Abstract
Microcosms capable of reductive dechlorination of polychlorinated dibenzo-p-dioxins/dibenzofurans (PCDD/Fs) were constructed in glass bottles by seeding them with a polluted river sediment and incubating them anaerobically with an organic medium. All of the PCDD/F congeners detected were equally reduced without the accumulation of significant amounts of less-chlorinated congeners as the intermediate or end products. Alternatively, large amounts of catechol and salicylic acid were produced in the upper aqueous phase. Thus, the dechlorination of PCDD/Fs and the oxidative degradation of the dechlorinated products seemed to take place simultaneously in the microcosm. Denaturing gel gradient electrophoresis and clone library analyses of PCR-amplified 16S rRNA genes from the microcosm showed that members of the phyla Firmicutes, Proteobacteria, and Bacteroidetes predominated. A significant number of Chloroflexi clones were also detected. Quantitative real-time PCR with specific primer sets showed that the 16S rRNA genes of a putative dechlorinator, “Dehalococcoides,” and its relatives accounted for 0.1% of the total rRNA gene copies of the microcosm. Most of the clones thus obtained formed a cluster distinct from the typical “Dehalococcoides” group. Quinone profiling indicated that ubiquinones accounted for 18 to 25% of the total quinone content, suggesting the coexistence and activity of ubiquinone-containing aerobic bacteria. These results suggest that the apparent complete dechlorination of PCDD/Fs found in the microcosm was due to a combination of the dechlorinating activity of the “Dehalococcoides”-like organisms and the oxidative degradation of the dechlorinated products by aerobic bacteria with aromatic hydrocarbon dioxygenases.
Remediation of environmental contamination with toxic chemicals, including polychlorinated dibenzo-p-dioxins (DD)/dibenzofurans (PCDD/Fs) and polychlorinated biphenyls (PCBs), is one of the most challenging problems in environmental science and technology. Biodegradation and biotransformation of PCDD/Fs by microorganisms have received extensive study in connection with their potential utilization in bioremediation of polluted environments. Fundamental data on the mechanism of microbial dioxin degradation, as well as on the biodiversity of dioxin-degrading microorganisms, has been accumulated, in particular during the past decade (for reviews, see references 8, 35, 56, and 69). The available information indicates that the ring structures of dibenzo-p-dioxin, dibenzofuran (DF), and related compounds are degraded by aerobic bacteria containing aromatic hydrocarbon dioxygenases having broad substrate specificity. In addition to this type of biodegradation, at least two other modes of dioxin biotransformation have so far been recognized: reductive dechlorination by anaerobic microorganisms and fungal degradation with lignin peroxidase or laccase.
Microbial reductive dechlorination of PCDD/Fs has increasingly attracted attention, because this activity may work against highly chlorinated congeners hardly attacked by aromatic hydrocarbon dioxygenases, and it is possibly the initial step of the transformation of the polychlorinated pollutants in the environment. The biological significance and potential for biotechnological applications of microbial reductive dehalogenation and dehalorespiration have been well reviewed (31, 40, 41, 53). Reductive dechlorination of PCDD/Fs has been demonstrated in anaerobic mixed cultures from polluted sediments, sludge, and soils (1-3, 9, 10-12, 15, 27, 28, 36, 45, 65).
Recent noteworthy findings are that the strictly anaerobic bacteria designated “Dehalococcoides” sp. strain CBDB1 and “Dehalococcoides ethenogenes” strain 195, which were initially reported to dechlorinate chlorobenzene (4) and chloroethene (52), respectively, have also been shown to be able to dechlorinate selected PCDD/F congeners (16, 25). Members of the “Dehalococcoides” group were reported to be putative dechlorinators in anaerobic enrichment cultures capable of dechlorinating a tetrachlorinated congener of PCBs (18, 71), chlorobenzene (70), and trichlorodibenzo-p-dioxin (10). To date, only scattered reports are available on the physiology and ecology of “Dehalococcoides,” and its taxonomic name does not yet have standing in nomenclature. Nevertheless, the detection of 16S rRNA gene sequences affiliated with “Dehalococcoides” and its relatives in polluted environments and dechlorinating enrichment cultures (20, 22, 24, 30, 32, 33, 42, 46, 50, 51, 57, 63) indicates that members of the “Dehalococcoides” group are widely distributed in nature and play major roles in the transformation of environmental organohalide pollutants.
The microbial-community analysis of PCDD/F-transforming environments is of primary importance not only for a comprehensive understanding of pollution microbial ecology but also for bioremediation purposes. However, the study of this subject has just begun (10, 28), and much less information is available on the significance and population dynamics of the “Dehalococcoides” group in PCDD/F-contaminated environments than in those polluted with aliphatic chlorinated compounds. In this study, a microbial community capable of dechlorinating PCDD/Fs was studied by using laboratory scale microcosms. The microbial community was characterized by rRNA gene sequence-based culture-independent methods, including PCR cloning and sequencing of environmental clones, denaturing gel gradient electrophoresis (DGGE), real-time PCR, and fluorescence in situ hybridization. Quinone profiling, a lipid biomarker approach to community analysis, was performed in addition.
MATERIALS AND METHODS
Samples and microcosm construction.
Sediment samples were taken from a highly PCDD/F-polluted river in Japan. The concentrations of PCDD/Fs detected in these samples were 1.8 to 3.3 nmol g−1 (dry weight) (920 to 1,800 pg TEQ [toxic equivalent] g−1). The samples were collected in polyethylene bottles and stored at 10°C for several weeks before being tested. Just before use, the samples had a moisture content of 64% and a pH of 6.2. For preliminary microcosm studies, one of the sediment samples (50 g [wet weight] each) was introduced into several screw-cap glass bottles (120-ml capacity), which were then filled with a culture medium designated OAM-1. The composition of OAM-1 medium was as follows (per liter): 0.5 g of NH4Cl; 0.5 g of KH2PO4; 0.2 g of NaCl; 0.2 g of MgCl2 · 6H2O; 0.05 g of CaCl2 · 2H2O; 1 ml of trace element solution SL8 (13); 1 ml of vitamin mixture PV1 (39); 100 ml of an organic acid mixture containing 10 mM each of acetate, benzoate, butyrate, formate, and pyruvate; and 900 ml of distilled water. The pH of the medium was adjusted to 7.0. Some bottles were purged with 100% N2 gas or H2 plus CO2 (1:1 [vol/vol]) to ensure anoxic conditions. To examine the effects of growth inhibitors, microcosms supplemented with either 0.1% paraformaldehyde, 0.01% chloramphenicol, 10 mM bromoethanesulfonate (BES), or 10 mM molybdate were also prepared. For the control test, the microcosms were filled with the mineral base of OAM-1 medium or distilled water in place of OAM-1 medium. All glass bottles were tightly sealed and settled at 25°C in darkness for 5 to 6 months. Every 4 weeks of incubation, sampling was performed, and the supernatant of the microcosm was replaced with fresh medium and mixed completely. The sediment slurry and supernatant samples collected were stored separately at −20°C until they were analyzed. For a long-term microcosm experiment, larger screw-cap glass bottles (660-ml capacity) were used. The bottles were seeded with 230 g of sediment, filled with OAM-1 medium, and incubated at 25°C for more than 1 year. Sampling and medium supplementation were performed as described for the smaller-scale microcosms.
Analyses of dioxins and relatives and abbreviations for congeners.
