Abstract
cDNA encoding a hemoprotein similar to the cytochrome domain of extracellular flavocytochrome cellobiose dehydrogenase (CDH) was cloned from the white-rot fungus Phanerochaete chrysosporium. The deduced amino acid sequence implies that there is a two-domain structure consisting of an N-terminal cytochrome domain and a C-terminal family 1 carbohydrate-binding module (CBM1) but that the flavin-containing domain of CDH is not present. The gene transcripts were observed in cultures in cellulose medium but not in cultures in glucose medium, suggesting that there is regulation by carbon catabolite repression. The gene was successfully overexpressed in Pichia pastoris, and the recombinant protein was designated carbohydrate-binding cytochrome b562 (CBCyt. b562). The resonance Raman spectrum suggested that the heme of CBCyt. b562 is 6-coordinated in both the ferric and ferrous states. Moreover, the redox potential measured by cyclic voltammetry was similar to that of the cytochrome domain of CDH. These results suggest that the redox characteristics may be similar to those of the cytochrome domain of CDH, and so CBCyt. b562 may have an electron transfer function. In a binding study with various carbohydrates, CBCyt. b562 was adsorbed with high affinity on both cellulose and chitin. As far as we know, this is the first example of a CBM1 connected to a domain without apparent catalytic activity for carbohydrate; this CBM1 may play a role in localization of the redox protein on the surface of cellulose or on the fungal sheath in vivo.
Wood, the most abundant biomaterial on earth, consists of cellulose, hemicellulose, and lignin. Of these constituents, lignin is the most recalcitrant polymer, and its decomposition requires a strong oxidative reaction. Although this property of lignin results in the high resistance of wood against microbial attack, a group of basidiomycetes known as white-rot fungi is able to destroy lignin through the reactions of various extracellular peroxidases, lignin peroxidases, manganese peroxidases, and versatile peroxidases (16, 33, 38, 54). Since hydrogen peroxide is required for activation of these enzymes, extracellular oxidases might participate in the redox networks leading to the fungal degradation of lignin (29, 30). However, mixtures of these enzymes have failed to degrade lignin in vitro.
In contrast to degradation of lignin, degradation of cellulose and hemicellulose has been thought to proceed only through the action of hydrolytic enzymes. However, Eriksson and coworkers have pointed out the importance of oxidative reactions in cellulose degradation (13, 14), and they isolated a cellobiose-oxidizing enzyme from a cellulolytic culture of the white-rot fungus Sporotrichum pulverulentum (Phanerochaete chrysosporium) (4). This enzyme contains one b-type heme and one flavin adenine dinucleotide in two separate domains (20, 40), and it can oxidize the reducing-end groups of cellobiose and higher cellooligosaccharides to the corresponding lactones (4). Although this enzyme was originally named cellobiose oxidase (EC 1.1.3.25), its name has been changed to cellobiose dehydrogenase (CDH) (EC 1.1.99.18) (5), since Fe(III)-containing compounds are much more effective electron acceptors than molecular oxygen (31, 47). The flavin domain derived by proteolysis of CDH (20, 58) has cellobiose-oxidizing ability in the presence of quinone but does not utilize Fe(III)-containing compounds as electron acceptors (17). Thus, the reduction of ferricytochrome c requires the cytochrome domain of CDH, suggesting the significance of the heme as an Fe(III) reductase (47).
Eriksson and coworkers first proposed a physiological function of CDH in lignin degradation by white-rot fungi, because this enzyme can prevent reoxidation of the degradation products with quinone, phenoxy, and aromatic radicals in the presence of cellobiose (56, 57). More recently, it has been hypothesized CDH has a role as a source of hydroxyl radical in wood degradation (22, 32). However, we previously reported the localization of CDH on cellulose surfaces in vivo (23); we showed that there is a synergistic interaction between CDH and cellobiohydrolase I (24), and we identified a kinetic advantage of CDH over extracellular β-glucosidase for cellobiose metabolism (26). Moreover, transcription of the cdh gene responded positively to cellobiose, whereas transcription of the bgl gene was repressed under the same conditions (62). Although these observations clearly indicate the importance of cellobiose oxidation by CDH, the overall picture of the redox network operating in cellulose degradation remains unclear.
Recently, the DoE Joint Genome Institute in the United States has disclosed the total genome sequence of P. chrysosporium (39; http://genome.jgi-psf.org/whiterot1/whiterot1.home.html). In order to find homologues of CDH and candidate electron acceptors, we searched the database of the P. chrysosporium genome and found one homologous region for the cytochrome domain of CDH. This gene encodes not only a protein similar to the cytochrome domain of CDH but also the family 1 carbohydrate-binding module (CBM1) signature in the C-terminal region instead of the flavin domain. In the present study, therefore, cDNA encoding the carbohydrate-binding cytochrome was cloned from P. chrysosporium grown on cellulose, and the recombinant protein was successfully produced in Pichia pastoris for characterization of its properties.
MATERIALS AND METHODS
Materials.
P. chrysosporium strain K-3 (27) was used as a source of target genes. Escherichia coli strain JM109 (Takara, Japan) and P. pastoris strain KM71H (Invitrogen, Carlsbad, CA) were used as the subcloning host and for the heterologous production of the recombinant protein, respectively.
Cloning and transcript analysis.
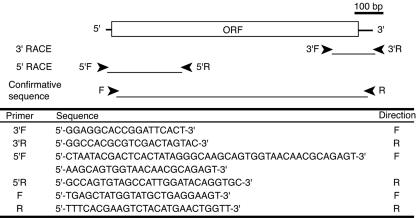
The cloning procedure used was the procedure described previously (61). Total RNA was extracted from P. chrysosporium grown in cellulose medium as described previously (18). The oligonucleotide primers were designed based on available genomic information, as illustrated in Fig. 1. The first-strand cDNA for 3′ rapid amplification of cDNA ends (RACE) was synthesized with a Ready-To-Go You-prime first-strand cDNA synthesis kit (Amersham Biosciences, Piscataway, NJ) using 3′ RACE adapter primer (Invitrogen, Carlsbad, CA), and the PCR was primed with 3′F and 3′R. The cDNA synthesized with a SMART RACE cDNA amplification kit (Clontech, Palo Alto, CA) was used for 5′ RACE as a template, and primers 5′F and 5′R were used for PCR. The nucleotide sequence, including the coding region, was confirmed by PCR amplification using specific primers F and R. For detection of transcripts, mycelia were grown in medium containing 2% glucose or cellulose as a sole carbon source, and the first-strand cDNA from each sample was used for reverse transcription PCR (RT-PCR) analysis with forward primer 5′-CGCTGGACTTCGGGCT-3′ and reverse primer 5′-CGCACTGACCCCACTCA-3′. The transcript level of the actin gene (accession no. AB115328), a housekeeping gene, was measured using primers actin-F (5′-GCCGTGTTCCCGTCCAT-3′) and actin-R (5′-CACTTGTAGATGGAGTTGTACGTCGT-3′).
