Abstract
Serotyping is the foundation of pathogenic Escherichia coli diagnostics; however, few laboratories have this capacity. We developed a molecular serotyping protocol that targets, genetically, the same somatic and flagellar antigens of E. coli O26:H11 used in traditional serotyping. It correctly serotypes strains untypeable by traditional methods, affording primary laboratories serotyping capabilities.
Human infections with Escherichia coli O26:H11 and O26:H-untypeable (H-) or H-nonmotile (NM) strains are associated with diarrhea, hemorrhagic colitis, and hemolytic uremic syndrome (2, 33). Depending on virulence factors, E. coli O26 is classified as enterohemorrhagic (EHEC) or enteropathogenic (EPEC). The E. coli O26 serotype, first reported to cause pediatric EPEC diarrhea (18), has been isolated from EHEC outbreaks in Europe (1, 13, 15, 31), South America (22, 28), and Japan (10, 11); from sporadic cases in Canada (12), Australia (7), and the United States (4); and from sick (14, 21) and healthy (20, 25, 32) livestock. E. coli O26 is the most frequently isolated non-O157 Shiga-toxigenic E. coli (STEC) associated with human clinical illness (3), and E. coli O26:H11 is the clinically most important and epidemiologically most predominant EPEC and EHEC O26 serotype (2, 33).
Conventional E. coli O:H serotyping by agglutination of somatic and flagellar antigens by the use of anti-E. coli polyclonal antiserum is time consuming, expensive, and available only in a small number of reference laboratories (3, 33). E. coli O:H serotyping of pathogens may be required, however, for proper diagnosis and treatment, to maximize an isolate's usefulness for surveillance and to determine overall disease trends (33). PCR-based methodologies to detect or identify pathogenic E. coli O26 strains have targeted virulence genes such as Shiga toxin (stx) (9) and intimin (eae) (12, 27), flagellar H-antigen genes fliC-fliA (17), and the O-antigen O26 wzx genes (5, 16, 23). PCR methods targeting O26 O-antigen or H11 fliC genes could substitute for serotyping (5, 24). We therefore developed a molecular serotyping method to target the somatic and flagellar antigens and to allow clinical laboratories to accurately serotype E. coli as O26 and/or H11 by multiplex PCR (mPCR).
A 12-kb region of the E. coli O26:NM O-antigen operon from bovine fecal isolate SB6629 was sequenced (accession number AY763106); this region is identical to the corresponding region of the O26 O-antigen operon sequence (AF529080) reported by D'Souza et al. (6). Twenty sets of E. coli O26 O-antigen specific primers were synthesized and evaluated against a small panel of E. coli O26 and non-O26 E. coli isolates. One primer set, wzx-wzyO26F and wzx-wzyO26R, targeting the O26 wzx-wzy genes (O-antigen flippase and polymerase, respectively), had 100% sensitivity and specificity and a suitable amplicon size.
The E. coli H7 flagellum gene sequence is heterogenous, and fliCH7 sequence of the O157 serogroup is distinct from fliCH7 sequence of other O serogroups (29). To determine H11 flagellum sequence heterogeneity across different O serogroups (35), the fliCH11 genes of 20 H11 E. coli isolates comprising five O serogroups were sequenced (accession numbers AY906918 to AY906937). Eighteen of these sequences were identical to those previously reported (34, 35). Two E. coli O26:H11 strains had one and two nonsynonymous substitutions, respectively, compared to the consensus fliCH11 sequence. Thus, we chose to use fliCH11 primers designed by Wang et al. (30) for our assay.
