Abstract
We describe a rapid and easily automated phylogenetic grouping technique based on analysis of bacterial genome single-nucleotide polymorphisms (SNPs). We selected 13 SNPs derived from a complete sequence analysis of 11 essential genes previously used for multilocus sequence typing (MLST) of 30 Escherichia coli strains representing the genetic diversity of the species. The 13 SNPs were localized in five genes, trpA, trpB, putP, icdA, and polB, and were selected to allow recovery of the main phylogenetic groups (groups A, B1, E, D, and B2) and subgroups of the species. In the first step, we validated the SNP approach in silico by extracting SNP data from the complete sequences of the five genes for a panel of 65 pathogenic strains belonging to different E. coli pathovars, which were previously analyzed by MLST. In the second step, we determined these SNPs by dideoxy single-base extension of unlabeled oligonucleotide primers for a collection of 183 commensal and extraintestinal clinical E. coli isolates and compared the SNP phylotyping method to previous well-established typing methods. This SNP phylotyping method proved to be consistent with the other methods for assigning phylogenetic groups to the different E. coli strains. In contrast to the other typing methods, such as multilocus enzyme electrophoresis, ribotyping, or PCR phylotyping using the presence/absence of three genomic DNA fragments, the SNP typing method described here is derived from a solid phylogenetic analysis, and the results obtained by this method are more meaningful. Our results indicate that similar approaches may be used for a wide variety of bacterial species.
Escherichia coli is a normal inhabitant of the gut flora of vertebrates, including humans, and the genetic variability of this species leads to differential colonization of hosts (14). However, it is also frequently associated with various intestinal (diarrhea) and extraintestinal (urinary tract infection, bacteremia, meningitis) diseases (10). In the United Kingdom, E. coli is the most frequent community-acquired pathogen in humans (13). The genetic population structure of E. coli is globally clonal (8, 24), and phylogenetic analysis has shown that this species is composed of five main phylogenetic groups (groups A, B1, E, D, and B2) (12, 16, 19). Several studies have demonstrated the link between the phylogeny (i.e., the evolutionary history) of the species and pathogenesis (2, 4, 11, 12). Virulent extraintestinal strains mainly belong to groups B2 and D (21), whereas severe diarrhea is due to strains that do not belong to these two groups (11). Finding an efficient phylogenic grouping technique has become a central goal for investigating the genetic relationship between clinical pathogenic strains and reference strains and for epidemiological surveillance and public health decisions. At present, several grouping techniques are available, such as multilocus enzyme electrophoresis (MLEE) (20), ribotyping (8), random amplified polymorphic DNA analysis (8), fluorescent amplified-fragment length polymorphism (FAFLP) analysis (1), PCR phylotyping using the presence/absence of three genomic DNA fragments (5), analysis of variation at mononucleotide repeats in intergenic sequences (9), and, more recently, multilocus sequence typing (MLST) (12, 19, 22). Only the MLST technique allows real phylogenetic analysis (i.e., reconstruction of the evolutionary history of the strains). MLST is now clearly the “gold standard” (25), but it is costly and time-consuming. A discriminative but simple, standardized, reproducible, and automated phylotyping technique would be an interesting tool for large-scale analysis of strain collections. In large-scale animal genotyping and medical diagnostics, the need for high-throughput genotyping technologies with simple steps and high accuracy that can be automated at a reasonable cost has led workers to focus on a new marker type, single-nucleotide polymorphism (SNP) (26). This marker type is just a point nucleotide change in a DNA sequence with the usual alternative of possible nucleotides at a given position. SNPs are known to be associated with inherited disorders, as well as susceptibility to various somatic diseases and microbial infections (15, 23).
Here, we developed a new, simple, rapid, cost-effective readout method for phylotyping based on single base changes that are informative for phylogeny. The phylogenetic SNPs were identified from a DNA sequence analysis of 11 essential genes (encoding seven metabolic proteins and four DNA polymerases), which allowed phylogenetic tree reconstruction for 30 E. coli reference strains (20) representing the genetic diversity of the species (12). In the first step, this phylogenetic typing method was validated in silico with 65 pathogenic strains. In the second step, it was applied to 183 commensal and extraintestinal pathogenic strains of E. coli and compared to several typing methods.
MATERIALS AND METHODS
Bacterial strains.
A total of 249 E. coli strains were chosen for analysis. These strains included the 72 strains of the E. coli reference (ECOR) collection established by Ochman and Selander (20), 61 isolates of the IAI collection (21), including 14 intestinal commensal and 47 extraintestinal pathogenic strains, 50 isolates of an extraintestinal pathogenic collection originating from Europe, North America, and Australia (Bertrand Picard, personal communication), 65 pathogenic strains representing the pathovar diversity of the species (enteroaggregative, diffusely adhering, Shiga toxin-producing, enterotoxigenic, enteroinvasive, enteropathogenic, and extraintestinal pathogenic) (11), and the E. coli K-12 strain.