FIG. 1.
Schematic diagram of cDNA encoding CBCyt. b562, showing locations and nucleotide sequences of primers used for PCR. F and R indicate the forward and reverse directions, respectively. ORF, open reading frame.
Sequence analysis.
The P. chrysosporium genome database (http://genome.jgi-psf.org/whiterot1/whiterot1.home.html) and the National Center for Biotechnology Information (NCBI) protein database (http://www.ncbi.nlm.nih.gov/BLAST/) were searched for amino acid sequences of CDH and carbohydrate-binding cytochrome b562 (CBCyt. b562) using the tblastn algorithm (1, 2). All searches were performed with standard settings and the BLOSUM 62 matrix. Multiple-alignment analysis was performed using ClustalW (53) at GenomeNet (http://clustalw.genome.jp/). The candidate genes were searched against the conserved domain database (37) at the NCBI (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and were scanned for the presence of signal peptides using the SignalP version 3.0 server (6, 43) at the Center for Biological Sequence Analysis (http://www.cbs.dtu.dk/services/SignalP/).
Production of the recombinant protein in P. pastoris and its purification.
The expression vector was constructed as described previously (60). The following two oligonucleotide primers were designed, based on the nucleotide sequence of the mature protein, for ligation into the XhoI and XbaI sites of the Pichia expression vector pPICZα (Invitrogen, Carlsbad, CA): primer 5′ (5′-TTTCTCGAGAAAAGACAGTCCAGCTCCCAGTTTTG-3′) and primer 3′ (5′-TTTTCTAGACTAAGCTTGGCACTGGTAGTAATAAGG-3′). Approximately 10 μg of the DNA construct in pPICZα was linearized with Bpu1102I (Takara, Japan) prior to transformation of P. pastoris. Electroporation, selection of the transformant, and production of recombinant protein were carried out according to the instructions in the manual of an EasySelect Pichia expression kit (version G; Invitrogen, Carlsbad, CA). The culture was centrifuged (30 min, 5,000 × g), and ammonium sulfate (70% saturation) was added to the cell-free culture medium to precipitate the protein. The precipitate was dissolved in 20 mM sodium acetate, pH 6.0, and the solution was then mixed with 5% (wt/vol) bentonite (Sigma Chemicals, St. Louis, MO), followed by incubation for 30 min at 26.5°C. The bentonite was removed by centrifugation (30 min, 10,000 × g), and the supernatant was dialyzed against 20 mM sodium acetate, pH 6.0, using a PM-10 ultrafiltration membrane (Millipore, Bedford, MA). The crude protein was fractionated on a QAE-Toyopearl 550C column (44 mm by 170 mm) equilibrated with 20 mM sodium acetate, pH 6.0. The protein was eluted from the column with a linear 0 to 0.5 M NaCl gradient in 1,000 ml. The fractions containing the recombinant protein were collected and equilibrated against 20 mM potassium phosphate buffer containing 1 M ammonium sulfate, pH 7.0. The solution was applied to a Phenyl-Toyopearl 650S column (16 mm by 120 mm) equilibrated with the same buffer. The protein was eluted with a 500-ml reverse gradient to 20 mM potassium phosphate buffer, pH 7.0. The fractions containing the recombinant protein were collected and equilibrated against 20 mM sodium acetate buffer, pH 5.5. Deglycosylation was performed using endo-β-N-acetylglucosaminidase H (endo-H) (New England Biolabs, Beverly, MA) as described previously (60). After overnight incubation, the solution was equilibrated against 20 mM sodium acetate buffer, pH 4.5, and the recombinant protein was finally purified on a DEAE-Toyopearl 650S column (16 mm x 170 mm) equilibrated with the same buffer. A linear 0 to 0.15 M NaCl gradient was used to elute the protein. The purity was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis.
Spectroscopic analysis.
All spectral analyses were carried out in 50 mM phosphate buffer, pH 7.0, at room temperature. The reduced form was prepared by addition of 1 mM ascorbic acid or sodium dithionite to obtain the working concentration. Optical absorption spectra were measured using a Shimadzu UV-1600PC spectrophotometer. Raman spectra were obtained with a JASCO NRS-1000 spectrometer using a Kaiser Optical holographic notch-plus filter and a liquid N2-cooled charge-coupled device detector with 4-cm−1 spectral resolution. Data were accumulated for 10 min. The excitation source was a Coherent Innova 90C Kr laser, and Raman spectra were collected using a backscattering geometry. Peak frequencies were calibrated relative to an indene standard and were accurate to ±1 cm−1.
Measurement of midpoint potential.
The redox reaction was analyzed by cyclic voltammetry as described previously (25) with an ALS 624A electrochemical analyzer. All measurements were obtained using a three-electrode cell containing a 2-mercaptoethanol-modified gold working electrode, an Ag-AgCl-3 M NaCl reference electrode (205 mV versus the normal hydrogen electrode at 25°C), and a Pt wire counter electrode at room temperature under an N2 atmosphere. The gold electrode modified with 2-mercaptoethanol was prepared by dipping the bare gold electrode in a 20.0 mM aqueous solution of 2-mercaptoethanol for 1 h. After dipping, the electrode was rinsed with water. The cyclic voltammogram was obtained in 50 mM sodium acetate buffer, pH 4.0, at 25°C.
Adsorption of the protein on carbohydrates.
Adsorption experiments were performed in 50 mM sodium acetate buffer, pH 4.0, using 0.1% cellulose, xylan, mannan, arabinan, or chitin. These insoluble carbohydrates were prepared as described elsewhere (3, 9, 44, 49, 52). The protein concentrations in the additive solutions were calculated from the absorbance at 420 nm (ɛ420 = 130 mM−1 cm−1), which corresponded to the Soret band of b-type heme, and they ranged from 1.6 μM to 23 μM. The mixtures of the protein and carbohydrates were incubated with rotation (diameter, 20 cm; 20 rpm) for 1 h at 30°C and then were centrifuged for 10 min at 16,100 × g. The supernatants were centrifuged again to remove the precipitates completely, and the protein concentrations in the supernatants were determined again. The equilibrium dissociation constant (Kd) and the adsorption maximum (Amax) were estimated as described previously (15).