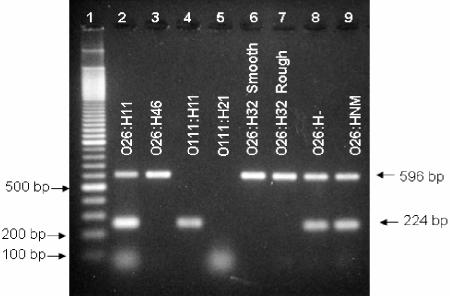
The primers for E. coli O26:H11 molecular serotyping were wzx-wzyO26F (5′-AAATTAGAAGCGCGTTCATC), wzx-wzyO26R (5′-CCCAGCAAGCCAATTATGACT), fliCRH11-1 (5′-ACTGTTAACGTAGATAGC) (30), and fliCRH11-2 (5′-TCAATTTCTGCAGAATATAC) (30). Cells were prepared by diluting overnight Trypticase soy broth cultures 1:10 with reagent grade water. The 20 μl PCR used 1 μl of template and 0.5 U HotStarTaq (QIAGEN, Valencia, CA). The PCR mix contained concentrations of 500 μM per nucleotide, 600 nM for each primer, and 4.5 mM MgCl2. Thermocycler conditions were 95°C for 15 min, 35 cycles of 94°C for 60 s, 56°C for 60 s, and 72°C for 60 s, and a final elongation of 72°C for 10 min. Amplicons were visualized by electrophoresis on a 2% agarose gel run at 100 V for 2 h, stained with ethidium bromide, and digitally photographed. Isolates positive for the O26 and H11 alleles displayed bands of 596 bp and 224 bp, respectively (Fig. 1). We applied the E. coli O26/H11 mPCR to three diverse bacterial isolate panels to evaluate assay performance. Panel I was composed of 344 isolates: 322 diverse E. coli of known O and/or H serotype (including 31 E. coli O26:H11 strains) of human, animal, insect, and environmental origins, and 22 non-E. coli. This panel was used to generate mPCR diagnostic sensitivity and specificity estimates. (A detailed list of the strains used is available at http://www.ars.usda.gov/sp2UserFiles/Place/54380570/AHRU/E.coli/AEM_Durso_isolate_list.pdf). The O26 and H11 serotype status of 42 E. coli strains comprising panel II was tested blindly (without knowledge of their conventional O:H serotype). Panel III, a subset of panel I composed of 35 E. coli O26:NM (34 eae positive, 1 eae negative) and 11 O26:H- (10 eae positive, 1 eae negative) strains, was used to estimate the proportion of E. coli O26:NM/H- strains that possess the fliCH11 gene without expressing the H11 antigen. Although all reactions were run in the multiplex format, assay performance was measured separately for the O26 and H11 primer sets. Additionally, E. coli O26 strains were serologically confirmed as O26 by enzyme immunoassay using an anti-E. coli O26 murine monoclonal antibody (26), and E. coli O26 and H11 strains were assayed for the presence of stx1, stx2, and eae genes as previously described (19) (Table 1).
FIG. 1.
Molecular serotyping of E. coli O26:H11 by multiplex PCR. Lane 1, molecular marker (100 bp); lane 2, H311b, O26 standard reference strain (E. coli O26:H11); lane 3, 5306-56 (E. coli O26:H46); lane 4, 88-4110 (E. coli O111:H11); lane 5, DEC 15C (E. coli O111:H21); lane 6, TWO7187a, O26 smooth (E. coli O26:H32); lane 7, TWO7187b, O26 rough (E. coli O26:H32); lane 8, 95.0256 (E. coli O26:H- untypeable); lane 9, TW04272 (E. coli O26:H nonmotile).
TABLE 1.
Sensitivity and specificity estimates of E. coli O26 and H11 mPCR compared to traditional serotyping for 344 panel I bacteria (322 E. coli and 22 non-E. coli)a
| Measure | No. of strains used | Estimate | 95% Confidence interval |
|---|---|---|---|
| O26 sensitivity (n = 81) | 48 E. coli O26 eae+, stx+ | 1.00 | 0.96-1.00 |
| 29 E. coli O26 eae+, stx− | |||
| 2 E. coli O26 eae−, stx+ | |||
| 2 E. coli O26 eae−, stx+ | |||
| O26 specificity (n = 260) | 238 non-O26 E. colib | 1.00 | 0.99-1.00 |
| 22 non-E. colic | |||
| H11 sensitivity (n = 38) | 26 E. coli H11 eae +, stx + | 1.00 | 0.91-1.00 |
| 10 E. coli H11 eae +, stx − | |||
| 2 E. coli H11 eae −, stx − | |||
| H11 specificity (n = 164) | 142 non-H11 E. colid | 1.00 | 0.98-1.00 |
| 22 non-E. colic |
Data compiled from multiplex PCR reactions. See isolate list for data on which strains were used in each analysis (http://www.ars.usda.gov/sp2UserFiles/Place/54380570/AHRU/E.coli/AEM_Durso_isolate_list.pdf).