Dioxins were extracted from microcosm samples by the Soxhlet method, fractionated by multiphase column chromatography and alumina column chromatography, and analyzed by gas-liquid chromatography-mass spectrometry as described previously (36). Gas-liquid chromatography-mass spectrometry analyses were performed by Sinto Environmental Measurement Center, Shintokogyo, Ltd. (Toyokawa, Japan). PCDD/F concentrations (as TEQ levels) were calculated based on the toxicity equivalent factors for different congeners redefined by the World Health Organization. In this study, mono-, di-, tri-, tetra-, penta-, hexa-, hepta-, and octachlorinated DD/DF congeners are abbreviated MoCDD/Fs, DiCDD/Fs, TrCDD/Fs, TCDD/Fs, PeCDD/Fs, HxCDD/Fs, HeCDD/Fs, and OCDD/Fs, respectively. PCDD/Fs denote tetra- to octachlorinated congeners of DD and DF, whereas the term 1-3CDD/Fs is used to indicate all congeners of MoCDD/Fs, DiCDD/Fs, and TrCDD/Fs. Aromatic compounds in the aqueous phase, including DD, DF, catechol, and salicylic acid, were extracted with ethyl acetate, concentrated under reduced pressure, and measured by reverse-phase high-performance liquid chromatography with external standards. The apparatus used was a Shimadzu model LC10A liquid chromatograph equipped with a Zorbax ODS column (4.6 by 250 mm) and a Shimadzu photodiode array detector. Samples were eluted with 100% methanol for DD and DF and 80% methanol for catechol and salicylic acid at a flow rate of 1 ml min−1 and monitored at 280 nm.
Epifluorescence microscopy.
For direct cell counting, sediment samples were sonicated and diluted with phosphate-buffered saline as described previously (55). Direct total counts of bacteria were measured by epifluorescence microscopy with ethidium bromide (EtBr) staining (58) with minor modifications (55). Fluorescence in situ hybridization (FISH) was performed according to the protocols described previously (7, 19). For this, the Cy3-labeled oligonucleotide probes EUB338 mix and ARC195, specific for the domains Bacteria and Archaea, respectively, and NONEUB for the negative control, were used. Hybridized specimens were counterstained with SYBR GREEN II (Molecular Probes, Eugene, OR). The stained cells were observed under an Olympus BX-50 epifluorescence microscope equipped with a Flovel model FD-120 digital charge-coupled device camera (Flovel Co., Tokyo, Japan). The microscopic images were analyzed with the WINROOF program (Flovel); 20 fields per sample and a total of 1,000 to 2,000 stained cells per sample were taken for counting.
Quinone analysis.
Microbial quinones from the sediment samples were extracted with an organic solvent mixture and fractionated into the menaquinone and ubiquinone fractions using Sep-Pak Vac silica gel cartridges (Waters, Milford, MA). The quinone components of each fraction were separated for identification and quantification by reverse-phase high-performance liquid chromatography and photodiode array and mass spectrometry detection with external standards. Detailed information on these analytical procedures has been given in previous studies (37, 38, 44). Ubiquinones and menaquinones with n isoprene units in their side chains are abbreviated as Q-n and MK-n, respectively. Partially hydrogenated menaquinones are expressed as MK-n(Hx), where x indicates the number of hydrogen atoms saturating the side chain.
DNA extraction and purification.
Bulk DNA was extracted from microcosm samples according to the protocol of Zhou et al. (72) with some modifications. One gram (wet weight) of the sediment pellet was suspended in 1 ml of extraction buffer (pH 7.0) containing 5 mM phosphate, 30 mM ammonium acetate, 4% yeast RNA, and 0.1% skim milk. This suspension was sonicated on ice for 20 s with 2-s intermittent bursts (20 kHz; output power, 10 W). The suspension was treated with lysozyme (80 mg ml−1) at 37°C for 30 min with gentle shaking and further incubated at 65°C for 30 min in the presence of 2.5% sodium dodecyl sulfate. Then, the sample was treated by being frozen at −80°C for 30 min and thawed at 65°C for 5 min. This freeze-thaw lysis procedure was repeated three times. The treated sample was centrifuged at 12,600 × g for 10 min to save the supernatant. Humic substances and cellular proteins in the supernatant were precipitated by adding 1% hexadecylmethylammonium bromide and removed by centrifugation (12,600 × g; 10 min). Crude DNA was precipitated by the addition of 0.8 volume of isopropanol and settling at room temperature for 2 h, collected by centrifugation, and resuspended in 500 μl TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0). DNA was further purified by a standard method including deproteinization with phenol-chloroform-isoamyl alcohol and RNase treatment (59). The DNA solution thus obtained was diluted as needed and used for PCR experiments.
PCR-DGGE.
16S rRNA gene fragments that corresponded to positions 341 to 537 in Escherichia coli numbering (14) were amplified with forward primer 357f (5′-CCTACGGGAGGCAGCAG-3′) with a GC clamp on the 5′ terminus and reverse primer 517r (5′-ATTACCGCGGCTGCTGG-3′) as described previously (54). The reaction mixture contained 50 ng of the purified DNA as the template, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 1 μM (each) of the primers, 2 ng of bovine serum albumin, and 5 units of Taq DNA polymerase in a total volume of 100 μl. PCR was performed by preheating the mixture at 94°C for 2 min, followed by 35 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 1.5 min. DGGE was performed according to the standard protocol (54) using the Bio-Rad Dcode system (Bio-Rad Laboratories, Hercules, CA). The denaturing gradient gel was prepared to contain 0.5× TAE buffer (20 mM Tris, 10 mM acetate, 0.5 mM Na2-EDTA, pH 8.0), 6% acrylamido-bis-acrylamide, and 40 to 60% denaturant (100% denaturant contains 7 M urea and 40% formamide). Electrophoresis was performed at 200 V for 3 h in 0.5× TAE buffer at 61°C. PCR products separated on the gel were stained with EtBr for 30 min and then photographed under illumination by UV light. Major DGGE fragments were cut from the gel, purified using a GENECLEAN Spin kit (Bio 101, Vista, CA), and subcloned with a pTBlue Perfectly Blunt cloning kit (Novagen, Madison, WI). Transformation of E. coli competent cells was carried out according to a standard manual of molecular cloning (61). Plasmid DNA was isolated and purified by using the Wizard Plus Minipreps DNA Purification System (Promega Inc., Madison, WI) according to the manufacturer's instructions.
16S rRNA gene clone library construction.
16S rRNA gene fragments from the DNA purified from microcosms were PCR amplified with a pair of bacterial consensus primers, 27f and 1492r (68). The components of the reaction mixture other than PCR primers were the same as described above. The cycle profile consisted of preheating at 95°C for 2 min and 30 cycles of denaturation at 95°C for 1 min, annealing at 50°C for 1 min, and extension at 72°C for 1.5 min; the final step was followed by postextension for 5 min. The PCR products were separated by agarose gel electrophoresis, purified, and then subcloned as described above.
Sequencing and phylogenetic analysis.
Cloned 16S rRNA genes were sequenced with a SequiTherm Long Read cycle-sequencing kit (Epicentre Technologies, Madison, WI) and analyzed with a Pharmacia ALFexpress DNA sequencer. Sequence data were compiled by the GENETYX-MAC program (Software Development Co., Tokyo, Japan), analyzed for chimera detection with the CHIMERA_CHECK program version 2.7, and compared with those retrieved from the Ribosomal Database Project II (17). Multiple alignment of sequences and calculation of the nucleotide substitution rate (Knuc) by Kimura's two-parameter model (47) were performed using the CLUSTAL W program (64). Distance matrix trees were constructed by the neighbor-joining method (60), and the topology of the trees was evaluated by bootstrapping with 1,000 resamplings (23). Alignment positions with gaps were excluded from the calculations.
Real-time PCR.