General strategy of SNP phylotyping.
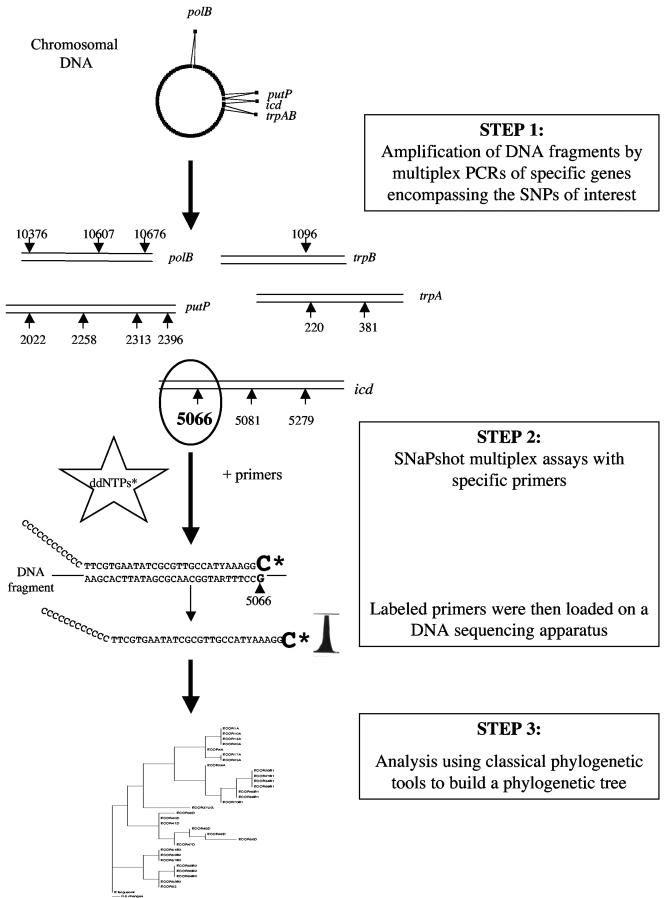
Multiple PCRs were first required to amplify the specific genes of interest encompassing the SNPs (step 1) (Fig. 1). Then dideoxy single-base extensions of unlabeled oligonucleotide primers were prepared with specific primers (SNaPshot multiplex assays), and the reaction products were loaded on a DNA sequencing apparatus (step 2). Finally, the SNP data obtained were analyzed as informative site data for phylogenetic analysis using classical phylogenetic tools (step 3).
FIG.1.
General strategy used for SNP phylotyping. In step 1, chromosomal DNA was used as a template for amplification of five genes (trpA, trpB, putP, icdA, and polB) in multiplex PCRs. In step 2, after purification of PCR fragments, dideoxy single-base extensions were added to specific oligonucleotide primers with fluorescent dideoxynucleoside triphosphates (ddNTPs) by using the SNaPshot reaction protocol (Applied Biosystems). The labeled primers were loaded onto a DNA sequencing apparatus, and the results were determined. In step 3, the SNP data were used as informative sites for phylogenetic tree construction using a classical phylogenetic approach. Note that the numbering of SNPs is arbitrary and corresponds to the localization of the SNPs in the concatenated alignment of 11 genes (12).
Amplification of DNA fragments by multiplex PCRs.
PCR amplification of trpB and icdA coding sequences was performed as previously described by using 2.5 μl of bacterial lysate as the template and a Gibco PCR kit (final volume, 25 μl) (Table 1) (12). Amplification of trpA, putP, and polB coding sequences was performed using 5 pmol, 10 pmol, and 20 pmol of each primer pair, respectively, to obtain the same quantity of each fragment (Table 1). The PCR amplification program consisted of 5 min at 94°C, 35 cycles of 30 s at 94°C, 30 s at 50°C, and 1 min 30 s at 72°C, and then 7 min at 72°C. Amplification products were checked and quantified on a 1% agarose gel. All PCR products were purified by double digestion as recommended by the manufacturer (Amersham Biosciences). PCR products were incubated for 1 h at 37°C with 4 U of exonuclease I and 10 U of shrimp alkaline phosphatase I, and then both enzymes were inactivated by heating for 15 min at 75°C.
TABLE 1.