Non-O26 E. coli includes 238 isolates of 111 different O serotypes and excludes three isolates for which O serotype data were unavailable.
Non-E. coli includes 22 isolates of 17 closely related genera.
Non-H11 E. coli includes 142 isolates of 51 different H serotypes and excludes all strains for which H serotype data were missing, all strains that did not react with any of the standard H antisera, and all nonmotile strains.
The E. coli O26/H11 mPCR assay was 100% sensitive and specific based on reactions with the 344 panel I bacteria (Table 1). The assay correctly determined the O26 and H11 serotype of the 42 blind-panel E. coli strains (Table 2). All 35 E. coli O26:NM and 10 of 11 E. coli O26:H- strains in panel III were fliCH11 positive. These results are consistent with the findings of Zhang et al. (34), who noted that eae-positive E. coli O26:H- strains belong to an H11 clonal complex. The fliCH11-negative E. coli O26:H- strain was eae negative and therefore not part of the H11 clonal complex. The mPCR assay also identified seven strains that had apparently been misserotyped or mislabeled with respect to O26 and/or H11 serotype. The mPCR and serotyping results for these seven strains initially appeared discordant. However, when these isolates were reserotyped by the E. coli Reference Center (University Park, PA), the revised results agreed with those of the mPCR O26/H11 assay. Importantly, molecular serotyping methods generate genotypic information, in contrast to traditional serotyping, which generates phenotypic (surface antigen expression) information. Thus, our E. coli O26/H11 mPCR permits serotyping of “antigenically silent” strains which are O or H untypeable by conventional serotyping techniques. For example, our PCR assay serotyped rough lipopolysaccharide (O-antigen-free) colonies of blind-panel isolate TW07187 as O26, and identified the fliCH11 gene in many NM and H- E. coli strains, in both O26 and non-O26 STEC.
TABLE 2.
Comparison of E. coli O26 and H11 serotyping results obtained from traditional serotyping, O26-specific monoclonal antibody enzyme-linked immunosorbent assay, and molecular serotyping by mPCR for 42 E. coli strains tested in a blind panel (panel II)
| Straina | Result by traditional serotyping
|
Result by MAbb α-O26 | Result by molecular serotyping
|
||
|---|---|---|---|---|---|
| O | H | O26 | H11 | ||
| TW07867 | 15 | 11 | negd | neg | + |
| TW09156 | 26 | 11 | + | + | + |
| TW07154 | 26 | 11 | + | + | + |
| TW07622 | 26 | 11 | + | + | + |
| TW07872 | 26 | 11 | + | + | + |
| TW08637 | 26 | 11 | + | + | + |
| TW08998 | 26 | 11 | + | + | + |
| IBL 7922 | 26 | 11 | + | + | + |
| IBL 8179 | 26 | 11 | + | + | + |
| IBL 8479 | 26 | 11 | + | + | + |
| IBL 8907 | 26 | 11 | + | + | + |
| TW07705 | 26 | 11 | + | + | + |
| IBL 8419 | 26 | 11 | + | + | + |
| TW01209 | 26 | 32 | neg | + | neg |
| TW07187 | 26 | 32 | + | + | neg |
| TW01221 | 26 | 36 | + | + | neg |
| CP-ALLG | 45 | 2 | neg | neg | neg |
| CP-COU | 45 | 2 | neg | neg | neg |
| CP-BALL | 45 | 2 | neg | neg | neg |
| TW01686 | 70 | 11 | neg | neg | + |
| TW08640 | 103 | 2 | neg | neg | neg |
| TW08872 | 103 | 11 | neg | neg | + |
| IBL 6582 | 103 | 25 | neg | neg | neg |
| IBL 8234 | 111 | NMc | neg | neg | neg |
| IBL 8361 | 111 | NM | neg | neg | neg |
| TW07926 | 111 | 8 | neg | neg | neg |
| TW03810 | 111 | 11 | neg | neg | + |
| TW05355 | 111 | 11 | neg | neg | + |
| TW08114 | 118 | 16 | + | neg | neg |
| TW08868 | 121 | 19 | neg | neg | neg |
| IBL 5518 | 121 | 19 | neg | neg | neg |