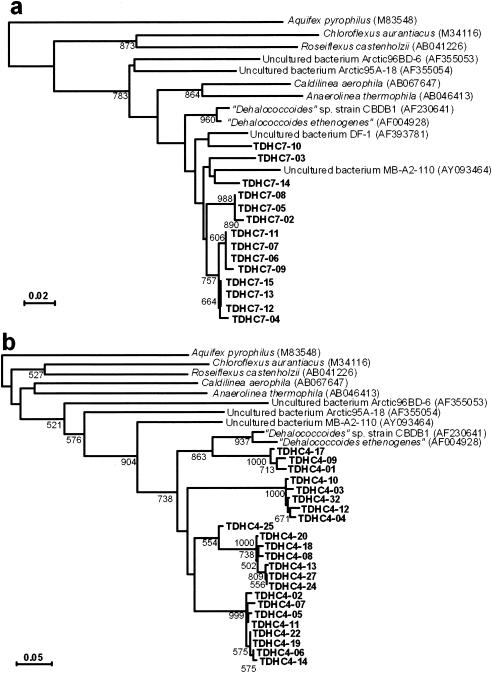
For quantitative detection of the “Dehalococcoides” group, two pairs of PCR primers were designed by using unique sequences of consensus variable regions in the “Dehalococcoides” 16S rRNA gene (GenBank accession numbers AF004928 and AF230641). One primer set was a combination of forward primer DHC793f (5′-GGGAGTATCGACCCTCTCTG-3′) and reverse primer DHC946r (5′-CGTTYCCCTTTCRGTTCACT-3′), which corresponded to positions 774 to 793 and 946 to 965 of the E. coli rRNA and which were modifications of primers DHC 774 and DHC 946, respectively, designed previously for the detection of “Dehalococcoides” (33). The other set consisted of forward primer DHC66f (5′-GGTCTTAAGCAATTAAGATAGTG-3′) and reverse primer DHC180r (5′-CACCAAGCRCCTTRCGGC-3′). Also, the bacterial universal primers 357f and 517r noted above were used for comparative amplification of total bacterial 16S rRNA genes. The specificities of all these primers were checked with the PROBE-MATCH program of the Ribosomal Database Project (17) and the BLAST program (6). They were also confirmed by the observation that all PCR experiments gave a single prominent band of an expected size upon agarose gel electrophoresis, except that PCR with primers DHC66f and DHC180r produced an additional weaker band in some cases. All real-time PCR assays were performed by the nested-PCR method, in which bacterial 16S rRNA genes amplified with universal primers 27f and 1492r from the microcosm DNA were used as the template. The reaction was performed using a LightCycler FastStart DNA Master SYBR GREEN I kit (Roche Molecular Biochemicals, Indianapolis, IN) and a LightCycler system (Roche Diagnostics, Mannheim, Germany) according to the manufacturers' instructions. The PCR profile consisted of preheating at 95°C for 10 min and 40 cycles of denaturation at 95°C for 15 s, annealing at 60°C for 10 s, and extension at 72°C for 20 s. The fluorescence signal was detected at 72°C at each cycle, and a melting curve was obtained by heating the product to 95°C and cooling it to 40°C. Calibration curves (log DNA concentration versus an arbitrarily set cycle threshold value) for the estimation of the total bacterial and “Dehalococcoides” rRNA genes were made by using serial dilutions of PCR-amplified products of the E. coli 16S rRNA gene and a “Dehalococcoides” clone, TDHC7-12, 16S rRNA gene (see Fig. 5a) of known concentration, respectively. The copy numbers of the amplified products were calculated using LightCycler software version 3.5 (Roche Diagnostics, Mannheim, Germany).
FIG. 5.
Neighbor-joining distance matrix tree of the 16S rRNA gene clones amplified with two different “Dehalococcoides”-specific primer sets, DHC793f and DHC946r (a) and DHC66f and DHC180r (b). The clones obtained in this study are shown in boldface. The sequence of Aquifex pyrophilus was used as an outgroup to root the tree. Scale = 2% (a) or 5% (b) base substitution (Knuc). The figures in parentheses following the species and clone names are DDBJ/EMBL/GenBank accession numbers. Bootstrap values (>600) with 1,000 resamplings are shown at branching points.
Nucleotide sequence accession numbers.
The nucleotide sequence data reported in this paper have been deposited under DDBJ accession numbers AB186751 to AB186774, AB186776 to AB186857, AB186860 to AB186895, AB187503 to AB188321, AB198668, and AB198669.
RESULTS
Preliminary microcosm studies.
Preliminary studies were performed to determine environmental factors affecting the dechlorination of PCDD/Fs in the microcosm. The glass bottles packed with a sediment sample and supplemented with different additives were incubated for 5 months, and the total amount of PCDD/Fs remaining was measured (Table 1). A significant decrease in PCDD/F concentrations was found only when the organic acid mixture, OAM-1 medium, was supplemented. Maintaining anoxic conditions by purging N2 or H2 plus CO2 had little effect on the reduction of PCDD/Fs. The addition of paraformaldehyde or chloramphenicol exerted inhibitory effects on PCDD/F reduction, whereas BES, a potent inhibitor of methanogenesis, had a minor effect. Molybdate had a moderately inhibitory effect on PCDD/F reduction. These observations suggested that the decrease in PCDD/F concentrations was due to the dechlorinating activity of a microbial consortium requiring organic compounds as the source of reducing power, in which members of the domain Bacteria were mainly involved.
TABLE 1.
Decrease in PCDD/F concentration with different additives during 210 days of incubation of microcosms
| Additive | Headspace | PCDD/Fs remaining (pmol g−1 [dry wt]) | Relative amt remaining (%) |
|---|---|---|---|
| None | Ambient | 840 | 100 |
| Minerals | Ambient | 740 | 88 |
| OAM-1 medium | Ambient | 570 | 68 |
| OAM-1 medium | N2 | 630 | 75 |
| OAM-1 medium | H2 + CO2 | 610 | 73 |
| OAM-1 medium + 0.1% paraformaldehyde | Ambient | 820 | 99 |
| OAM-1 medium + 0.01% chloramphenicol | Ambient | 780 | 93 |
| OAM-1 medium + 10 mM BES | Ambient | 630 | 75 |
| OAM-1 medium + 10 mM molybdate | Ambient | 700 | 83 |
To study the effects of the incubation temperature on PCDD/F reduction in the microcosm, the glass bottles packed with the sediment slurry and supplemented periodically with OAM-1 medium were incubated for 5 months at different temperatures ranging from 10 to 55°C. The reduction of PCDD/Fs was most remarkable at 25 to 37°C and much lower at higher and lower temperatures (data not shown). This observation indicates that the microbial consortium capable of PCDD/F reduction favored mesophilic conditions.
The release of PCDD/Fs and lower chlorinated congeners from the sediment into the upper aqueous phase in the microcosms noted above was checked every time the supernatant was exchanged for fresh medium. In any case, the concentrations of all dioxin congeners in the supernatant were less than 0.01% of those in the sediment on a molar basis (data not shown). Thus, the carryover of dioxin congeners by discarding the supernatant was negligible.
Long-term microcosm study.
Based on the foregoing results, we performed a long-term experiment with a laboratory microcosm which was periodically supplemented with OAM-1 medium and incubated at 25°C for more than 1 year. During 2 months of the startup period, the microcosm turned black with a strong odor of sulfide, indicating the activity of sulfate-reducing bacteria. The black color and sulfide odor disappeared gradually with time. The pH of the microcosm was lowered to 5.0 to 6.0 in every batch cycle of nutrient supplementation during the initial 2 months of incubation and became almost neutral thereafter.
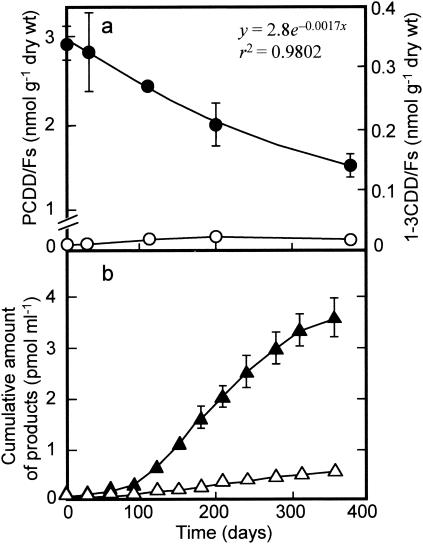
Changes in the concentrations of PCDD/Fs and 1-3CDD/Fs in the microcosm during 1 year of incubation are shown in Fig. 1a. After a lag time of a few weeks, the total concentration and TEQ level of PCDD/Fs in the sediment decreased during the overall period of incubation. The decrease rate was relatively fast at the beginning and weakened gradually with time, resulting in a reduction of 49% of the initial concentration on day 360. By assuming that the decrease took place logarithmically, as expressed by a regression equation (Fig. 1a), the half-reduction time was estimated to be approximately 14 months. Unexpectedly, all of the PCDD/F congeners detected were totally reduced, and only trace amounts of 1-3CDD/F congeners as the intermediate or end products accumulated during the period of the experiment (Fig. 1a). On day 360, the less-chlorinated congeners were produced in the following order: MoCDFs (35% of the total 1-3CDD/F content) > TrCDDFs (26%) > DiCDFs (16%) > DiCDDs (15%) > TrCDDs (9%).
