or dead cells can be proposed. Low CFU in seawater have been attributed to low plating efficiency, due to virus infection (34). However, since the inactive DAPI-stained particles represent >90% of the total community, the pool of virus-infected cells (10 to 50% of the community) cannot alone be responsible for the apparent low level of culturability. Also, as reported by Riemann and Middelboe, lysis of virus-infected cells of specific bacteria may result in complete disintegration of the cells (35). Thus, apart from virus infection, other factors must exist. One option to cope with stress could be survival mechanisms such as programmed cell death, a stress adaptation mechanism resulting in a coordinated mortality (19, 25). The evolutionary value of this behavior would be to protect the population from extinction (25), but the effect of this regulatory mechanism would significantly influence culturability, since inactive bacteria counted in the microscope cannot be distinguished from active bacteria.
A more trivial issue but nevertheless relevant to consider is the fact that the in vitro growth conditions in the DCA assays might limit or exclude growth of organisms with specific demands, whether sterile seawater or defined media were used. Against this concern, we argue that in our assays, we were able to account for bacteria such as members of the SAR11, Roseobacter, Rhodobacter, and Sphingomonas clades, previously shown to be common in the water column by cloning and 16S rRNA gene sequencing. This would indicate that the culturability estimates in this study include growth of the major groups of bacterioplankton.
Cloning data have shown that bacteria unable to form colonies on common culturing media are the dominant species in the water column (15). In the present study, we have shown that in certain environments colony-forming bacteria can be as numerous as non-colony-forming bacteria and speculated that the high DOC level in the Baltic supplies a substrate allowing a higher abundance of eutrophic bacteria. This is the first study showing that data obtained with culture-dependent techniques are congruous with culture-independent techniques, i.e., molecular data. Furthermore, our results support previous findings that >90% of DAPI counts are inactive particles, which leads to a low calculated percent culturable bacteria, when DAPI staining for total bacterial counts is used.
Acknowledgments
We thank Lasse Riemann and Jarone Pinhassi for critical reading of the manuscript, and we are grateful for the use of laboratory and sampling facilities at the Kristineberg Marine Research Station.
This work was supported by the Swedish Science Council (grant 621-2001-2814) and by the Faculty of Natural Sciences of Kalmar University.
REFERENCES
- 1.Azam, F. 1998. Microbial control of oceanic carbon flux: the plot thickens. Science 280:694-696. [Google Scholar]
- 2.Bernard, L., H. Schäfer, F. Joux, C. Courties, G. Muyzer, and P. Lebaron. 2000. Genetic diversity of total, actve and culturable marine bacteria in costal seawater. Aquat. Microb. Ecol. 23:1-11. [Google Scholar]
- 3.Blodgett, R. 2001. Appendix 2: most probable number from serial dilutions. Bacteriological analytical manual online, 8th ed. U.S. Department of Health and Human Services, U.S. Food & Drug Administration. [Online.] http://www.cfsan.fda.gov/∼ebam/bam-a2.html.
- 4.Bogosian, G., and E. V. Bourneuf. 2001. A matter of bacterial life and death. EMBO Rep. 2:770-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruns, A., H. Cypionka, and J. Overmann. 2002. Cyclic AMP and acyl homoserine lactones increase the cultivation efficiency of heterotrophic bacteria from the central Baltic Sea. Appl. Environ. Microbiol. 68:3978-3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Button, D. K., F. Schut, P. Quang, R. Martin, and B. R. Robertson. 1993. Viability and isolation of marine bacteria by dilution culture: theory, procedures, and initial results. Appl. Environ. Microbiol. 59:881-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Connon, S. A., and S. J. Giovannoni. 2002. High-throughput methods for culturing microorganisms in very-low-nutrient media yield diverse new marine isolates. Appl. Environ. Microbiol. 68:3878-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis, C. L. 1989. Uptake and incorporation of thymidine by bacterial isolates from an upwelling environment. Appl. Environ. Microbiol. 55:1267-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eguchi, M., M. Ostrowski, F. Fegatella, J. Bowman, D. Nichols, T. Nishino, and R. Cavicchioli. 2001. Sphingomonas alaskensis strain AFO1, an abundant oligotrophic ultramicrobacterium from the North Pacific. Appl. Environ. Microbiol. 67:4945-4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eilers, H., J. Pernthaler, F. O. Glockner, and R. Amann. 2000. Culturability and in situ abundance of pelagic bacteria from the North Sea. Appl. Environ. Microbiol. 66:3044-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eilers, H., J. Pernthaler, J. Peplies, F. O. Glockner, G. Gerdts, and R. Amann. 2001. Isolation of novel pelagic bacteria from the German bight and their seasonal contributions to surface picoplankton. Appl. Environ. Microbiol. 67:5134-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuhrman, J. A., and F. Azam. 1980. Bacterioplankton secondary production estimates for coastal waters of British Columbia, Antarctica, and California. Appl. Environ. Microbiol. 39:1085-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuhrman, J. A., and F. Azam. 1982. Thymidine incorporation as a measure of heterotrophic bacterioplankton production in marine surface waters: evaluation and field results. Mar. Biol. 66:109-120. [Google Scholar]
- 14.Giovannoni, S. J. 1991. The polymerase chain reaction, p. 177-203. In E. Stackebrandt and M. Goodfellow (ed.), Sequencing and hybridization techniques in bacterial systematics. John Wiley & Sons, Inc., New York, N.Y.