| IBL 6507 | 121 | 19 | neg | neg | neg |
| TW01110 | 124 | 30 | neg | neg | neg |
| IBL 6940 | 145 | NM | neg | neg | neg |
| IBL 8235 | 145 | NM | neg | neg | neg |
| IBL 7606 | 146 | 21 | neg | neg | neg |
| IBL 7427 | 157 | 7 | neg | neg | neg |
| IBL 8257 | 157 | 7 | neg | neg | neg |
| IBL 8434 | 157 | 7 | neg | neg | neg |
| IBL 8463 | 157 | 7 | neg | neg | neg |
| IBL 8827 | 157 | 7 | neg | neg | neg |
| IBL 9488 | 157 | 7 | neg | neg | neg |
Strains with TW prefix provided by T. Whittam, Michigan State University, East Lansing, MI; Strains with CP prefix provided by C. Park, Inova Fairfax Hospital, Falls Church, VA; strains with IBL prefix provided by V. Lockary, State of Idaho Bureau of Laboratories, Boise, ID.
Enzyme-linked immunosorbent assay reactivity with anti-O26 monoclonal antibody (MAb) 12F5 (26).
NM denotes nonmotile strain.
neg, negative.
In conclusion, the E. coli O26/H11 mPCR assay accurately determined O26 and H11 serotypes of 364 conventionally serotyped E. coli strains by targeting the genes that encode the same somatic and flagellar antigens used in traditional serotyping. Furthermore, the mPCR assay serotyped E. coli isolates (such as rough O26 and nonmotile H11 strains) which were untypeable by conventional methods. Third, we found that 45 out of 46 STEC and non-STEC O26:NM/H- strains were fliCH11 positive, analogous to the case of STEC O157:NM strains, which almost always possess the fliCH7 gene (8). Finally, the assay allowed for confirmation of E. coli O26 and H11 serotype.
Traditional E. coli O:H serotyping, which detects surface antigen expression, will remain important, but application of molecular serotyping techniques that detect their genetic analogues (counterparts) have some relative advantages. While molecular E. coli O:H serotyping may not be appropriate for all bacteriologic investigations, it is well suited for clinical diagnostic, research laboratory, or epidemiologic investigations. The E. coli O26/H11 mPCR assay described here should allow laboratories to generate E. coli O26 and H11 serotype data using routine PCR techniques and readily available equipment, since few laboratories have the capacity for serotyping non-O157 STEC (33). The assay was designed to allow for the addition of other PCR targets, such as stx or eae. We optimized this assay for use with a pure bacterial culture; as with traditional serotyping, it is not appropriate for application to complex sample matrices with mixed microbial flora.
Nucleotide sequence accession numbers.
The nucleotide sequence for E. coli O26 for the NM O-antigen operon from bovine fecal isolate SB6629 is deposited in GenBank as AY763106. Nucleotide sequences for 20 E. coli fliCH11 strains are as follows: strain ATCC 35401, O serotype 78, AY906918; strain DEC 10J, O serotype 70, AY906919; strain P1331, O serotype 26, AY906920; strain H311B, O serotype 26, AY906921; strain 89-491, O serotype 26, AY906922; strain 88-353, O serotype 26, AY906923; strain 88-157, O serotype 26, AY906924; strain H19, O serotype 26, AY906925; strain DEC 9E, O serotype 26, AY906926; strain DEC 9D, O serotype 26, AY906927; strain DEC 9A, O serotype 26, AY906928; strain DEC 10E, O serotype 26, AY906929; strain DEC 10D, O serotype 26, AY906930; strain DEC 10C, O serotype 26, AY906931; strain DEC 10B, O serotype 26, AY906932; strain Su 4321, O serotype 13, AY906933; strain 88-41, O serotype 111, AY906934; strain DEC 10A, O serotype 26, AY906935; strain DEC 8D, O serotype 26, AY906936; strain CL5, O serotype 26, AY906937.