Nucleotide sequence accession number.
The nucleotide sequence of cDNA encoding CBCyt. b562 has been deposited in the DDBJ database under accession number AB193288.
RESULTS
Nucleotide and amino acid sequences.
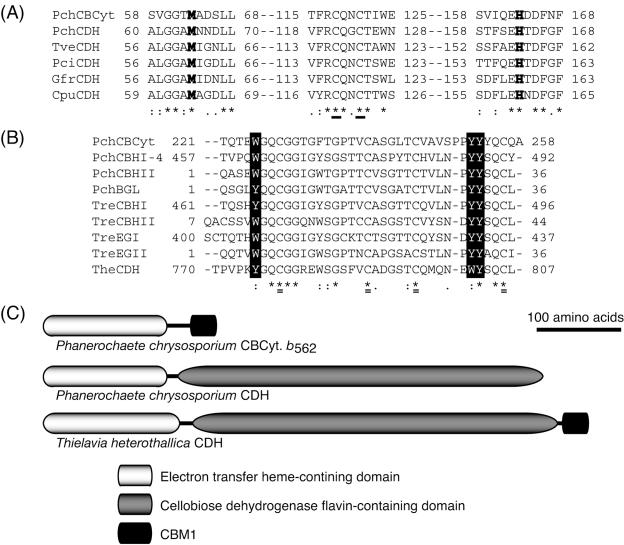
During the search of the P. chrysosporium genome database, a gene similar to the gene encoding the cytochrome domain of CDH was found at positions 110991 to 111711 in scaffold 87. The nucleotide sequence of the cDNA was determined by using RT-PCR and RACE with total RNA of P. chrysosporium K-3, and it was identical to that of the cellulose-binding iron reductase gene (cir1) recently described by Cullen (accession number AY682742). The N-terminal 19-amino-acid sequence was predicted to be a signal peptide by the SignalP program, suggesting that the mature protein consists of 258 amino acid residues with a molecular mass of 27.0 kDa and a pI of 3.95. The amino acid sequence contains two possible sites of N glycosylation and numerous possible sites of O glycosylation. In the sequence comparisons of the cDNA and cir1 and of the deduced amino acid sequences of the mature protein, eight nucleotides and three amino acids (S4T, T195A, and A258V) were different.
In homology searches of several public databases (PIR, Swiss-Prot, DAD, PDB, and PRF) at NCBI, the N-terminal 188 amino acids of the mature protein exhibited 44 to 48% identity with the cytochrome domains of basidiomycete CDHs and 31 to 33% identity with cytochrome domains of ascomycete CDHs. The sequence was aligned with the cytochrome domain of basidiomycete CDHs by ClustalW, and two possible heme ligands (Met 63 and His 163) and two cysteine residues forming a disulfide bond (Cys 118 and Cys 121) were conserved among these proteins, as shown in Fig. 2A. The C-terminal CBM1 contains three aromatic residues (W225, Y252, and Y253), which might contribute to binding on the surface of carbohydrates (Fig. 2B). As shown in Fig. 2C, the domain structure of CBCyt. b562 includes a C-terminal CBM1 but no flavin-containing domain. It thus differs significantly from the domain structure of Thielavia heterothallica CDH, which includes both CBM1 and a flavin domain, and from the domain structure of P. chrysosporium CDH, which includes a flavin domain but no CBM1.
FIG. 2.
(A) Multiple alignments of the cytochrome domains of CBCyt. b562 and basidiomycetes CDHs. Boldface type indicates methionine and histidine residues that are possible heme ligands, and conserved cysteines used for disulfide bond formation are underlined. PchCBCyt, CBCyt. b562 from P. chrysosporium; PchCDH, CDH from P. chrysosporium (accession no. X88897); TveCDH, CDH from Trametes versicolor (AF029668); PciCDH, CDH from Pycnoporus cinnabarinus (AF081574); GfrCDH, CDH from Grifola frondosa (AB083245); CpuCDH, CDH from Coniophora puteana (AB161046). (B) Multiple alignments of CBCyt. b562 CBM1 with other known CBM1s. Aromatic residues that are candidates for carbohydrate binding are indicated by solid boxes, and conserved cysteines used for disulfide bond formation are double underlined. PchCBHI-4, cellobiohydrolase I-4 from P. chrysosporium (accession no. L22656); PchCBHII, cellobiohydrolase II from P. chrysosporium (S76141); PchBGL, β-glucosidase from P. chrysosporium (AB081121); TreCBHI, cellobiohydrolase I from Trichoderma reesei (P62694); TreCBHII, cellobiohydrolase II from T. reesei (M16190); TreEGI, endoglucanase I from T. reesei (P07981); TreEGII, endoglucanase II from T. reesei (P07982); TheCDH, CDH from Thielavia heterothallica (AF074951). (C) Domain organization of CBCyt. b562 and CDH from P. chrysosporium and CBM1-carrying CDH from T. heterothallica.
Effect of carbon sources on expression of the CBCyt. b562 gene.
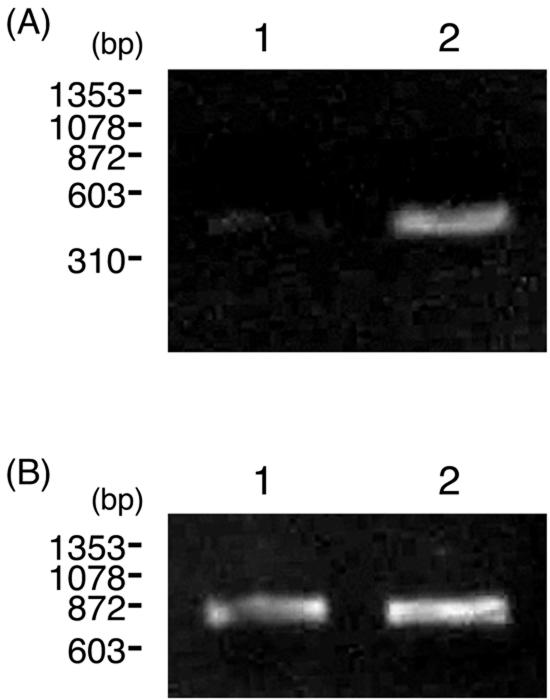
P. chrysosporium was grown on glucose or cellulose medium for 5 days, and RT-PCR analysis was carried out to detect expression of the CBCyt. b562 gene, using total RNA extracted from the mycelia. As shown in Fig. 3, transcripts of the CBCyt. b562 gene were detected in cellulose medium but not in glucose medium, as was the case for other carbohydrate-degrading enzymes, suggesting that transcription of the CBCyt. b562 gene is regulated by carbon catabolite repression.