Primers used for the amplification of trpA, trpB, putP, icdA, and polB and primers used for SNaPshot analysisa
| Primer | Sequence | Length (b) | Fragment length (bp) |
|---|---|---|---|
| Amplification primers | |||
| trpA-F | 5′-ATGGAACGCTACGAATCTCTGTTTGCCC-3′ | ||
| trpA-R | 5′-TCGCCGCTTTCATCGGTTGTACAAA-3′ | 796 | |
| trpB-F | 5′-ACAATGACAAGATTACTTAACCCCT-3′ | ||
| trpB-R | 5′-TTTCCCCCTCGTGCTTTCAAAATATC-3′ | 1,191 | |
| put-int-F | 5′-GCGACGATCCTTTACACCTTTATTG-3′ | ||
| put-int-R | 5′-CGCATCGGCCTCGGCAAAGCG-3′ | 9,58 | |
| icdA-5′ | 5′-GAAAGTAAAGTAGTTGTTCCGG-3′ | ||
| icdA-3′ | 5′-GATGATCGCGTCACCAAAYTC-3′ | 1,236 | |
| polB-5′ | 5′-TGGAAAAACTCAACGCCTGGT-3′ | ||
| polB-3′ | 5′-TGGTTGGCATCAGAAAACGGC-3′ | 1,202 | |
| SNaPshot reaction 1 primers | |||
| SNP-10676 | 5′-CCYAATCTCGGCGAAGTGCC-3′ | 20 | |
| SNP-10376 | 5′-CCCTCTCTTCATTCTCGCTGGAAAC-3′ | 25 | |
| SNP-10607 | 5′-CCCCCCCCGSGGAAAATAGAGATGACCAAA-3′ | 30 | |
| SNP-2396 | 5′-CCCCCCCCCCCCTCCAGTGCGATTACYGAAGATTT-3′ | 35 | |
| SNP-2313 | 5′-CCCCCCCCCCCCCCCGTTTAACCCGTGGATTGCYGGGATT-3′ | 40 | |
| SNP-2258 | 5′-CCCCCCCCCCCCCCCCAGAATTTGYGCCAGTTCRATAAACACRCG-3′ | 45 | |
| SNaPshot reaction 2 primers | |||
| SNP-381 | 5′-AACGCGGCCTGGCGGAAGGG-3′ | 20 | |
| SNP-5279 | 5′-CCCGCAGGAATTTAATCACTTTCTC-3′ | 25 | |
| SNP-1096 | 5′-CCCCCCCACGTCTTTGGCMCCCATATAAAT-3′ | 30 | |
| SNP-220 | 5′-CCCCCCCCCGAGYTTCTGGCGAATSAGTGCCAGCA-3′ | 35 | |
| SNP-5066 | 5′-CCCCCCCCCCCCTTCGTGAATATCGCGTTGCCATYAAAGG-3′ | 40 | |
| SNP-2022 | 5′-CCCCCCCCCCCCCCCCCATCARYGAGATAATGGCRACAAARTTCA-3′ | 45 | |
| SNP-5081 | 5′-CCCCCCCCCCCCCCCCCCCCCCCCCCCAGAGARCGAATACCGCCACCAWC-3′ | 50 |
Two SNaPshot reactions were performed, SNaPshot reaction 1 and SNaPshot reaction 2. Primers longer than 30 bases (b) were purified by HPLC.
SNaPshot multiplex reaction.
Primers longer than 30 bases were purified by high-performance liquid chromatography (HPLC). Two SNaPshot reactions (ABI Prism SNaPshot multiplex kit; Applied Biosystems) were performed by following the manufacturer's recommendations. SNaPshot reaction 1 was performed with a multiplex of six primers, as follows. Five microliters of SNaPshot multiplex Ready Reaction mixture (Applied Biosystems) was mixed with 0.4 μM of primers SNP-10607, SNP-10376, SNP-2313, and SNP-2258, 0.2 μM of primer SNP-10676, and 0.1 μM of primer SNP-2396 (Table 1). PCR products of the putP, polB and trpA genes were added before the thermal cycling, which consisted of 25 cycles of 96°C for 10 s, 53°C for 5 s, and 60°C for 30 s. SNaPshot reaction 2 with a multiplex of seven primers was performed with 0.025 μM of SNP-2022, 0.05 μM of SNP-5279, 0.1 μM of primer SNP-381, 0.2 μM of SNP-5081, 0.3 μM of SNP-220, 0.6 μM of SNP-5066, and 1.6 μM of SNP-1096 (Table 1) and PCR products of five genes (putP, polB, trpA, trpB, and icdA). The annealing temperature was 54°C. To make sure that unincorporated fluorescent dideoxynucleoside triphosphates did not interfere with fragments of interest, a postextension treatment was included that consisted of adding 1 U of shrimp alkaline phosphatase I and incubating the preparation for 1 h at 37°C. The enzyme was inactivated by incubation for 15 min at 75°C.