FIG. 1.
Changes in the concentrations of PCDD/Fs and lower chlorinated congeners (1-3CDD/Fs) (a) and cumulative amounts of catechol and salicylic acid produced (b) during 1 year of incubation of the microcosm. Closed circles, PCDD/Fs; open circles, 1-3CDD/Fs; closed triangles, catechol; open triangles, salicylic acid. The data show the averages with standard deviations (shown by error bars) of three different determinations. A regression equation and a correlation coefficient for the relationships between the concentration of PCDD/Fs and time are shown in panel a.
The total amount of PCDD/Fs and less-chlorinated congeners in the aqueous phase detected at each sampling time was negligible, as noted above (less than 0.01% of that in the sediment). On the other hand, each aqueous sample contained much larger amounts of catechol and salicylic acid (Fig. 1b). The cumulative amounts of catechol and salicylic acid produced from the microcosm were 3.6 and 0.49 nmol liter−1, respectively, on day 360. The total amount of these products corresponds to 3.5% of the PCDD/Fs removed on a molar basis. Thus, it is likely that the reductive dechlorination of PCDD/Fs and the oxidative degradation of the dechlorinated products took place simultaneously in the microcosm.
DGGE analysis.
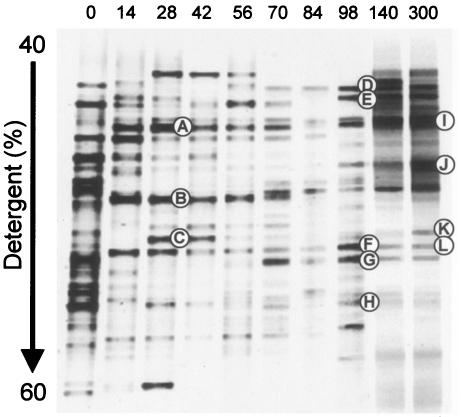
The microbial succession in the microcosm noted above was monitored by PCR-DGGE profiling. The DGGE analysis showed a drastic change in microbial community structure during the startup acclimation (Fig. 2). Namely, the DGGE pattern changed during the first 1 month of incubation and became stable after 2 months of incubation. This change seemed to take place along with the changes in the pH and appearance of the microcosm noted above. In view of these collective data, the microbial community in the microcosm steadied after 2 months of incubation.
FIG. 2.
Changes in DGGE patterns of PCR-amplified 16S rRNA gene fragments during the startup operation of the microcosm. The DGGE bands designated A to H were purified and subjected to sequence analysis (see Table 2).
Twelve major DGGE products shown in Fig. 2 were purified and sequenced. These DGGE clones were assigned to members of the phyla Bacteroidetes, Firmicutes, Proteobacteria, and Spirochaetes (Table 2). Interestingly, most of the major DGGE clones were most similar to uncultured clones obtained previously from dechlorinating microbial consortia or polluted environments containing chlorobenzene and dichloropropane.
TABLE 2.
Phylogenetic assignment of DGGE clones isolated from microcosm
| DGGE band(s)a | Phylum/class assigned | Closest relativeb | Accession no. | Similarity (%) |
|---|---|---|---|---|
| A | Deltaproteobacteria | Uncultured clone WCHB1-67 from a polluted aquifer | AF050536 | 99.0 |
| B, J | Firmicutes | Trichococcus collinsiium strain 37AN3 | AJ306612 | 100 |
| Uncultured clone IIA-2 from a chlorobenzene-dechlorinating consortium | AJ488083 | 100 | ||
| C | Firmicutes | Uncultured clone TANB01 from a TCE-dechlorinating ground water | AY667266 | 95.2 |
| D | Bacteroidetes | Uncultured clone 1A-16 from a chlorobenzene-dechlorinating consortium | AJ488070 | 94.7 |
| E | Unidentified | Uncultured clone SHA-4 from a 1,2-DCP-dechlorinating consortium | AJ306785 | 95.9 |
| F, L | Bacteroidetes | Uncultured clone SHA-94 from a 1,2-DCP-dechlorinating consortium | AJ306738 | 99.5 |
| G | Spirochaetes | Uncultured clone WCHB1-40 from a polluted aquifer | AF050549 | 96.9 |
| H | Firmicutes | Uncultured clone A41 from a tetrachloroethylene-dechlorinating consortium | AF158719 | 100 |
| I | Gammaproteobacteria | Pseudomonas putida strain DSM 3601 | AF509330 | 97.9 |
| K | Alphaproteobacteria | Methylosinus trichosporium strain OB3b | Y18947 | 100 |
Corresponding DGGE bands are shown in Fig. 2.
TCE, trichloroethene; DCP, dichlorophenol.
Clone library analysis.
Two 16S rRNA gene clone libraries were constructed from the microcosm on days 140 and 300. All of the 16S rRNA gene clones thus obtained were sequenced over 1,400 bp and assigned to members of 10 established phyla and one unnamed phylum (OP11 division) of the domain Bacteria (Table 3). There seemed to be no fundamental differences in the phylogenetic compositions of the two clone libraries. Nevertheless, the detection frequencies of different phyla were somewhat different between the two. In the library constructed from the 140-day-old microcosm, clones belonging to the phylum Firmicutes predominated (38%) and Bacteroidetes was the second most abundant group. Some classes of the phylum Proteobacteria, such as Betaproteobacteria and Deltaproteobacteria, also occurred in significant proportions. This result agreed fairly well with that of the DGGE analysis noted above. On the other hand, clones in the 300-day-old microcosm were more widely distributed among several phylogenetic groups, including Bacteroidetes, Betaproteobacteria, Chloroflexi, and Firmicutes.
TABLE 3.
Phylogenetic compositions of the two 16S rRNA gene clone libraries constructed from the PCDD/F-transforming microcosm
| Phylum/class | No. of clones detected in microcosm
|
|
|---|---|---|
| Day 140a | Day 300 | |
| Acidobacteria | 0 | 2 (3.3) |
| Actinobacteria | 1 (1.8) | 0 |
| Bacteroidetes | 9 (16) | 6 (10) |
| Chlorobi | 1 (1.8) | 3 (5.0) |
| Chloroflexi | 3 (5.4) | 7 (12) |
| Firmicutes | 21 (38) | 9 (15) |
| Nitrospira | 1 (1.8) | 5 (8.3) |
| Proteobacteria | ||
| Alphaproteobacteria | 1 (1.8) | 4 (6.7) |
| Betaproteobacteria | 8 (14) | 9 (15) |
| Gammaproteobacteria | 3 (5.4) | 5 (8.3) |
| Deltaproteobacteria | 5 (8.9) | 5 (8.3) |
| Epsilonproteobacteria | 1 (1.8) | 0 |
| Spirochaetes | 0 | 1 (1.7) |
| OP11 division | 1 (1.8) | 0 |
| Unidentified | 1 (1.8) | 4 (6.7) |
| Total | 60 | 56 |
Figures in parentheses indicate the percentages of clones.