FIG. 3.
RT-PCR analysis of CBCyt. b562 gene (A) and actin gene (B) expression in mycelium grown on different carbon sources. Lane 1, glucose medium; lane 2, cellulose medium. The PCR products were separated on a 2% agarose gel.
Production and purification of the recombinant protein.
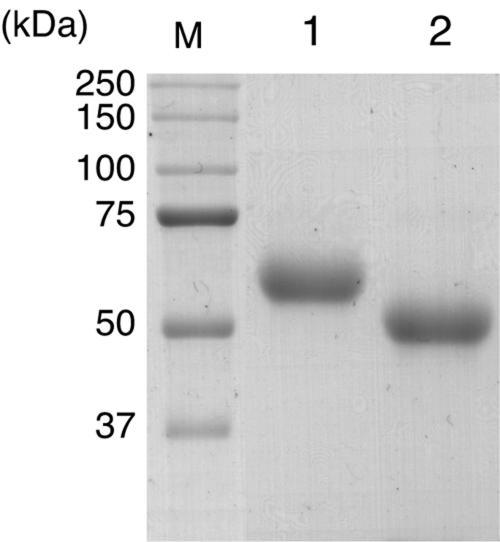
Recombinant CBCyt. b562 (rCBCyt. b562) was successfully produced in the expression system of P. pastoris and was purified from the culture solution by three-step column chromatography; the product gave a single band on sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis, as shown in Fig. 4. The actual molecular mass of the protein is higher than that calculated from the amino acid sequence. The N-terminal amino acid sequence (Gln-Ser-Ser-Ser-Gln-Phe-X-Asp-Ser-Ser) was identical to the predicted N-terminal sequence, and treatment with endo-H resulted in a considerable decrease in the molecular mass, suggesting that the higher-than-expected molecular mass mainly reflects N glycosylation of the recombinant protein. However, since the molecular mass of the deglycosylated protein (49 kDa) is still higher than that calculated from the amino acid sequence, the rCBCyt. b562 might also have undergone O glycosylation.
FIG. 4.
Effect of deglycosylation on purified rCBCyt. b562. Lane M, molecular mass standard; lane 1, rCBCyt. b562; lane 2, rCBCyt. b562 after incubation with endo-H. Two micrograms of each sample was separated on a 12% polyacrylamide gel.
Spectral analysis of the recombinant protein.
As shown in Fig. 5, the absorption spectrum of the oxidized form had broad peaks around 533 nm and 570 nm and a sharp peak at 420 nm. Upon reduction by addition of ascorbic acid or sodium dithionite, peaks appeared at 562, 533, and 430 nm, corresponding to the α-, β- and γ (Soret)-bands, respectively.
FIG. 5.
Absorption spectra of rCBCyt. b562. Solid line, oxidized form; dotted line, reduced form obtained by addition of sodium dithionite; dashed line, reduced form obtained by addition of ascorbic acid. All determinations were carried out in 50 mM phosphate buffer, pH 7.0, at room temperature.
For further characterization of the heme in CBCyt. b562, resonance Raman spectra were measured for both oxidized and reduced rCBCyt. b562 with excitation at 413 nm. The oxidation state marker, υ4, and core size markers, υ2 and υ3, of the oxidized form were present at 1,373 cm−1, 1,578 cm−1, and 1,504 cm−1, indicating the presence of 6-coordinated, low-spin heme. These values shifted to 1,365 cm−1, 1,582 cm−1, and 1,494 cm−1, respectively, in the reduced form prepared by addition of ascorbic acid. Therefore, the ferrous heme in CBCyt. b562 is also in a 6-coordinated, low-spin state (10). As shown in Table 1, the resonance Raman spectra of rCBCyt. b562 were almost identical to those of cytCDHs. These spectral analyses indicated that there is environmental similarity around the heme cofactor, including coordination and spin state, and, therefore, CBCyt. b562 should have properties similar to those of heme cofactors for the cytochrome domain of CDHs.
TABLE 1.
Resonance Raman frequencies of CBCyt. b562 and cytochrome domain of CDH
| Assignment | CBCyt.b562(cm−1)
|
Cytochrome domain of CDH (cm−1)a
|
||
|---|---|---|---|---|
| Ferric | Ferrous | Ferric | Ferrous | |
| υ10 | 1,619 | 1,618 | 1,622 | 1,618 |
| υ2 | 1,578 | 1,582 | 1,576 | 1,583 |
| υ38 | 1,565 | 1,556 | 1,561 | 1,555 |
| υ3 | 1,504 | 1,494 | 1,504 | 1,494 |
| 1,431 | 1,433 | 1,431 | 1,450 | |
| υ20 | 1,389 | 1,390 | ||
| υ4 | 1,373 | 1,365 | 1,370 | 1,362 |
| 1,310 | 1,312 | 1,307 | 1,311 | |
| 1,231 | 1,226 | 1,228 | 1,224 | |
| 1,172 | 1,1173 | |||
| υ6 + υ8 | 1,126 | 1,129 | 1,126 | 1,129 |
| 996 | 1,002 | 991 | 994 | |
Data from reference 10.
Electrochemical analysis of the recombinant protein.
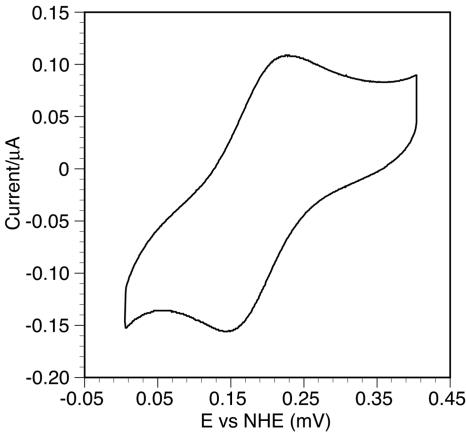
As shown in Fig. 6, apparent electron transfer was observed between CBCyt. b562 and the electrode without any mediator, and the midpoint potential of CBCyt. b562 was estimated to be 194 mV at pH 4.0. This value is slightly more positive than that for the cytochrome domain of CDH (180 mV) measured by the same technique (25).
FIG. 6.
Cyclic voltammogram of rCBCyt. b562. The midpoint potentials were determined from the average of the cathodic and anodic peaks of the voltammogram.