Samples were analyzed with an ABI PRISM 3100 genetic analyzer (Applied Biosystems). One microliter of a SNaPshot reaction mixture was added to 14 μl of Hi-Di formamide. Samples were denatured at 100°C for 5 min and then placed on ice until they were loaded onto the analyzer. The parameters used were dye set E and the SNP36_POP4 default module (Applied Biosystems). After runs, the results were read using the GeneScan software (Applied Biosystems). Note that the numbering of SNPs is arbitrary and corresponds to their localization in the concatenated alignment of 11 genes (12) (Fig. 2B).
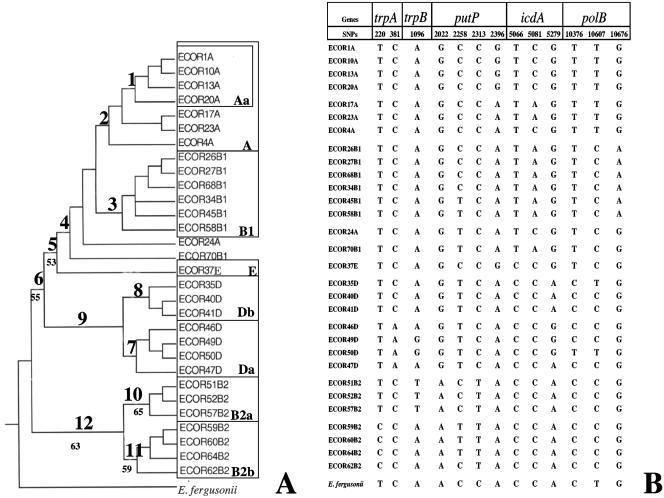
FIG. 2.
Phylogenetic SNPs used for E. coli typing. (A) Phylogenetic tree of 30 strains from the ECOR collection (20) reconstructed by parsimony using the 13 informative phylogenetic SNPs extracted from the complete sequences of five genes and rooted with E. fergusonii. The 12 nodes were numbered as follows. Nodes 1 and 2 correspond to subgroup Aa (SNP 2396) and group A (SNP 10607), respectively. Node 3 (SNPs 5081 and 10676) corresponds to the B1 group. Nodes 4 (SNP 5066), 5 (SNP 10376), and 6 (SNP 2022) correspond to the ancestor groups of groups A and B1, groups A, B1, and E, and groups A, B1, E, and D, respectively. Node 9 corresponds to group D (SNP 2258), and nodes 7 and 8 correspond to subgroups Da (SNPs 381) and Db (SNP 10607). Node 12 corresponds to group B2 (SNP 2313), and nodes 10 and 11 correspond to subgroups B2a (SNP 1096) and B2b (SNPs 220 and 2258). The phylogenetic group to which a strain belongs is indicated after the designation of the strain and was based on MLEE data (16) in most cases; the exception was the ECOR37 strain, which was considered a group E strain (12) (tree length = 24, consistency index = 0.58, retention index = 0.90). The numbers below the nodes are bootstrap percentages calculated from 1,000 iterations. Only values that are ≥50% are indicated. (B) SNP combinations for five genes (trpA, trpB, putP, icdA, and polB) for the 30 E. coli strains and E. fergusonii. The nucleotides at the 13 SNP positions are indicated for each strain.
Phylogenetic analysis.
Phylogenetic SNPs were selected from previously published sequences (12) using the PAUP*4.0 (Sinauer Associates, Inc., Sunderland, Mass.) and MacClade (Maddison & Maddison) softwares. Phylogenetic grouping was then performed by tree reconstruction using the 13 phylogenetic SNPs localized in the trpA, trpB, putP, polB, and icdA genes as informative sites. Fifty percent majority rule consensus trees were obtained using Wagner parsimony as the optimality criterion with the heuristic search of the PAUP*4.0 software. Parsimony was chosen as an optimality criterion to analyze the SNP data because the SNPs were selected as informative, based on a parsimony analysis of the 11 genes (12).
RESULTS AND DISCUSSION
Choice of SNPs informative for phylogeny by in silico analysis.