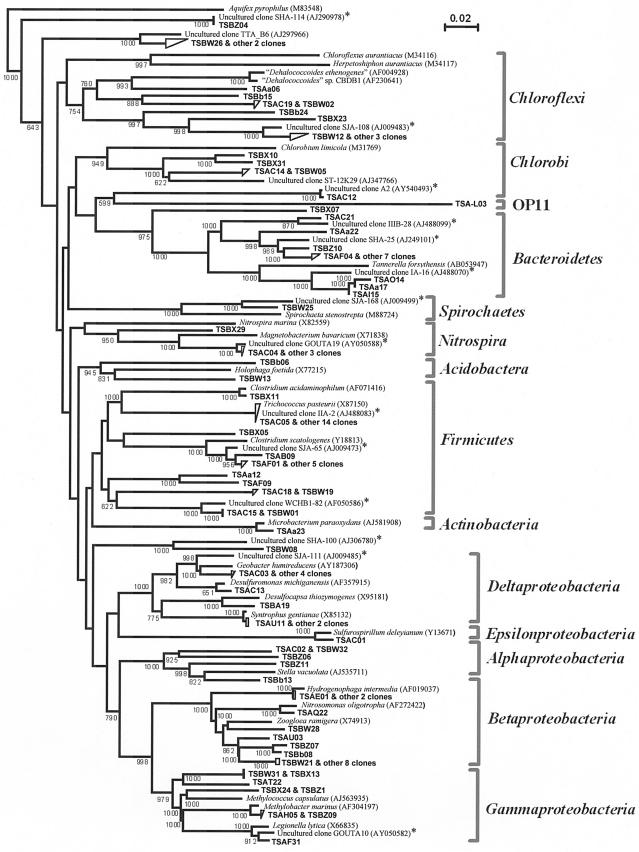
A phylogenetic tree of the 16S rRNA gene clones detected and their relatives retrieved from the databases is shown in Fig. 3. It was noteworthy that a number of the clones belonging to Bacteroidetes, Chloroflexi, Firmicutes, and Proteobacteria (Delta- and Gammaproteobacteria) were clustered most closely with those previously isolated from contaminated environments or microbial consortia capable of dechlorinating chlorinated pollutants. Some of the 16S rRNA gene clones (clone TSC05 and 14 other clones of Firmicutes, and Bacteroidetes clones TSA14, TSAa17, TSAI15, and TSAa22) corresponded to the DGGE clones shown in Table 2. No clones assignable to the “Dehalococcoides” group were detected, and the Chloroflexi clones obtained had only 74.9 to 86.8% similarities to this group.
FIG. 3.
Neighbor-joining distance matrix tree of 16S rRNA gene clones detected from the microcosm and their phylogenetic relatives. The sequence of Aquifex pyrophilus was used as an outgroup to root the tree. Scale = 10% base substitution (Knuc). The clones obtained in this study are shown in boldface, and those with TSA and TSB numbers were obtained from the microcosm on days 140 and 300, respectively. The asterisks indicate those clones obtained previously from dechlorinating mixed cultures and chlorinated-compound-polluted environments (5, 21, 62, 67). The figures in parentheses following the species and clone names are DDBJ/EMBL/GenBank accession numbers. Bootstrap values (>600) with 1,000 resamplings are shown at branching points.
Population dynamics and diversity of the “Dehalococcoides” group.
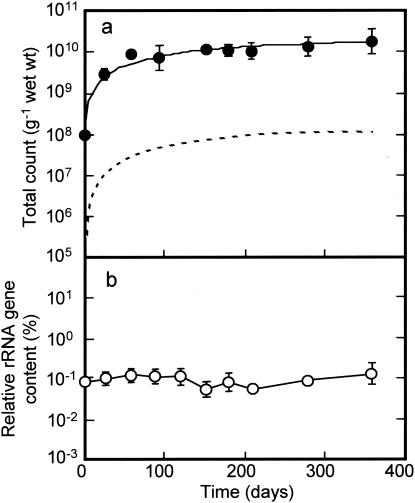
The population dynamics of the “Dehalococcoides” group as a putative dechlorinator in the microcosm were monitored by quantitative real-time PCR. Changes in total bacterial counts and the proportion of the “Dehalococcoides” 16S rRNA gene to the total rRNA gene content of the microcosm are shown in Fig. 4. The total count in the microcosm as measured by EtBr staining changed from 1.3 × 108 ± 0.1 × 108 to 6.3 × 109 ± 3.6 × 109 cells g−1 (wet weight) of sediment during the first 3 months of incubation. Thereafter, the total population remained constant at an order of magnitude of 1010 cells g−1 (wet weight) (Fig. 4a). The proportion of the “Dehalococcoides” rRNA gene to the total bacterial rRNA gene content remained relatively unchanged during the period of incubation, accounting for approximately 0.1% of the total content (Fig. 4b). By assuming that the copy number of the rRNA gene per genome in “Dehalococcoides” was 1 (from the “D. ethenogenes” sequence in the database) versus the average copy number in bacteria of 3.8 (26), the proportion of the “Dehalococcoides” population to the total population in the microcosm was estimated to range from 0.2 to 0.5% (Fig. 4a), corresponding to a population range of 8.0 × 107 to 1.1 × 108 cells g−1 (wet weight) of sediment in the steady state.
FIG. 4.
Changes in total bacterial and “Dehalococcoides” populations during 1 year of incubation of the microcosm. (a) Total counts by EtBr staining (solid line) and the “Dehalococcoides” population assumed based on the relative rRNA gene copies (broken line); (b) the ratio of “Dehalococcoides” rRNA gene copies to the total rRNA gene copies as measured by real-time PCR using a specific primer set of DHC793f and DHC946r. The data show the geometric means with standard deviations (shown by error bars) of three different determinations.
To confirm whether the sequences detected by the real-time PCR method were actually derived from the “Dehalococcoides” group, the PCR products were subcloned, sequenced, and phylogenetically analyzed. The clones detected with a primer set of DHC793f and DHC946r showed similarity levels of 90.6 to 95.3% to “Dehalococcoides” sp. strain CBDB1 and “D. ethenogenes” strain 195 and fell into three major clusters (Fig. 5a). One of the clones formed a cluster with the uncultured bacterium DF-1, known as a putative PCB dechlorinator (70) (Fig. 5a). To check whether such diverse groups of the putative dechlorinators existed in the microcosm, an additional real-time PCR assay was performed with another primer set, DHC66f and DHC180r. This PCR assay gave similar results, and the clones obtained were also divided into the three major groups, giving a similar topography of the phylogenetic tree (Fig. 5b).
FISH and quinone profiling.
The microbial community in the microcosm under steady state (on days 300 to 400) was further studied by FISH and quinone profiling. The populations stained with probes EUB338 mix and ARC915 accounted for 60.4 ± 9.8% and 8.5 ± 0.5% of the total count, indicating that members of the domain Bacteria occurred in much larger populations than those of Archaea. Respiratory quinones were detected at a concentration of 6.0 to 9.8 nmol g−1 (wet weight) of sediment. A previous report showed that 1 nmol of total respiratory quinones corresponds to 2.5 × 109 cells of respiratory bacteria (34). If this relationship is taken into account, it can be estimated that the microcosm contained 1.5 × 1010 to 2.5 × 1010 cells of respiratory bacteria per g (wet weight). This range of respiratory bacterial populations corresponded to 38 to 84% of the total population as measured by epifluorescence microscopy. Interestingly, the high-redox-potential quinones, ubiquinones, accounted for 15 to 21 mol% of the total quinone content, with Q-8 predominating (11 to 15%) and Q-10 as the second most abundant component (3.1 to 3.7%). The major components of the remainder (>5 mol%) were menaquinone species, MK-7 (18 to 21%), MK-6 (12 to 14%), MK-9(H2) (7.7 to 9.4%), MK-8(H2) (6.5 to 8.2%), and MK-7(H2) (5.5 to 6.1%).