Acknowledgments
This work was supported, in part, by USDA CSREES NRI grant 2002-2239 (awarded to J.E.K.). We thank Sandy Fryda-Bradley, Ron Mlejnek, and Liz Ossian for outstanding technical assistance; Kevin Tennill for photographic assistance; Joan Rosch for secretarial assistance; and Tom Wittum from The Ohio State University, Columbus, OH, Greg Siragusa from the USDA, ARS, Russell Research Center, Athens, GA, and anonymous reviewers for helpful comments on the manuscript. Bacterial isolates were kindly provided by C. Park, Inova Fairfax Hospital, Falls Church, VA; D. Francis, South Dakota State University, Brookings, SD; M. Doyle, University of Georgia, Athens, GA; D. Acheson, Food and Drug Administration, College Park, MD; J. Johnson, University of Minnesota Veterans Hospital, Minneapolis, MN; L. Riley, University of California, Berkeley, CA; T. Whittam, Michigan State University, East Lansing, MI; Centers for Disease Control and Prevention, Atlanta, GA; Texas Department of Health, Austin, TX; National Animal Disease Center, Ames, IA; and V. Lockary, State of Idaho Bureau of Laboratories, Boise, ID.
Product names are necessary to report factually on available data; however, the USDA neither guarantees nor warrants the standard of the product, and the use of the name by USDA implies no approval of the product to the exclusion of others that may also be suitable.
REFERENCES
- 1.Allerberger, F., A. W. Friedrich, K. Grif, M. P. Dierich, H. J. Dornbusch, C. J. Mache, E. Nachbaur, M. Freilinger, P. Rieck, M. Wagner, A. Caprioli, H. Karch, and L. B. Zimmerhackl. 2003. Hemolytic-uremic syndrome associated with enterohemorrhagic Escherichia coli O26:H infection and consumption of unpasteurized cow's milk. Int. J. Infect. Dis. 7:42-45. [DOI] [PubMed] [Google Scholar]
- 2.Anjum, M. F., S. Lucchini, A. Thompson, J. C. D. Hinton, and M. J. Woodward. 2003. Comparative genomic indexing reveals the phylogenomics of Escherichia coli pathogens. Infect. Immun. 71:4674-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bettelheim, K. A. 2003. Non-O157 verotoxin-producing Escherichia coli: a problem, paradox, and paradigm. Exp. Biol. Med. 228:333-344. [DOI] [PubMed] [Google Scholar]
- 4.Bopp, C. A., K. D. Greene, F. P. Downes, E. G. Sowers, J. G. Wells, and I. K. Wachsmuth. 1987. Unusual verotoxin-producing Escherichia coli associated with hemorrhagic colitis. J. Clin. Microbiol. 25:1486-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DebRoy, C., E. Roberts, J. Kundrat, M. A. Davis, C. E. Briggs, and P. M. Fratamico. 2004. Detection of Escherichia coli serogroups O26 and O113 by PCR amplification of the wzx and wzy genes. Appl. Environ. Microbiol. 70:1830-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D'Souza, J. M., L. Wang, and P. Reeves. 2002. Sequence of the Escherichia coli O26 O antigen gene cluster and identification of O26 specific genes. Gene 297:123-127. [DOI] [PubMed] [Google Scholar]
- 7.Elliott, E. J., R. M. Robins-Browne, E. V. O'Loughlin, V. Bennett-Wood, J. Bourke, P. Henning, G. G. Hogg, J. Knight, H. Powell, D. Redmond, and Contributors to the Australian Paediatric Surveillance Unit. 2001. Nationwide study of haemolytic uraemic syndrome: clinical, microbiological, and epidemiological features. Arch. Dis. Child. 85:125-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fields, P. I., K. Blom, H. J. Hughes, L. O. Helsel, P. Feng, and B. Swaminathan. 1997. Molecular characterization of the gene encoding H antigen in Escherichia coli and development of a PCR-restriction fragment length polymorphism test for identification of E. coli O157:H7 and O157:NM. J. Clin. Microbiol. 