- 15.Giovannoni, S. J., and M. S. Rappe. 2000. Evolution, diversity, and molecular ecology of marine procaryotes, p. 47-84. In D. L. Kirschman (ed.), Microbial ecology of the oceans. Wiley, New York, N.Y.
- 16.Gonzalez, J. M., and M. A. Moran. 1997. Numerical dominance of a group of marine bacteria in the alpha-subclass of the class Proteobacteria in coastal seawater. Appl. Environ. Microbiol. 63:4237-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grossart, H. P., T. Kiorboe, K. Tang, and H. Ploug. 2003. Bacterial colonization of particles: growth and interactions. Appl. Environ. Microbiol. 69:3500-3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heissenberger, A., G. G. Leppard, and G. J. Herndl. 1996. Relationship between the intracellular integrity and the morphology of the capsular envelope in attached and free-living marine bacteria. Appl. Environ. Microbiol. 62:4521-4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hochman, A. 1997. Programmed cell death in prokaryotes. Crit. Rev. Microbiol. 23:207-214. [DOI] [PubMed] [Google Scholar]
- 20.Jannasch, H. W., and G. E. Jones. 1959. Bacteria populations in sea water as determined by different methods of enumeration. Limnol. Oceanogr. 4:128-139. [Google Scholar]
- 21.Jeffrey, W. H., and J. H. Paul. 1990. Thymidine uptake, tymidine incorporation, and tymidine kinase activity in marine bacterium isolates. Appl. Environ. Microbiol. 56:1367-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karner, M., and J. A. Fuhrman. 1997. Determination of active marine bacterioplankton: a comparison of universal 16S rRNA probes, autoradiography, and nucleoid staining. Appl. Environ. Microbiol. 63:1208-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kiorboe, T., H. P. Grossart, H. Ploug, and K. Tang. 2002. Mechanisms and rates of bacterial colonization of sinking aggregates. Appl. Environ. Microbiol. 68:3996-4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirchman, D. L., E. K'nees, and R. Hodson. 1985. Leucine incorporation and its potential as a measure of protein synthesis by bacteria in natural waters. Appl. Environ. Microbiol. 49:599-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewis, K. 2000. Programmed death in bacteria. Microbiol. Mol. Biol. Rev. 64:503-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morris, R. M., M. S. Rappe, S. A. Connon, K. L. Vergin, W. A. Siebold, C. A. Carlson, and S. J. Giovannoni. 2002. SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420:806-810. [DOI] [PubMed] [Google Scholar]
- 27.Nyström, T. 1998. To be or not to be: the ultimate decision of the growth arrested bacterial cell. FEMS Microbiol. Rev. 21:283-290. [Google Scholar]
- 28.Pinhassi, J., and T. Berman. 2003. Differential growth response of colony-forming α- and γ-Proteobacteria in dilution culture and nutrient addition experiments from Lake Kinneret (Israel), the eastern Mediterranean Sea, and the Gulf of Eilat. Appl. Environ. Microbiol. 69:199-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pinhassi, J., U. L. Zweifel, and A. Hagström. 1997. Dominant marine bacterioplankton species found among colony-forming bacteria. Appl. Environ. Microbiol. 63:3359-3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Porter, K. G., and Y. S. Feig. 1980. The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 25:943-948. [Google Scholar]
- 31.Proctor, L. M., and J. A. Fuhrman. 1990. Viral mortality of marine bacteria and cyanobacteria. Nature 343:60-62. [Google Scholar]