DISCUSSION
As reported here, a PCDD/F-dechlorinating microcosm that exhibited an apparent half-reduction rate of 14 months was successfully constructed. One of the most striking observations in this microcosm is that PCDD/Fs were totally dechlorinated without the accumulation of significant amounts of 1-3CDD/Fs as the intermediate or end products. Thus, the complete dechlorination of PCDD/Fs seemingly took place in the microcosm. This observation contrasts with those in previous studies of dechlorinating microbial consortia, in which PCDD/Fs decreased with the significant accumulation of less-chlorinated DD and DF (1, 9, 11, 12, 15, 65). The discrepancy between the previous results and ours might be due to differences in the conditions used for dechlorination. The previous studies demonstrated the reductive dechlorination of PCDD/Fs under strictly anaerobic conditions that were achieved by purging an anoxic gas, such as nitrogen or carbon dioxide. On the other hand, the anaerobiosis of our microcosm was made only by filling up the bottles with the medium, with the chance of having a small atmospheric headspace in every exchange of supernatant for fresh medium, and thus, of allowing growth of aerobic dioxin-transforming bacteria. In fact, catechol and salicylate, which are the oxidatively degraded products of DD and DF, respectively (35, 69), accumulated in the microcosm. Although there is a possibility that the catechol and salicylate were produced from the added benzoate and other sources (e.g., humic acid), it is reasonable to consider the involvement of dioxin-oxidizing bacteria with aromatic hydrocarbon dioxygenases. Therefore, it is most likely that the apparent complete dechlorination of PCDD/Fs in our microcosm was due to a combination of the reductive dechlorination of PCDD/Fs and the oxidative degradation of the dechlorinated products. However, there remains another possibility, that PCDD/F is completely transformed into nonchlorinated products by a single organism prior to oxidative degradation by others, although such a biological process has not yet been demonstrated. A question of whether abiotic reductive dechlorination of PCDD/Fs (2, 49) is involved should also be clarified by further study.
The results of FISH probing indicate that members of the domain Bacteria predominated but that those of the domain Archaea were much less significant in our microcosm. PCR-DGGE and clone library analyses of the bacterial community revealed that members of the phyla Bacteroidetes, Chloroflexi, Firmicutes, and Proteobacteria were the major constituents of the total population at steady state. This community structure is similar to those found previously in a chlorinated-solvent-contaminated aquifer (21) and in microbial consortia capable of reductive dechlorination of 1,2-dichloropropane (62), trichlorobenzene (5, 67), TCDD (10), and PCB (42, 48). Nearly half of the clones detected in our microcosm were most closely related to the uncultured bacterial clones obtained from organohalide-polluted freshwater environments (5, 21, 62, 67). Schlötelburg et al. (62) reported that 16S rRNA gene clones found in a 1,2-dichloropropane-dechlorinating microcosm with river sediment as the seed were phylogenetically similar to inhabitants in other reductively dechlorinating freshwater consortia but not in marine dechlorinating consortia. In view of these previous data and ours, it is likely that reductively dechlorinating microbial communities in freshwater environments have similar phylogenetic structures irrespective of the kinds of organic chlorinated compounds acting as electron acceptors for dehalorespiration.
A characteristic feature of the microbial community in our microcosm was the presence of possibly aerobic bacteria affiliated with Alpha-, Beta-, and Gammaproteobacteria, as shown by the clone library analysis. This is supported by the detection of a significant amount of ubiquinones, which are the high-redox-potential respiratory quinones specific to those classes of Proteobacteria among prokaryotes (34). Also, the detection of partially hydrogenated menaquinones suggests the presence of aerobic bacteria belonging to Actinobacteria (34), although the clone library produced only a few clones affiliated with this phylum. As mentioned above, aerobic bacteria of the above-mentioned phyla are probably responsible for the oxidative degradation of dechlorinated products in the microcosm. We have isolated a number of DF-degrading aerobic bacteria from the microcosm, as well as from PCDD/F-polluted soil exhibiting dechlorinating activity (29, 36). Detailed information about the population level and the functional and taxonomic characteristics of aerobic DF-degrading bacteria in the dechlorinating microcosm will be reported elsewhere.
The detection of the “Dehalococcoides” group in the microcosm, as well as in the river sediment used as the seed, supports the previous supposition (33) that this group of bacteria is distributed worldwide in organohalide-polluted environments. A previous study showed that “Dehalococcoides ethenogenes” strain 195 grew up to 109 cells ml−1 by dehalorespiration with 5 to 7 mM tetrachloroethene (52). Also, “Dehalococcoides” sp. strain CBDB1 was shown to grow up to 106 cells ml−1 by complete dechlorination of 15 μM 1,2,3-trichlorobenzene (4). In the present study, similar levels of the “Dehalococcoides” population (107 to 108 cells g−1 [wet weight]) were detected in the microcosm in the steady state. These levels of the “Dehalococcoides” population were much lower than the coexisting total population but seem to be reasonable because the concentrations of PCDD/Fs in the microcosm usable as the terminal electron acceptor for dehalorespiration were low (1 to 3 nmol g−1 [dry weight]). Therefore, this group of bacteria might actually be responsible for the dechlorination of PCDD/Fs in the microcosm. The detection of “Dehalococcoides” genes coding for different types of dehalogenases (43, 66) and their expression in the microcosm would provide more direct evidence for the involvement of the organism in dechlorination. It is noteworthy that most of the “Dehalococcoides” clones obtained in this study formed phylogenetic clusters different from those of typical “Dehalococcoides” strains, such as “D. ethenogenes” strain 195 and “Dehalococcoides” sp. strain CBDB1. This fact suggests the possibility that members of the “Dehalococcoides” group more diverse than those described so far exist and play roles in the dechlorination of PCDD/Fs in the environment. In addition, since dehalorespiration is a widespread property among different phyla of bacteria (31, 40, 41, 53), there is a need to elucidate whether bacteria belonging to phyla other than Chloroflexi are involved in the dechlorination of PCDD/Fs.
In summary, anaerobic bacteria capable of reductive dechlorination of PCDD/Fs and aerobic bacteria oxidizing the dechlorinated products with aromatic hydrocarbon dioxygenases probably coexist under certain PCDD/F-dechlorinating conditions, as described here. As already pointed out (35), understanding how to create environmental conditions that allow such a coexistence of the different physiological groups of bacteria may be important in order to facilitate the apparently complete dechlorination of PCDD/Fs for bioremediation purposes.
Acknowledgments
We are grateful to H. Nihei and T. Nakatani of Mitsui Engineering & Shipbuilding Co. for providing sediment samples.
This work was carried out as part of the Project for Development of Technologies for Analyzing and Controlling the Mechanism of Biodegrading and Processing, which was supported by the New Energy and Industrial Technology Development Organization (NEDO). The work was also carried out as part of the 21st Century COE Program “Ecological Engineering and Homeostatic Human Activities” founded by the Ministry of Education, Culture, Sports, Science and Technology, Japan.