35:1066-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gannon, V. P. J., R. K. King, J. Y. Kim, and E. J. Golsteyn Thomas. 1992. Rapid and sensitive method for detection of Shiga-like toxin-producing Escherichia coli in ground beef using the polymerase chain reaction. Appl. Environ. Microbiol. 58:3809-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hiruta, N., T. Murase, and N. Okamura. 2001. An outbreak of diarrhoea due to multiple antimicrobial-resistant Shiga toxin-producing Escherichia coli O26:H11 in a nursery. Epidemiol. Infect. 127:221-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoshina, K., A. Itagaki, R. Seki, K. Yamamoto, S. Masuda, T. Muku, and N. Okada. 2001. Enterohemorrhagic Escherichia coli O26 outbreak caused by contaminated natural water supplied by facility owned by local community. Jpn. J. Infect. Dis. 54:247-248. [PubMed] [Google Scholar]
- 12.Louie, M., S. Read, A. E. Simor, J. Holland, L. Louie, K. Ziebell, J. Brunton, and J. Hii. 1998. Application of multiplex PCR for detection of non-O157 verocytotoxin-producing Escherichia coli in bloody stools: identification of serogroups O26 and O111. J. Clin. Microbiol. 36:3375-3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McMaster, C., E. A. Roch, G. A. Willshaw, A. Doherty, W. Kinnear, and T. Cheasty. 2001. Verocytotoxin-producing Escherichia coli serotype O26:H11 outbreak in an Irish creche. Eur. J. Clin. Microbiol. Infect. Dis. 20:430-432. [DOI] [PubMed] [Google Scholar]
- 14.Mercado, E. C., S. M. Rodríguez, A. M. Elizondo, G. Marcoppido, and V. Parreño. 2004. Isolation of Shiga toxin-producing Escherichia coli from a South American camelid (Lama guanicoe) with diarrhea. J. Clin. Microbiol. 42:4809-4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Misselwitz, J., H. Karch, M. Bielazewska, U. John, F. Ringelmann, G. Ronnefarth, and L. Patzer. 2003. Cluster of hemolytic-uremic syndrome caused by Shiga toxin-producing Escherichia coli O26:H11. Pediatr. Infect. Dis. J. 22:349-354. [DOI] [PubMed] [Google Scholar]
- 16.Murinda, S. E., S. D. Baston, L. T. Nguyen, B. E. Gillespie, and S. P. Oliver. 2004. Phenotypic and genetic markers for serotype-specific detection of Shiga toxin-producing Escherichia coli O26 strains from North America. Foodborne Path. Dis. 1:125-135. [DOI] [PubMed] [Google Scholar]
- 17.O'Hanlon, K. A., T. M. G. Catarame, G. Duffy, I. S. Blair, and D. A. McDowell. 2004. RAPID detection and quantification of E. coli O157/O26/O111 in minced beef by real-time PCR. J. Appl. Microbiol. 96:1013-1023. [DOI] [PubMed] [Google Scholar]
- 18.Orskov, F. 1951. On the occurrence of E. coli belonging to O-group 26 in cases of infantile diarrhoea and white scours. Acta Pathol. Microbiol. Scand. 29:373-378. [PubMed] [Google Scholar]
- 19.Paton, A. W., and J. C. Paton. 1988. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J. Clin. Microbiol. 36:598-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pearce, M. C., C. Jenkins, L. Vali, A. W. Smith, H. I. Knight, T. Cheasty, H. R. Smith, G. J. Gunn, M. E. J. Woolhouse, S. G. B. Amyes, and G. Frankel. 2004. Temporal shedding patterns and virulence factors of Escherichia coli serogroups O26, O103, O111, O145, and O157 in a cohort of beef calves and their dams. Appl. Environ. Microbiol. 70:1708-1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pearson, G. R., K. J. Bazeley, J. R. Jones, R. F. Gunning, M. J. Green, A. Cookson, and M. J. Woodward. 1999. Attaching and effacing lesions in the large intestine of an eight-month-old heifer associated with Escherichia coli O26 infection in a group of animals with dysentery. Vet. Rec. 145:370-373. [DOI] [PubMed] [Google Scholar]