Previous MLST analyses reconstructed the phylogenetic history of 30 ECOR strains based on the coding sequences of 11 essential genes (encoding seven metabolic proteins and four polymerases) (12), which were shown to exhibit low levels of horizontal gene transfer (7, 12, 19). The tree was rooted with the closest relative of E. coli, Escherichia fergusonii (18). From previously work (12), we identified 12 robust nodes based on bootstrap values. These nodes included one node for each of four major phylogenetic groups (groups A, B1, D, and B2; nodes 2, 3, 9, and 12, respectively); three nodes for the ancestor of groups A and B1 (node 4), the ancestor of groups A, B1, and E (node 5), and the ancestor of groups A, B1, E, and D (node 6); two internal nodes in groups D and B2 (nodes 7 and 8 defining subgroups Da and Db and nodes 10 and 11 defining subgroups B2a and B2b, respectively); and one internal node in group A (node 1 defining subgroup Aa) (Fig. 2A). These 12 nodes are supported by 374 informative sites distributed over the 11 genes. We then selected 13 of these informative sites that were (i) located in a limited number of genes (a tradeoff was made between the risk of horizontal gene transfers to scramble the phylogeny if too few genes were selected and a reasonable number of PCRs to perform), (ii) located in an otherwise conserved region to avoid primer mismatches, (iii) specific for each node, and (iv) sufficient to reconstruct a phylogenetic tree with a topology similar to that of the tree based on the 11 essential gene sequences (12). These informative sites, which we called phylogenetic SNPs, are localized in the trpA (two SNPs, SNPs 220 and 381), trpB (one SNP, SNP 1096), putP (four SNPs, SNPs 2022, 2258, 2313, and 2396), icdA (three SNPs, SNPs 5066, 5081, and 5279), and polB (three SNPs, SNPs 10376, 10607, and 10676) genes (Fig. 2B). The tree obtained from the phylogenetic reconstruction of the 13 SNPs using parsimony for the 30 ECOR strains and rooted with E. fergusonii (Fig. 2A) is consistent with the E. coli phylogenetic history (12).
In silico analysis of a collection of E. coli strains representing the pathovar diversity of the species.
To validate in silico the choice of these SNPs, our SNP phylotyping technique was first applied to a collection of 65 E. coli strains representing all the intestinal and extraintestinal pathovars previously studied by MLST (11) and the E. coli K-12 strain (3). The 13 SNPs were extracted from the complete sequences of five genes (trpA, trpB, putP, icdA, and polB) (11, 12). The SNP data for these 66 strains were added individually for each strain to the data of the 30 ECOR reference strains for phylogenetic reconstruction of a 50% majority rule consensus tree using parsimony in PAUP*4.0. The data for 48 (73%) of the 66 strains did not disturb the topology of the phylogenic tree and allowed easy grouping within the defined groups and subgroups. For 16 strains (23%), addition of the SNP data disturbed the topology of the phylogenic tree, but the strains in groups and subgroups remained closely related. In these cases, the E. coli strain tested could also be assigned to a phylogenetic group or subgroup since the strain clustered with a reference strain. Finally, one strain, DAEC213, could not be assigned to any group or subgroup (see Fig. S1A in the supplemental material). This could have resulted from horizontal transfers in the genes studied affecting the tree reconstructed from the SNP data but not the tree reconstructed from the MLST data, as in this case the horizontally transferred genes were swamped in the remaining informative sites. Results obtained from the SNP phylotyping were then compared to previous phylogenic MLST typing done with this collection (12). Excluding the strain that we were unable to group by SNP typing, 60 of 65 strains (92.3%) belonged to identical groups as determined by both approaches. The five differences were due to mistyping involving strains that in the MLST analysis (11) were in a basal position with low bootstrap values in the A, B1, and D groups, indicating weak support of the phylogenetic positions of the strains. As expected, the K-12 strain was assigned to the A group.
These results show that the information for the 13 SNPs could be sufficient to reconstruct the phylogenic history of E. coli and that the SNP phylotyping approach could be suitable and robust for phylogenic analyses of new collections of strains.
Oligonucleotide design and SNaPshot validation.