REFERENCES
- 1.Adriaens, P., and D. Grbíc-Galíc. 1994. Reductive dechlorination of PCDD/F by anaerobic cultures and sediments. Chemosphere 29:2253-2259. [Google Scholar]
- 2.Adriaens, P., P. R. Chang, and A. L. Barkovskii. 1996. Dechlorination of PCDD/F by organic and inorganic electron transfer molecules in reduced environments. Chemosphere 32:433-441. [Google Scholar]
- 3.Adriaens, P., Q. Fu, and D. Grbíc-Galíc. 1995. Bioavailability and transformation of highly chlorinated dibenzo-p-dioxins and dibenzofurans in anaerobic soils and sediments. Environ. Sci. Technol. 29:2252-2260. [DOI] [PubMed] [Google Scholar]
- 4.Adrian, L., U. Szewzyk, J. Wecke, and H. Görisch. 2000. Bacterial dehalorespiration with chlorinated benzenes. Nature 408:580-583. [DOI] [PubMed] [Google Scholar]
- 5.Alfreider, A., C. Vogt, and W. Babel. 2002. Microbial diversity in an in situ reactor system treating monochlorobenzene contaminated groundwater as revealed by 16S ribosomal DNA analysis. Syst. Appl. Microbiol. 25:232-240. [DOI] [PubMed] [Google Scholar]
- 6.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amann, R. I., L. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Armengaud, J., and K. N. Timmis. 1997. Biodegradation of dibenzo-p-dioxin and dibenzofuran by bacteria. J. Microbiol. 35:241-252. [Google Scholar]
- 9.Ballerstedt, H., A. Kraus, and U. Lechner. 1997. Reductive dechlorination of 1,2,3,4-tetrachlorodibenzo-p-dioxin and its products by anaerobic mixed cultures from Saale River sediment. Environ. Sci. Technol. 31:1749-1753. [Google Scholar]
- 10.Ballerstedt, H., J. Hantke, M. Bunge, B. Werner, J. Gerritse, J. R. Andreesen, and U. Lechner. 2004. Properties of a trichlorodibenzo-p-dioxin-dechlorinating mixed culture with a Dehalococcoides as putative dechlorinating species. FEMS Microbiol. Ecol. 47:223-234. [DOI] [PubMed] [Google Scholar]
- 11.Barkovskii, A. L., and P. Adriaens. 1996. Microbial dechlorination of historically present and freshly spiked chlorinated dioxins and diversity of dioxin-dechlorinating populations. Appl. Environ. Microbiol. 62:4556-4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beurskens, J. E. M., M. Toussaint, J. de Wolf, J. M. D. van der Steen, P. C. Slot, L. C. M. Commandeur, and J. R. Parsons. 1995. Dehalogenation of chlorinated dioxins by an anaerobic microbial consortium from sediment. Environ. Toxicol. Chem. 14:939-943. [Google Scholar]
- 13.Biebl, H., and N. Pfennig. 1978. Growth yields of green sulfur bacteria in mixed cultures with sulfur and sulfate reducing bacteria. Arch. Microbiol. 117:9-16. [Google Scholar]
- 14.Brosius, J., M. L. Palmer, P. J. Kennedy, and H. F. Noller. 1978. Complete nucleotide sequence of a 16S rRNA gene from Escherichia coli. Proc. Natl. Acad. Sci. USA 75:4801-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bunge, M., H. Ballerstedt, and U. Lechner. 2001. Regiospecific dechlorination of spiked tetra- and trichlorodibenzo-p-dioxins by anaerobic bacteria from PCDD/F-contaminated Spittelwasser sediments. Chemosphere 43:675-681. [DOI] [PubMed] [Google Scholar]
- 16.Bunge, M., L. Adrian, A. Kraus, M. Opel, W. G. Lorenz, J. R. Andreesen, H. Gorisch, and U. Lechner. 2003. Reductive dehalogenation of chlorinated dioxins by an anaerobic bacterium. Nature 421:357-360. [DOI] [PubMed] [Google Scholar]
- 17.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cutter, L. A., J. E. Watts, K. R. Sowers, and H. D. May. 2001. Identification of a microorganism that links its growth to the reductive dechlorination of 2,3,5,6-chlorobiphenyl. Environ. Microbiol. 3:699-709. [DOI] [PubMed] [Google Scholar]
- 19.Daims, H., A. Brühl, R. Amann, K.-H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 20.Dennis, P. C., B. E. Sleep, R. R. Fulthorpe, and S. N. Liss. 2003. Phylogenetic analysis of bacterial populations in an anaerobic microbial consortium capable of degrading saturation concentrations of tetrachloroethylene. Can. J. Microbiol. 49:15-27. [DOI] [PubMed] [Google Scholar]
- 21.Dojka, M. A., P. Hugenholtz, S. K. Haak, and N. R. Pace. 1998. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl. Environ. Microbiol. 64:3869-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duhamel, M., K. Mo, and E. A. Edwards. 2004. Characterization of a highly enriched Dehalococcoides-containing culture that grows on vinyl chloride and trichloroethylene. Appl. Environ. Microbiol. 70:5538-5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 24.Fennell, D. E., A. B. Carroll, J. M. Gossett, and S. H. Zinder. 2001. Assessment of indigenous reductive dechlorinating potential at a TCE-contaminated site using microcosms, polymerase chain reaction analysis, and site data. Environ. Sci. Technol. 35:1830-1839. [DOI] [PubMed] [Google Scholar]
- 25.Fennell, D. E., I. Nijenhuis, S. F. Wilson, S. H. Zinder, and M. M. Häggblom. 2004. Dehalococcoides ethenogenes strain 195 reductively dechlorinates diverse chlorinated aromatic pollutants. Environ. Sci. Technol. 38:2075-2081. [DOI] [PubMed] [Google Scholar]
- 26.Fogel, G. B., C. R. Collins, J. Li, and C. F. Brunk. 1999. Prokaryotic genome size and SSU rDNA copy number: estimation of microbial relative abundance from a mixed population. Microb. Ecol. 38:93-113. [DOI] [PubMed] [Google Scholar]
- 27.Fu, Q., A. L. Barkovskii, and P. Adriaens. 2001. Dioxin cycling in aquatic sediments: the Passaic River Estuary. Chemosphere 43:643-648. [DOI] [PubMed] [Google Scholar]
- 28.Fu, Q., A. L. Barkovskii, and P. Adriaens. 2005. Microbial dechlorination of dioxins in estuarine enrichment cultures: effects of respiratory conditions and priming compound on community structure and dechlorination patterns. Mar. Environ. Res. 59:177-195. [DOI] [PubMed] [Google Scholar]
- 29.Futamata, H., T. Uchida, N. Yoshida, Y. Yonemitsu, and A. Hiraishi. 2004. Distribution of dibenzofuran-degrading bacteria in soils polluted with different levels of polychlorinated dioxins. Microbes Environ. 19:172-177. [Google Scholar]
- 30.Gu, A. Z., B. P. Hedlund, J. T. Staley, S. E. Strand, and H. D. Stensel. 2004. Analysis and comparison of the microbial community structures of two enrichment cultures capable of reductively dechlorinating TCE and cis-DCE. Environ. Microbiol. 6:45-54. [DOI] [PubMed] [Google Scholar]
- 31.Haggblom, M. M., Y. B. Ahn, D. E. Fennell, L. J. Kerkhof, and S. K. Rhee. 2003. Anaerobic dehalogenation of organohalide contaminants in the marine environment. Adv. Appl. Microbiol. 53:61-84. [DOI] [PubMed] [Google Scholar]
- 32.He, J., K. M. Ritalahti, M. R. Aiello, and F. E. Loffler. 2003. Complete detoxification of vinyl chloride by an anaerobic enrichment culture and identification of the reductively dechlorinating population as a Dehalococcoides species. Appl. Environ. Microbiol. 69:996-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hendrickson, E. R., J. A. Payne, R. M. Young, M. G. Starr, M. P. Perry, S. Fahnestock, D. E. Ellis, and R. C. Ebersole. 2002. Molecular analysis of Dehalococcoides 16S ribosomal DNA from chloroethene-contaminated sites throughout North America and Europe. Appl. Environ. Microbiol. 68:485-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hiraishi, A. 1999. Isoprenoid quinones as biomarkers of microbial populations in the environment. J. Biosci. Bioeng. 88:449-460. [DOI] [PubMed] [Google Scholar]
- 35.Hiraishi, A. 2003. Biodiversity of dioxin-degrading microorganisms and potential utilization in bioremediation. Microbes Environ. 18:105-125. [Google Scholar]