- 22.Peixoto, J. C. C., S. Y. Bando, J. A. G. Ordonez, B. A. Botelho, L. R. Trabulsi, and C. A. Moreira-Filho. 2001. Genetic differences between Escherichia coli O26 strains isolated in Brazil and in other countries. FEMS Microbiol. Lett. 196:239-244. [DOI] [PubMed] [Google Scholar]
- 23.Perelle, S., F. Dilasser, J. Grout, and P. Fach. 2004. Detection by 5′-nuclease PCR of Shiga-toxin producing Escherichia coli O26, O55, O91, O103, O111, O113, O145 and O157:H7, associated with the world's most frequent clinical cases. Mol. Cell. Probes 18:185-192. [DOI] [PubMed] [Google Scholar]
- 24.Prager, R., U. Strutz, A. Fruth, and H. Tschäpe. 2003. Subtyping of pathogenic Escherichia coli strains using flagellar (H)-antigens: serotyping versus fliC polymorphisms. Int. J. Med. Microbiol. 292:477-486. [DOI] [PubMed] [Google Scholar]
- 25.Renter, D. G., S. L. Checkley, J. Campbell, and R. King. 2004. Shiga toxin-producing Escherichia coli in the feces of Alberta feedlot cattle. Can. J. Vet. Res. 68:150-153. [PMC free article] [PubMed] [Google Scholar]
- 26.Rivera-Betancourt, M., and J. E. Keen. 2000. Murine monoclonal antibodies specific for lipopolysaccharide of Escherichia coli O26 and O111. Appl. Environ. Microbiol. 66:4124-4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma, V. K. 2002. Detection and quantitation of enterohemorrhagic Escherichia coli O157, O111, and O26 in beef and bovine feces by real-time polymerase chain reaction. J. Food Prot. 65:1371-1380. [DOI] [PubMed] [Google Scholar]
- 28.Vaz, T. M. I., K. Irino, M. A. M. F. Kato, Â. M. G. Dias, T. A. T. Gomes, M. I. C. Medeiros, M. M. M. Rocha, and B. E. C. Guth. 2004. Virulence properties and characteristics of Shiga toxin-producing Escherichia coli in São Paulo, Brazil, from 1976 through 1999. J. Clin. Microbiol. 42:903-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang, L., D. Rothemund, H. Curd, and P. R. Reeves. 2000. Sequence diversity of the Escherichia coli H7 fliC genes: implication for a DNA-based typing scheme for E. coli O157:H7. J. Clin. Microbiol. 38:1786-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, L., D. Rothemund, H. Curd, and P. R. Reeves. 2003. Species-wide variation in the Escherichia coli flagellin (H-antigen) gene. J. Bacteriol. 185:2936-2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Werber, D., A. Fruth, A. Liesegang, M. Littmann, U. Buchholz, R. Prager, H. Karch, T. Breuer, H. Tschäpe, and A. Ammon. 2002. A multistate outbreak of Shiga toxin-producing Escherichia coli O26:H11 infections in Germany, detected by molecular subtyping surveillance. J. Infect. Dis. 186:419-422. [DOI] [PubMed] [Google Scholar]
- 32.Widiasih, D. A., N. Ido, K. Omoe, S. Sugii, and K. Shinagawa. 2003. Duration and magnitude of faecal shedding of Shiga toxin-producing Escherichia coli from naturally infected cattle. Epidemiol. Infect. 132:67-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.World Health Organization. 1999. Zoonotic non-O157 Shiga toxin-producing Escherichia coli (STEC). W.H.O. Scientific Working Group Meeting, Berlin, Germany.
- 34.Zhang, W. L., M. Bielaszewska, J. Bockemuhl, H. Schmidt, F. Scheutz, and H. Karch. 2000. Molecular analysis of H antigens reveals that human diarrheagenic Escherichia coli O26 strains that carry the eae gene belong to the H11 clonal complex. J. Clin. Microbiol. 38:2989-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang, W. L., M. Bielaszewska, A. Liesegang, H. Tschape, H. Schmidt, M. Bitzan, and H. Karch. 2000. Molecular characteristics and epidemiological significance of Shiga toxin-producing Escherichia coli O26 strains. J. Clin. Microbiol. 38:2134-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]