Carbohydrate-binding properties of CBM1 in CBCyt. b562.
The amount of CBCyt. b562 adsorbed on insoluble carbohydrates was measured at various protein concentrations, and the calculated values for Amax and Kd are shown in Table 2. Although the Amax value of CBCyt. b562 for bacterial microcrystalline cellulose was the highest Amax value for the insoluble glycans tested, the binding efficiency (Amax/Kd) for chitin was highest and was approximately threefold higher than that for cellulose. Such binding behavior is often observed with CBM1 (e.g., in the case of Trichoderma reesei cellobiohydrolase II) (8, 35).
TABLE 2.
Binding parameters of CBCyt. b562 for various carbohydratesa
| Carbohydrate | Organism | Kd (μM) | Amax (μmol/g) | Amax/ Kd |
|---|---|---|---|---|
| Cellulose | Acetobacter xylinius | 5.16 | 7.48 | 1.45 |
| Chitin | Thalassiosira weissflogii | 0.54 | 2.34 | 4.33 |
| Xylan | Betula papyrifera | 4.29 | 0.38 | 0.09 |
| Mannan | Phytelephs macrocarpa | 6.06 | 1.63 | 0.27 |
| Arabinan | Beta vulgaris | 3.50 | 1.04 | 0.30 |
The parameters were derived from adsorption data plotted as described in Materials and Methods.
DISCUSSION
Many carbohydrate-acting enzymes contain carbohydrate-binding modules (CBMs), which are defined as noncatalytic polysaccharide-recognizing modules (7). CBMs are classified into 42 families based on the similarity of amino acid sequences and the three-dimensional structures in the CAZY database (http://afmb.cnrs-mrs.fr/CAZY/), and they display diverse ligand specificities for various carbohydrates. Among them, family 1 CBMs (CBM1s), are often found in fungal carbohydrate-acting enzymes, and they are thought to contribute to binding to crystalline cellulose. Removal of the module causes a significant decrease in both binding and catalytic activity against cellulose (36, 50, 55), suggesting that the CBM1 is directly related to the catalytic function of these enzymes. Most CDHs do not have this type of module, although it is known that CDHs also bind to cellulose and that this ability is due to part of the flavin domain (20, 21, 46, 48). Since cellulose-bound CDH retains cellobiose-oxidizing activity (20) and the enzyme is localized on cellulose surfaces in vivo (23), the cellulose-binding ability of CDH helps its catalytic activity, as is the case for many CBM-containing enzymes. In the present study, we found that CBCyt. b562 has a unique structure; i.e., the protein lacks the cellobiose-oxidizing flavin domain of CDH but carries a C-terminal CBM1 instead. As far as we know, this is the first example of a CBM1 connected to a domain without apparent catalytic activity toward carbohydrate. The CBM1 in CBCyt. b562 nevertheless presumably contributes to the localization of the protein. In this study, we tested the binding ability of the protein not only for cellulose but also for other polysaccharides, and we observed a high affinity of CBCyt. b562 for chitin, as well as cellulose. Although the surface condition of these glycans strongly depends on the source organism and it is not easy to compare binding parameters, the results clearly suggest the possibility of localization of CBCyt. b562 on the fungal sheath, as well as on the surface of cellulose. Further cytochemical analysis is required to identify the localization of CBCyt. b562 in vivo.
The cytochrome domain of CBCyt. b562 shows significant homology with that of CDHs, and residues essential to ligate heme (19) are well conserved. Moreover, the Raman spectrum showed that the heme of CBCyt. b562 is 6-coordinated in both the ferric and ferrous states, suggesting that CBCyt. b562 has an electron transfer function, like that of the cytochrome domain of CDH. In RT-PCR analysis, the CBCyt. b562 gene transcripts were detected in cellulose medium but not in glucose medium, as is the case for cdh (34). This result led us to suspect that the function of CBCyt. b562 is as a physiological electron acceptor of CDH. Indeed, when CBCyt. b562 was mixed with CDH in vitro, it was partially reduced by addition of cellobiose (data not shown). However, the similar values for the midpoint potentials of the cytochrome domains of CBCyt. b562 and CDH would be thermodynamically disadvantageous for accepting electrons from CDH. Recently, Martinez and coworkers found various unknown genes encoding extracellular oxidoreductases in the P. chrysosporium genome (39). Therefore, some of these enzymes are candidates for the natural electron donor of CBCyt. b562. On the other hand, functional and structural similarities between CBCyt. b562 and CDH indicate that the heme in CBCyt.b562 may have Fe(III)-reducing ability. Therefore, Fe(III)-containing compounds might be candidates for the possible electron acceptor for CBCyt. b562.
To date, the cytochrome domain of CDH is the only hemoprotein with an immunoglobulin-like β-sandwich fold in the Structural Classification of Proteins (SCOP) (11, 42). During BLAST searches of various genome databases, however, we found many genes that may encode similar hemoproteins which share the putative ligands of heme and the two cysteine residues for forming a disulfide bond (19). A comparison of the deduced amino acid sequences indicated that some of these proteins have an N-terminal signal peptide and some do not, as shown in Table 3. In addition to the flavin-binding domain or CBM1, moreover, there are hemoproteins carrying both the flavin domain and CBM1 (Gibberella zeae, Magnaporthe grisea, Neurospora crassa, and Thielavia heterothallica), a Rieske [2Fe-2S] center (Aspergillus nidulans), or a transmembrane domain (M. grisea, Neurospora crassa) in the C-terminal region. These results clearly suggest that these proteins might have evolved by fusion of ancestral functional domains in order to acquire various redox systems, and it seems likely that the hemoproteins function not only extracellularly but also in the intracellular region or membrane. The occurrence of such hemoproteins, as well as CBCyt. b562 as described here, may provide unique insight into the novel redox networks in filamentous fungi.
TABLE 3.