- 32.Proctor, L. M., A. Okubo, and J. A. Fuhrman. 1993. Calibrating estimates of phage-induced mortality in marine bacteria; ultrastructural studies of marine bacteriophage development from one-step growth experiments. Microb. Ecol. 25:161-182. [DOI] [PubMed] [Google Scholar]
- 33.Rappe, M. S., S. A. Connon, K. L. Vergin, and S. J. Giovannoni. 2002. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418:630-633. [DOI] [PubMed] [Google Scholar]
- 34.Rehnstam, A.-S., S. Bäckman, D. C. Smith, F. Azam, and Å. Hagström. 1993. Blooms of sequence-specific culturable bacteria in the sea. FEMS Microbiol. Ecol. 102:161-166. [Google Scholar]
- 35.Riemann, L., and M. Middelboe. 2002. Viral lysis of maine bacterioplankton: implications for organic matter cycling and bacterial clonal composition. Ophelia 56:57-68. [Google Scholar]
- 36.Schut, F., E. J. de Vries, J. C. Gottschal, B. R. Robertson, W. Harder, R. A. Prins, and D. K. Button. 1993. Isolation of typical marine bacteria by dilution cultures: growth, maintenance, and characteristics of isolates under laboratory conditions. Appl. Environ. Microbiol. 59:2150-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schut, F., R. A. Prins, and J. C. Gottschal. 1997. Oligotrophy and pelagic marine bacteria: facts and fiction. Aquat. Microb. Ecol. 12:177-202. [Google Scholar]
- 38.Simu, K., and Å. Hagström. 2004. Oligotrophic bacterioplankton with a novel single-cell life strategy. Appl. Environ. Microbiol. 70:2445-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith, D. C., and F. Azam. 1992. A simple, economical method for measuring bacterial protein synthesis rates in seawater using 3H-leucine. Mar. Microb. Food Webs 6:107-114. [Google Scholar]
- 40.Staley, J. T., and A. Konopka. 1985. Measurments of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestial habitats. Annu. Rev. Microbiol. 39:321-346. [DOI] [PubMed] [Google Scholar]
- 41.Suzuki, M. T., C. M. Preston, F. P. Chavez, and E. F. Delong. 2001. Quantitative mapping of bacterioplankton population in seawater: field test across an upwelling plume in Monterey bay. Aquat. Microb. Ecol. 24:117-127. [Google Scholar]
- 42.Suzuki, M. T., M. S. Rappe, Z. W. Haimberger, H. Winfield, N. Adair, J. Strobel, and S. J. Giovannoni. 1997. Bacterial diversity among small-subunit rRNA gene clones and cellular isolates from the same seawater sample. Appl. Environ. Microbiol. 63:983-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vosjan, J. H., and G. van Noort. 1998. Enumerating nucleoid-visible marine bacterioplankton: bacterial abundance determined after storage of formalin fixed samples agrees with isopropanol rinsing method. Aquat. Microb. Ecol. 14:149-154. [Google Scholar]
- 44.Wedborg, M., A. Skoog, and E. Fogelqvist. 1994. Organic carbon and humic substances in the Baltic Sea, Kattegat, and the Skagerrak, p. 917-924. In N. Sense and T. M. Miano (ed.), Humic substances in the global environment and implication on human health. Elsevier, Amsterdam, The Netherlands.
- 45.Weinbauer, M. G., and M. G. Höfle. 1998. Distribution and life strategies of two bacterial populations in a eutrophic lake. Appl. Environ. Microbiol. 64:3776-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weinbauer, M. G., and C. A. Suttle. 1996. Potential significance of lysogeny to bacteriophage production and bacterial mortality in coastal waters of the Gulf of Mexico. Appl. Environ. Microbiol. 62:4374-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zweifel, U. L., and Å. Hagström. 1995. Total counts of marine bacteria include a large fraction of non-nucleoid-containing bacteria (ghost). Appl. Environ. Microbiol. 61:2180-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]