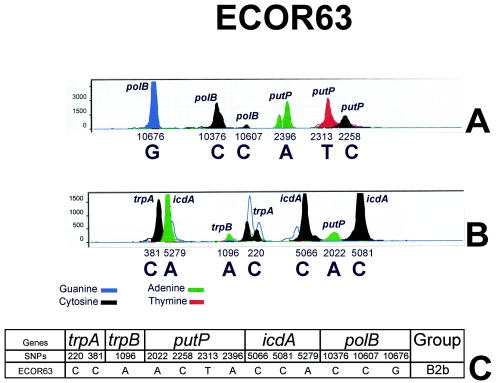
To use SNaPshot analysis, primers for each of the 13 phylogenetic SNPs were designed so that they exactly adjoined the variable nucleotide with the constraint of homogeneity at the annealing temperature (Table 1). The 30 ECOR strains that were used to determine the phylogenetic SNPs were then analyzed by the SNaPshot approach. The reproducibility of the method was assessed by using a panel of five ECOR strains belonging to different phylogenetic groups (ECOR4, ECOR26, ECOR27, ECOR40, and ECOR47). Identical results for a given strain were obtained when SNaPshot reactions were performed (i) with the same PCR products or (ii) with PCR products resulting from different DNA amplifications (data not shown). An example of an electropherogram of two SNaPshot reactions for strain ECOR63 is shown in Fig. 3. For all strains, double peaks were observed with primer SNP-2396 in the first SNaPshot reaction (Fig. 3A) and with primer SNP-220 in the second SNaPshot reaction (Fig. 3B). Although these primers were purified by HPLC, mass spectrometry analysis showed that each oligonucleotide primer (SNP-2396 and SNP-220) is a mixture of molecules of different lengths, one corresponding to the expected size (35 bases) and another of 34 bases (data not shown). This explains the two peaks observed experimentally. Moreover, another double blue (guanine) peak, as well as a weak green (adenine) peak in SNaPshot reaction 2 with the SNP-220 primer, were observed (Fig. 3B). These peaks were considered artifacts and were not taken in consideration since they were also present with only the SNP-220-primer in the absence of PCR-amplified DNA (data not shown). Such peaks were probably due to a potential 7-nucleotide annealing site between two molecules of the SNP-220 primer, as predicted by bioinformatics analysis (data not shown). This annealing led to addition of an adenine or a guanine at the end of the SNP-220 primer during the SNaPshot reaction.
FIG. 3.
SNaPshot analysis migration profile of strain ECOR63. (A) SNaPshot reaction 1. (B) SNaPshot reaction 2. The migration order corresponds to the primer lengths. The double peaks obtained in SNaPshot reaction 1 with the SNP-2396 primer (putP) (A) and in SNaPshot reaction 2 with the SNP-220 primer (trpA) (B) were due to a mixture of primers of different lengths (34- and 35-mers). Unfilled peaks with SNP-220 (trpA) are obtained in the presence of the primer without template DNA and thus were not taken into account in the analysis and were considered artifacts. The SNP combination defining ECOR63 as a subgroup B2b strain is indicated in panel C. Note that the numbering of SNPs is arbitrary and corresponds to the localization of the SNPs in the concatenated alignment of 11 genes (12).
As expected, the results obtained experimentally were in perfect agreement with the results obtained with the complete sequences of the genes.
Validation of SNP phylotyping by analysis of the complete ECOR collection.
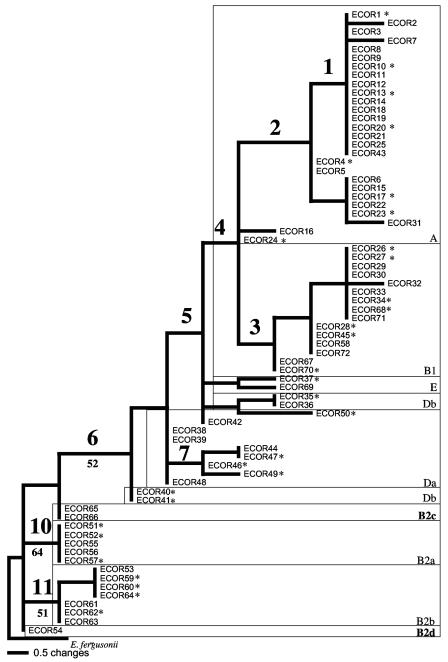
To determine whether the new method of phylotyping was suitable for the diversity of the species E. coli, we extended the analysis to the 42 remaining strains of the ECOR collection (20). The SNPs were determined for these 42 additional strains by SNaPshot reactions and were used to construct a tree with the SNPs of the 30 ECOR strains, which was rooted with E. fergusonii (Fig. 4). Whatever optimal criterion was chosen to reconstruct the tree (parsimony, unweighted-pair group method using average linkages, or neighbor joining), the global topology of the tree was in accordance with our current knowledge concerning the phylogeny of the species. Only the tree constructed with parsimony as the optimal criterion is presented in Fig. 4. The B2 group strains are basal strains. There are two additional B2 subgroups, one consisting of the ECOR54 strain and the other intermediate between the B2 and D groups consisting of strains ECOR65 and ECOR66. It should be noted that the ECOR66 strain is the most external of the group B2 strains in the MLEE tree (16) and that it has a group D profile as determined by ribotyping (6). Group D strains are located between group B2 and groups A, B1, and E. They include the ECOR42 strain considered “ungrouped” in the MLEE analysis (16) but classified as a group D strain by PCR phylotyping (5). The ECOR69 group B1 strain is clustered with the ECOR37 strain in group E, outside groups A and B1. This strain has an atypical ribotype profile that is different from that of group B1 strains (6). Groups A and B1 are sister groups. Group B1 is monophyletic. Group A also is monophyletic except for strains ECOR24 and ECOR16. ECOR31 and ECOR43, which were also considered “ungrouped” in the MLEE analysis (16), were grouped with the group A strains. The ECOR43 strain was also grouped as a group A strain by PCR phylotyping (5), FAFLP analysis (1), and MLST (Escobar-Páramo and Denamur, unpublished data). Strain ECOR24 is an atypical group A strain as it exhibits numerous virulence factors (17) and has a group B1 profile as determined by ribotyping (6). Strain ECOR16 also is not a group A strain as determined by FAFLP analysis (1).