Genes encoding CBCyt. b562, CDH, and unknown hemoproteins similar to the cytochrome domains of CBCyt. b562 and CDH
| Protein | Organism | Accession no. | Feature | Reference |
|---|---|---|---|---|
| CBCyt. b562 | Phanerochaete chrysosporium | AB193288 | Extracellular cytochrome, carries CBM1 | This study |
| CDH | Phanerochaete chrysosporium | X88897 | Extracellular flavocytochrome | 45 |
| Trametes versicolor | AF029668 | Extracellular flavocytochrome | 12 | |
| Pycnoporus cinnabarinus | AF081574 | Extracellular flavocytochrome | 41 | |
| Grifola frondosa | AB083245 | Extracellular flavocytochrome | 61 | |
| Athelia rolfsii | AY187232 | Extracellular flavocytochrome | Databasea | |
| Coniophora puteana | AB161046 | Extracellular flavocytochrome | 28 | |
| Irpex lacteus | AB187223 | Extracellular flavocytochrome | Databasea | |
| Thielavia heterothallica | AF074951 | Extracellular flavocytochrome, carries CBM1 | 51 | |
| Humicola insolens | AF257654 | Extracellular flavocytochrome | 59 | |
| Aspergillus nidulans | XM_411367 | Extracellular flavocytochrome | Databasea | |
| Gibberella zeae | XM_389261 | Extracellular flavocytochrome, carries a CBM1 | Databasea | |
| Gibberella zeae | XM_383918 | Extracellular flavocytochrome | Databasea | |
| Gibberella zeae | XM_385048 | Extracellular flavocytochrome | Databasea | |
| Magnaporthe grisea | XM_360402 | Extracellular flavocytochrome, carries CBM1 | Databasea | |
| Neurospora crassa | XM_325777 | Extracellular flavocytochrome | Databasea | |
| Neurospora crassa | XM_322291 | Extracellular flavocytochrome, carries a CBM1 | Databasea | |
| Unknown oxidoreductase | Aspergillus nidulans | XM_408099 | Extracellular oxidoreductase, carries Rieske [2Fe-2S] domain | Databasea |
| Unknown cytochrome | Gibberella zeae | XM_382527 | Extracellular cytochrome, Shows short sequence at C-terminus | Databasea |
| Magnaporthe grisea | XM_368045 | Intracellular cytochrome, shows short sequence at C terminus | Databasea | |
| Magnaporthe grisea | XM_365000 | Intracellular cytochrome | Databasea | |
| Magnaporthe grisea | XM_369170 | Extracellular cytochrome | Databasea | |
| Neurospora crassa | XM_329062 | Extracellular cytochrome | Databasea | |
| Neurospora crassa | XM_325449 | Intracellular cytochrome | Databasea | |
| Magnaporthe grisea | XM_363329 | Membrane-bound cytochrome, carries a transmembrane domain | Databasea | |
| Neurospora crassa | XM_330633 | Membrane-bound cytochrome, carries a transmembrane domain | Databasea |
GenBank/EMBL/DDBJ/PDB.
Acknowledgments
We are grateful to Toshio Iwasaki (Department of Biochemistry and Molecular Biology, Nippon Medical School) for valuable discussions about cytochrome b562.
This research was supported by grant-in-aid for scientific research 14360094 (to M.S.) and by research fellowship 08446 (to M.Y.) from the Japan Society for the Promotion of Science.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Araki, J., M. Wada, S. Kuga, and T. Okano. 1998. Flow properties of microcrystalline cellulose suspension prepared by acid treatment of native cellulose. Colloids Surf. A 142:75-82. [Google Scholar]
- 4.Ayers, A. R., S. B. Ayers, and K.-E. Eriksson. 1978. Cellobiose oxidase, purification and partial characterization of a hemoprotein from Sporotrichum pulverulentum. Eur. J. Biochem. 90:171-181. [DOI] [PubMed] [Google Scholar]
- 5.Bao, W., S. N. Usha, and V. Renganathan. 1993. Purification and characterization of cellobiose dehydrogenase, a novel extracellular hemoflavoenzyme from the white-rot fungus Phanerochaete chrysosporium. Arch. Biochem. Biophys. 300:705-713. [DOI] [PubMed] [Google Scholar]
- 6.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 7.Boraston, A. B., D. N. Bolam, H. J. Gilbert, and G. J. Davies. 2004. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382:769-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrard, G., and M. Linder. 1999. Widely different off rates of two closely related cellulose-binding domains from Trichoderma reesei. Eur. J. Biochem. 262:637-643. [DOI] [PubMed] [Google Scholar]
- 9.Chanzy, H. D., A. Grosrnaud, R. Voung, and W. Mackie. 1984. The crystal polymorphism of mannan in plant cell walls and after recrystallization. Planta 161:320-329. [DOI] [PubMed] [Google Scholar]
- 10.Cohen, J. D., W. Bao, V. Renganathan, S. S. Subramaniam, and T. M. Loehr. 1997. Resonance Raman spectroscopic studies of cellobiose dehydrogenase from Phanerochaete chrysosporium. Arch. Biochem. Biophys. 341:321-328. [DOI] [PubMed] [Google Scholar]
- 11.Conte, L. L., S. E. Brenner, T. J. P. Hubbard, C. Chothia, and A. G. Murzin. 2002. SCOP database in 2002: refinements accommodate structural genomics. Nucleic Acids Res. 30:264-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dumonceaux, T. J., K. A. Bartholomew, T. C. Charles, S. M. Moukha, and F. S. Archibald. 1998. Cloning and sequencing of a gene encoding cellobiose dehydrogenase from Trametes versicolor. Gene 210:211-219. [DOI] [PubMed] [Google Scholar]
- 13.Eriksson, K.-E., B. Pettersson, and U. Westermark. 1974. Oxidation: an important enzyme reaction in fungal degradation of cellulose. FEBS Lett. 49:282-285. [DOI] [PubMed] [Google Scholar]
- 14.Eriksson, K.-E. 1978. Enzyme mechanisms involved in cellulose hydrolysis by the rot fungus Sporotrichum pulverulentum. Biotechnol. Bioeng. 70:317-332. [Google Scholar]
- 15.Gilkes, N. R., E. Jervis, B. Henrissat, B. Tekant, R. C. Miller, Jr., R. A. J. Warren, and D. G. Kilburn. 1992. The adsorption of a bacterial cellulose and its two isolated domains to crystalline cellulose. J. Biol. Chem. 267:6743-6749. [PubMed] [Google Scholar]