- 36.Hiraishi, A., H. Miyakoda, B.-R. Lim, H.-Y. Hu, K. Fujie, and J. Suzuki. 2001. Toward the bioremediation of dioxin-polluted soil: structural and functional analyses of in situ microbial populations by quinone profiling and culture-dependent methods. Appl. Microbiol. Biotechnol. 57:248-256. [DOI] [PubMed] [Google Scholar]
- 37.Hiraishi, A., Y. Ueda, J. Ishihara, and T. Mori. 1996. Comparative lipoquinone analysis of influent sewage and activated sludge by high-performance liquid chromatography and photodiode array detection. J. Gen. Appl. Microbiol. 42:457-469. [Google Scholar]
- 38.Hiraishi, A., Y. Yamanaka, and T. Narihiro. 2000. Seasonal microbial community dynamics in a flowerpot-using personal composting system for disposal of household biowaste. J. Gen. Appl. Microbiol. 46:133-146. [DOI] [PubMed] [Google Scholar]
- 39.Hiraishi, A., Y. Yonemitsu, M. Matsushita, Y. K. Shin, H. Kuraishi, and K. Kawahara. 2002. Characterization of Porphyrobacter sanguineus sp. nov., an aerobic bacteriochlorophyll-containing bacterium capable of degrading biphenyl and dibenzofuran. Arch. Microbiol. 178:45-52. [DOI] [PubMed] [Google Scholar]
- 40.Holliger, C., and G. Schraa. 1994. Physiological meaning and potential for application of reductive dechlorination by anaerobic bacteria. FEMS Microbiol. Rev. 15:297-305. [DOI] [PubMed] [Google Scholar]
- 41.Holliger, C., G. Wohlfarth, and G. Diekert. 1998. Reductive dechlorination in the energy metabolism of anaerobic bacteria. FEMS Microbiol. Rev. 22:383-398. [Google Scholar]
- 42.Holoman, T. R., M. A. Elberson, L. A. Cutter, H. D. May, and K. R. Sowers. 1999. Characterization of a defined 2,3,5,6-tetrachlorobiphenyl-ortho-dechlorinating microbial community by comparative sequence analysis of genes coding for 16S rRNA. Appl. Environ. Microbiol. 64:3359-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hölscher, T., R. Krajmalnik-Brown, K. M. Ritalahi, F. von Wintzingerode, H. Görisch, F. E. Löffler, and L. Adrian. 2004. Multiple nonidentical reductive-dehalogenase-homologous genes are common in Dehalococcoides. Appl. Environ. Microbiol. 70:5290-5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iwasaki, M., and A. Hiraishi. 1998. A new approach to numerical analysis of microbial quinone profiles in the environment. Microbes Environ. 13:67-76. [Google Scholar]
- 45.Kao, C. M., S. C. Chen, J. K. Liu, and M. J. Wu. 2001. Evaluation of TCDD biodegradability under different redox conditions. Chemosphere 44:1447-1454. [DOI] [PubMed] [Google Scholar]
- 46.Kassenga, G., J. H. Pardue, W. M. Moe, and K. S. Bowman. 2004. Hydrogen thresholds as indicators of dehalorespiration in constructed treatment wetlands. Environ. Sci. Technol. 38:1024-1030. [DOI] [PubMed] [Google Scholar]
- 47.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 48.LaMontagne, M. G., G. J. Davenport, L. H. Hou, and S. K. Dutta. 1998. Identification and analysis of PCB dechlorinating anaerobic enrichments by amplification: accuracy of community structure based on restriction analysis and partial sequencing of 16S rRNA genes. J. Appl. Microbiol. 84:1156-1162. [DOI] [PubMed] [Google Scholar]
- 49.Lee, W., and B. Batchelor. 2004. Abiotic reductive dechlorination of chlorinated ethylenes by soil. Chemosphere 55:705-713. [DOI] [PubMed] [Google Scholar]
- 50.Löffler, F. E., Q. Sun, J. Li, and J. M. Tiedje. 2000. 16S rRNA gene-based detection of tetrachloroethene-dechlorinating Desulforomonas and Dehalococcoides species. Appl. Environ. Microbiol. 66:1369-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Major, D. W., M. L. McMaster, E. E. Cox, E. A. Edwards, S. M. Dworatzek, E. R. Hendrickson, M. G. Starr, J. A. Payne, and L. W. Buonamici. 2002. Field demonstration of successful bioaugmentation to achieve dechlorination of tetrachloroethene to ethene. Environ. Sci. Technol. 36:5106-5116. [DOI] [PubMed] [Google Scholar]
- 52.Maymo-Gatell, X., Y.-T. Chien, J. M. Gossett, and S. H. Zinder. 1997. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science 276:1568-1571. [DOI] [PubMed] [Google Scholar]
- 53.Mohn, W. W., and J. M. Tiedje. 1992. Microbial reductive dehalogenation. Microbiol. Rev. 56:482-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Narihiro, T., Y. Yamanaka, and A. Hiraishi. 2003. High culturability of bacteria in commercially available personal composters for fed-batch treatment of household biowaste. Microbes Environ. 18:94-99. [Google Scholar]
- 56.Nojiri, H., and T. Omori. 2002. Molecular bases of aerobic bacterial degradation of dioxins: involvement of angular dioxygenation. Biosci. Biotechnol. Biochem. 66:2001-2016. [DOI] [PubMed] [Google Scholar]
- 57.Richardson, R. E., V. K. Bhupathiraju, D. L. Song, T. A. Goulet, and L. Alvarez-Cohen. 2002. Phylogenetic characterization of microbial communities that reductively dechlorinate TCE based upon a combination of molecular techniques. Environ. Sci. Technol. 36:2652-2662. [DOI] [PubMed] [Google Scholar]
- 58.Roser, D. J. 1980. Ethidium bromide: a general purpose fluorescent stain for nucleic acid in bacteria and eucaryotes and its use in microbial ecology studies. Soil Biol. Biochem. 12:329-336. [Google Scholar]
- 59.Saito, H., and K. I. Miura. 1963. Preparation of transforming deoxyribonucleic acid by phenol treatment. Biochim. Biophys. Acta 72:619-629. [PubMed] [Google Scholar]
- 60.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 61.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 62.Schlötelburg, C., F. von Wintzingerode, R. Hauck, W. Hegemann, and U. B. Göbell. 2000. Bacteria of an anaerobic 1,2-dichloropropane-dechlorinating mixed culture are phylogenetically related to those of other anaerobic dechlorinating consortia. Int. J. Syst. Evol. Microbiol. 50:1505-1511. [DOI] [PubMed] [Google Scholar]
- 63.Smits, T. H., C. Devenoges, K. Szynalski, J. Maillard, and C. Holliger. 2004. Development of a real-time PCR method for quantification of the three genera Dehalobacter, Dehalococcoides, and Desulfitobacterium in microbial communities. J. Microbiol. Methods 57:369-378. [DOI] [PubMed] [Google Scholar]
- 64.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vargas, C., D. E. Fennell, and M. M. Haggblom. 2001. Anaerobic reductive dechlorination of chlorinated dioxins in estuarine sediments. Appl. Microbiol. Biotechnol. 57:786-790. [DOI] [PubMed] [Google Scholar]
- 66.Villemur, R., M. Saucier, A. Gauthier, and R. Beaudet. 2002. Occurrence of several genes encoding putative reductive dehalogenases in Desulfitobacterium hafniense/frappieri and Dehalococcoides ethenogenes. Can. J. Microbiol. 48:697-706. [DOI] [PubMed] [Google Scholar]
- 67.von Wintzingerode, F., B. Selent, W. Hegemann, and U. B. Göbel. 1999. Phylogenetic analysis of an anaerobic, trichlorobenzene-transforming microbial consortium. Appl. Environ. Microbiol. 65:283-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wittich, R.-M. 1998. Degradation of dioxin-like compounds by microorganisms. Appl. Microbiol. Biotechnol. 49:489-499. [DOI] [PubMed] [Google Scholar]
- 70.Wu, Q., C. E. Milliken, G. P. Meier, J. E. Watts, K. R. Sowers, and H. D. May. 2002. Dechlorination of chlorobenzenes by a culture containing bacterium DF-1, a PCB dechlorinating microorganism. Environ. Sci. Technol. 36:3290-3294. [DOI] [PubMed] [Google Scholar]
- 71.Wu, Q., J. E. Watts, K. R. Sowers, and H. D. May. 2002. Identification of a bacterium that specifically catalyzes the reductive dechlorination of polychlorinated biphenyls with doubly flanked chlorines. Appl. Environ. Microbiol. 68:807-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou, J., M. A. Bruns, and J. M. Tiedje. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316-322. [DOI] [PMC free article] [PubMed] [Google Scholar]