FIG. 4.
Fifty percent majority rule consensus tree (phylogram), obtained by using parsimony, based on simultaneous analysis of 13 SNPs of the 72 ECOR strains and rooted with E. fergusonii. SNP data were obtained from SNaPshot analysis. The 30 E. coli strains used to reconstruct the tree in Fig. 2 are indicated by asterisks. The numbers above the nodes correspond to conserved nodes in the tree of the 30 ECOR strains rooted with E. fergusonii (Fig. 2). Subgroups B2c and B2d defined by SNP analysis are indicated by boldface type (tree length = 32, consistency index = 0.47, retention index = 0.92). The numbers below the nodes are bootstrap percentages calculated from 1,000 iterations. Only values that are ≥50% are indicated. The SNP phylotyping is in agreement with the MLEE typing (16) except for the “ungrouped” ECOR31, ECOR37, ECOR42, and ECOR43 strains, which appear in groups A, E, D, and A, respectively, and the group B1 strain ECOR69, which appears in group E.
Thus, the SNP phylotyping approach can be applied to the genetic diversity of the species represented by the complete ECOR collection. Strains ECOR65 and ECOR66 and strain ECOR54, which represented two nodes of the B2 group (subgroups B2c and B2d, respectively) (Fig. 4), were added to the 30 reference strains to provide a phylogenetic framework for the species for further analyses.
Analysis of a collection of commensal and extraintestinal pathogenic E. coli strains.
A total of 111 commensal and pathogenic extraintestinal E. coli strains originating from Europe, America, and Australia were analyzed by SNP phylotyping. These strains were classified in the four main phylogenetic groups by PCR phylotyping (5) for this work (C. Amorin and E. Denamur, unpublished data). According to this typing approach, 37%, 4.5%, 10.8%, and 47.7% of them belong to the A, B1, D, and B2 phylogenetic groups, respectively. SNP data for each strain were added one by one to the SNP data set for the 32 ECOR strains representative of the phylogenetic diversity of the species. From these data, a 50% majority rule consensus tree was then reconstructed using parsimony in PAUP*4.0. The phylogenetic position of each strain was determined by comparison with the 32 reference strains (see Fig. S1B in the supplemental material). Ninety-three strains (84%) were easily classified in a phylogenetic group since the topology of the constructed phylogenic tree was conserved after addition of the SNP strain data. For 13 strains (12%), the tree topology was not conserved when the SNP data were added, but they could also be easily classified in a phylogenetic group. Indeed, as stated above, the main phylogenic groups and subgroups were still closely related in the nonconserved topology trees. Finally, five strains (4%) remained ungrouped by SNP phylotyping analysis (see Fig. S1B in the supplemental material).The five genes (trpA, trpB, putP, icdA, and polB) of strains that did not conserve the phylogenetic topology of the reference tree or were ungrouped were sequenced to see if the SNaPshot technology failed in the identification of the SNPs. The sequencing results confirmed the results obtained by SNaPshot analysis (data not shown). Of the 111 strains analyzed, 24.3% belonged to group A (node 2), with three-fourths of them in internal group Aa (node 1), 10% belonged to group B1 (node 3), and 14.4% belonged to group D (node 9); the majority of the group D strains were in internal group Da (node 7). Forty-five percent of the strains analyzed belonged to group B2 (node 12), with 28%, 42%, 10%, and 8% of them belonging to subgroups B2a (node 10), B2b (node 11), B2c (ECOR66), and B2d (ECOR54), respectively, and 1.8% (including ECOR37) belonged to group E. We then compared the SNP phylotyping results with the results obtained by PCR phylotyping (5) (see Table S1 in the supplemental material). More than 80% of the strains in this E. coli strain collection were grouped identically by PCR typing and SNaPshot analysis. In contrast to PCR typing, the new approach could identify strains belonging to group E. Indeed, many of the differences observed between PCR phylotyping and SNP phylotyping were due to group E strains that were mistyped as group D or B2 strains by PCR phylotyping. Other observed differences were mainly due to strains localized in group A by PCR typing and in group B1 by the SNP phylotyping approach or localized in group B2 by PCR typing and in group D by the SNP approach. Discrepancies between groups A and B2 were rarely observed (see Table S1 in the supplemental material).