- 16.Glenn, J. K., M. A. Morgan, M. B. Mayfield, M. Kuwahara, and M. H. Gold. 1983. An extracellular H2O2-requiring enzyme preparation involved in lignin biodegradation by the white rot basidiomycete Phanerochaete chrysosporium. Biochem. Biophys. Res. Commun. 114:1077-1083. [DOI] [PubMed] [Google Scholar]
- 17.Habu, N., M. Samejima, J. F. Dean, and K.-E. L. Eriksson. 1993. Release of the FAD domain from cellobiose oxidase by proteases from cellulolytic cultures of Phanerochaete chrysosporium. FEBS Lett. 327:161-164. [DOI] [PubMed] [Google Scholar]
- 18.Habu, N., K. Igarashi, M. Samejima, B. Pettersson, and K.-E. L. Eriksson. 1997. Enhanced production of cellobiose dehydrogenase in cultures of Phanerochaete chrysosporium supplemented with bovine calf serum. Biotechnol. Appl. Biochem. 26:97-102. [PubMed] [Google Scholar]
- 19.Hallberg, B. M., T. Bergfors, K. Backbro, G. Pettersson, G. Henriksson, and C. Divne. 2000. A new scaffold for binding haem in the cytochrome domain of the extracellular flavocytochrome cellobiose dehydrogenase. Structure 8:79-88. [DOI] [PubMed] [Google Scholar]
- 20.Henriksson, G., G. Pettersson, G. Johansson, A. Ruiz, and E. Uzcategui. 1991. Cellobiose oxidase from Phanerochaete chrysosporium can be cleaved by papain into two domains. Eur. J. Biochem. 196:101-106. [DOI] [PubMed] [Google Scholar]
- 21.Henriksson, G., A. Salumets, C. Divne, and G. Pettersson. 1997. Studies of cellulose binding by cellobiose dehydrogenase and a comparison with cellobiohydrolase 1. Biochem. J. 324:833-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henriksson, G., G. Johansson, and G. Pettersson. 2000. A critical review of cellobiose dehydrogenases. J. Biotechnol. 78:93-113. [DOI] [PubMed] [Google Scholar]
- 23.Igarashi, K., M. Samejima, Y. Saburi, N. Habu, and K.-E. L. Eriksson. 1997. Localization of cellobiose dehydrogenase in cellulose-grown cultures of Phanerochaete chrysosporium. Fungal Genet. Biol. 21:214-222. [DOI] [PubMed] [Google Scholar]
- 24.Igarashi, K., M. Samejima, and K.-E. L. Eriksson. 1998. Cellobiose dehydrogenase enhances Phanerochaete chrysosporium cellobiohydrolase I activity by relieving product inhibition. Eur. J. Biochem. 253:101-106. [DOI] [PubMed] [Google Scholar]
- 25.Igarashi, K., M. F. J. M. Verhagen, M. Samejima, M. Schulein, K.-E. L. Eriksson, and T. Nishino. 1999. Cellobiose dehydrogenase from the fungi Phanerochaete chrysosporium and Humicola insolens. J. Biol. Chem. 274:3338-3344. [DOI] [PubMed] [Google Scholar]
- 26.Igarashi, K., T. Tani, R. Kawai, and M. Samejima. 2003. Family 3 β-glucosidase from cellulose-degrading culture of the white-rot fungus Phanerochaete chrysosporium is a glucan 1,3-β-glucosidase. J. Biosci. Bioeng. 95:572-576. [DOI] [PubMed] [Google Scholar]
- 27.Johnsrud, S. C., and K.-E. Eriksson. 1985. Cross-breeding of selected and mutated homokaryotic strains of Phanerochaete chrysosporium K-3: new cellulase deficient strains with increased ability to degrade lignin. Appl. Microbiol. Biotechnol. 21:320-327. [Google Scholar]
- 28.Kajisa, T., M. Yoshida, K. Igarashi, A. Katayama, T. Nishino, and M. Samejima. 2004. Characterization and molecular cloning of cellobiose dehydrogenase from the brown-rot fungus Coniophora puteana. J. Biosci. Bioeng. 98:57-63. [DOI] [PubMed] [Google Scholar]
- 29.Kelley, R. L., and C. A. Reddy. 1986. Purification and characterization of glucose oxidase from Ligninolytic cultures of Phanerochaete chrysosporium. J. Bacteriol. 166:269-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kersten, P. J., and T. K. Kirk. 1987. Involvement of a new enzyme, glyoxal oxidase, in extracellular H2O2 production by Phanerochaete chrysosporium. J. Bacteriol. 169:2195-2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kremer, S. M., and P. M. Wood. 1992. Evidence that cellobiose oxidase from Phanerochaete chrysosporium is primarily an Fe(III) reductase. Eur. J. Biochem. 205:133-138. [DOI] [PubMed] [Google Scholar]
- 32.Kremer, S. M., and P. M. Wood. 1992. Production of Fenton's reagent by cellobiose oxidase from cellulolytic cultures of Phanerochaete chrysosporium. Eur. J. Biochem. 208:807-814. [DOI] [PubMed] [Google Scholar]
- 33.Kuwahara, M., J. K. Glenn, M. A. Morgan, and M. H. Gold. 1984. Separation and characterization of two extracellular H2O2-dependent oxidases from ligninolytic cultures of Phanerochaete chrysosporium. FEBS Lett. 169:247-250. [Google Scholar]
- 34.Li, B., S. R. Nagalla, and V. Renganathan. 1996. Cloning of a cDNA encoding cellobiose dehydrogenase, a hemoflavoenzyme from Phanerochaete chrysosporium. Appl. Environ. Microbiol. 62:1329-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Linder, M., I. Salovuori, L. Ruohonen, and T. T. Teeri. 1996. Characterization of a double cellulose-binding domain. J. Biol. Chem. 271:21268-21272. [DOI] [PubMed] [Google Scholar]
- 36.Linder, M., and T. T. Teeri. 1997. The role and function of cellulose-binding domains. J. Biotechnol. 57:15-28. [DOI] [PubMed] [Google Scholar]
- 37.Marchler-Bauer, A., J. B. Anderson, C. DeWeese-Scott, N. D. Fedorova, L. Y. Geer, S. He, D. I. Hurwitz, J. D. Jackson, A. R. Jacobs, C. J. Lanczycki, C. A. Liebert, C. Liu, T. Madej, G. H. Marchler, R. Mazumder, A. N. Nikolskaya, A. R. Panchenko, B. S. Rao, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, S. Vasudevan, Y. Wang, R. A. Yamashita, J. J. Yin, and S. H. Bryant. 2003. CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res. 31:383-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martinez, A. T. 2002. Molecular biology and structural-function of lignin-degrading heme peroxidases. Enzyme Microb. Technol. 30:425-444. [Google Scholar]
- 39.Martinez, D., L. F. Larrondo, N. Putnam, M. D. S. Gelpke, K. Huang, J. Chapman, K. G. Helfenbein, P. Ramaiya, J. C. Detter, F. Larimer, P. M. Coutinho, B. Henrissat, R. Berka, D. Cullen, and D. Rokhsar. 2004. Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat. Biotechnol. 22:695-700. [DOI] [PubMed] [Google Scholar]
- 40.Morpeth, F. F. 1985. Some properties of cellobiose oxidase from the white-rot fungus Sporotrichum pulverulentum. Biochem. J. 228:557-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moukha, S. M., T. J. Dumonceaux, E. Record, and F. S. Archibald. 1999. Cloning and analysis of Pycnoporus cinnabarinus cellobiose dehydrogenase. Gene 234:23-33. [DOI] [PubMed] [Google Scholar]
- 42.Murzin, A. G., S. E. Brenner, T. Hubbard, and C. Chothia. 1995. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 247:536-540. [DOI] [PubMed] [Google Scholar]
- 43.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 44.Noishiki, Y., Y. Nishiyama, M. Wada, S. Okada, and S. Kuga. 2003. Inclusion complex of β-chitin and aliphatic amines. Biomacromolecules 4:944-949. [DOI] [PubMed] [Google Scholar]
- 45.Raices, M., E. Paifer, J. Cremata, R. Montesino, J. Stahlberg, C. Divne, I. J. Szabo, G. Henriksson, G. Johansson, and G. Pettersson. 1995. Cloning and characterization of a cDNA encoding a cellobiose dehydrogenase from the white rot fungus Phanerochaete chrysosporium. FEBS Lett. 369:233-238. [DOI] [PubMed] [Google Scholar]
- 46.Renganathan, V., S. N. Usha, and F. Lindenburg. 1990. Cellobiose-oxidizing enzymes from the lignocellulose-degrading basidiomycete Phanerochaete chrysosporium: interaction with microcrystalline cellulose. Appl. Microbiol. Biotechnol. 32:609-613. [Google Scholar]
- 47.Samejima, M., and K.-E. L. Eriksson. 1992. A comparison of the catalytic properties of cellobiose quinone oxidoreductase and cellobiose oxidase from Phanerochaete chrysosporium. Eur. J. Biochem. 207:103-107. [DOI] [PubMed] [Google Scholar]
- 48.Samejima, M., T. Ohkubo, K. Igarashi, A. Isogai, S. Kuga, J. Sugiyama, and K.-E. L. Eriksson. 1997. The behavior of Phanerochaete chrysosporium cellobiose dehydrogenase on adsorption to crystalline and amorphous celluloses. Biotechnol. Appl. Biochem. 25:135-141. [Google Scholar]
- 49.Seto, A., Y. Kojima, N. Tonouchi, T. Tsukada, and F. Yoshinaga,. 1997. Screening of bacterial cellulose-producing Acetobacter strains suitable for sucrose as a carbon source. Biosci. Biotechnol. Biochem. 61:735-736. [Google Scholar]
- 50.Stahlberg, J., G. Johansson, and G. Pettersson. 1993. Trichoderma reesei has no true exo-cellulase: all intact and truncated cellulases produce new reducing end groups on cellulose. Biochim. Biophys. Acta 1157:107-113. [DOI] [PubMed] [Google Scholar]
- 51.Subramaniam, S. S., S. R. Nagalla, and V. Renganathan. 1999. Cloning and characterization of a thermostable cellobiose dehydrogenase from Sporotrichum thermophile. Arch. Biochem. Biophys. 365:223-230. [DOI] [PubMed] [Google Scholar]
- 52.Sugiyama, J., J. Persson, and H. Chanzy. 1991. Combined infrared and electron diffraction study of the polymorphism of native celluloses. Macromolecules 24:2461-2466. [Google Scholar]
- 53.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tien, M., and T. K. Kirk. 1984. Lignin-degrading enzyme from Phanerochaete chrysosporium: purification, characterization, and catalytic properties of a unique H2O2-requiring oxygenase. Proc. Natl. Acad. Sci. USA 81:2280-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tomme, P., H. van Tilbeurgh, G. Pettersson, J. van Damme, J. Vandekerckhove, J. Knowles, T. T. Teeri, and M. Claeyssens. 1988. Studies of the cellulolytic system of Trichoderma reesei QM 9414. Analysis of domain function in two cellobiohydrolases by limited proteolysis. Eur. J. Biochem. 170:575-581. [DOI] [PubMed] [Google Scholar]
- 56.Westermark, U., and K.-E. Eriksson. 1974. Carbohydrate-dependent enzyme quinone reduction during lignin degradation. Acta Chem. Scand. Ser. B 28:204-208. [Google Scholar]
- 57.Westermark, U., and K.-E. Eriksson. 1974. Cellobiose:quinone oxidoreductase, a new wood-degrading enzyme from white-rot fungi. Acta Chem. Scand. Ser. B 28:209-214. [Google Scholar]
- 58.Wood, J. D., and P. M. Wood. 1992. Evidence that cellobiose:quinone oxidoreductase from Phanerochaete chrysosporium is a breakdown product of cellobiose oxidase. Biochim. Biophys. Acta 1119:90-96. [DOI] [PubMed] [Google Scholar]
- 59.Xu, F., E. J. Golightly, K. R. Duke, S. F. Lassen, B. Knusen, S. Christensen, K. M. Brown, S. H. Brown, and M. Schulein. 2001. Humicola insolens cellobiose dehydrogenase: cloning, redox chemistry, and “logic gate”-like dual functionality. Enzyme Microb. Technol. 28:744-753. [DOI] [PubMed] [Google Scholar]
- 60.Yoshida, M., T. Ohira, K. Igarashi, H. Nagasawa, K. Aida, B. M. Hallberg, C. Divne, T. Nishino, and M. Samejima. 2001. Production and characterization of recombinant Phanerochaete chrysosporium cellobiose dehydrogenase in the methylotrophic yeast Pichia pastoris. Biosci. Biotechnol. Biochem. 65:2050-2057. [DOI] [PubMed] [Google Scholar]
- 61.Yoshida, M., T. Ohira, K. Igarashi, H. Nagasawa, and M. Samejima. 2002. Molecular cloning and characterization of a cDNA encoding cellobiose dehydrogenase from the wood-rotting fungus Grifola frondosa. FEMS Microbiol. Lett. 217:225-230. [DOI] [PubMed] [Google Scholar]
- 62.Yoshida, M., K. Igarashi, R. Kawai, K. Aida, and M. Samejima. 2004. Differential transcription of β-glucosidase and cellobiose dehydrogenase genes in cellulose degradation by the basidiomycete Phanerochaete chrysosporium. FEMS Microbiol. Lett. 235:177-182. [DOI] [PubMed] [Google Scholar]