The presence of strains outside previously defined groups or subgroups could have resulted from horizontal transfers, as stated above. Disagreements between PCR typing and SNP typing could have been due to horizontal transfers that scrambled the SNP phylogeny, especially when the disagreement was between closely related groups, such as groups A and B1 or groups D and B2. Alternatively, as the PCR typing method is based on only three genomic DNA fragments, recent mobilization of these fragments with acquisition or loss could be a pitfall of this approach (O. Clermont and E. Denamur, unpublished data). Furthermore, one of the DNA fragments, chuA, is involved in the iron capture process and so cannot be considered neutral.
Taken together, these results confirmed the validity of the new phylogenic typing method.
Concluding remarks.
We developed a new and easily automated phylogenetic grouping method similar to MLST, which was based on SNP analysis. This new method was validated with E. coli, which had one advantage: the availability of a reference collection representative of the diversity of the species (20) previously studied by a wide range of typing methods, including MLST. Although the new approach is less discriminative than MLST with the number of SNPs that we analyzed, the resolution of this approach could be increased by increasing the number of SNPs studied. Our results indicate that similar approaches may be used for a wide variety of bacterial species.
Supplementary Material
Acknowledgments
We are grateful to Stéphanie Gouriou and Bertrand Picard for providing pathogenic extraintestinal strains, to Julien Grassot for help with PAUP, to Christine Amorin for the PCR phylotyping, to Olivier Clermont and Bernard Grandchamp for critical advice, and to Elisabeth Gadreau for reading the manuscript.
This work was partially supported by grants from the Programme de Recherche Fondamentale en Microbiologie et Maladies Infectieuses et Parasitaires (MENRT) and from the Fondation pour la Recherche Médicale.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Arnold, C., L. Metherell, G. Willshaw, A. Maggs, and J. Stanley. 1999. Predictive fluorescent amplified-fragment length polymorphism analysis of Escherichia coli: high-resolution typing method with phylogenetic significance. J. Clin. Microbiol. 37:1274-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bingen, E., B. Picard, N. Brahimi, S. Mathy, P. Desjardins, J. Elion, and E. Denamur. 1998. Phylogenetic analysis of Escherichia coli strains causing neonatal meningitis suggests horizontal gene transfer from a predominant pool of highly virulent B2 group strains. J. Infect. Dis. 177:642-650. [DOI] [PubMed] [Google Scholar]
- 3.Blattner, F., G. Plunkett 3rd, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 4.Boyd, E., and D. Hartl. 1998. Chromosomal regions specific to pathogenic isolates of Escherichia coli have a phylogenetically clustered distribution. J. Bacteriol. 180:1159-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microb. 66:4555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clermont, O., C. Cordevant, S. Bonacorsi, A. Marecat, M. Lange, and E. Bingen. 2001. Automated ribotyping provides rapid phylogenetic subgroup affiliation of clinical extraintestinal pathogenic Escherichia coli strains. J. Clin. Microbiol. 39:4549-4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denamur, E., G. Lecointre, P. Darlu, O. Tenaillon, C. Acquaviva, C. Sayada, I. Sunjevaric, R. Rothstein, J. Elion, F. Taddei, M. Radman, and I. Matic. 2000. Evolutionary implications of the frequent horizontal transfer of mismatch repair genes. Cell 103:711-721. [DOI] [PubMed] [Google Scholar]
- 8.Desjardins, P., B. Picard, B. Kaltenbock, J. Elion, and E. Denamur. 1995. Sex in Escherichia coli does not disrupt the clonal structure of the population: evidence from random amplified polymorphic DNA and restriction-fragment-length polymorphism. J. Mol. Evol. 41:440-448. [DOI] [PubMed] [Google Scholar]
- 9.Diamant, E., Y. Palti, R. Gur-Arie, H. Cohen, E. Hallerman, and Y. Kashi. 2004. Phylogeny and strain typing of Escherichia coli, inferred from variation at mononucleotide repeat loci. Appl. Environ. Microbiol. 70:2464-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donnenberg, M. 2002. Escherichia coli virulence mechanisms of versatile pathogen. Elsevier Science, San Diego Calif.
- 11.Escobar-Páramo, P., O. Clermont, A. Blanc-Potard, H. Bui, C. Le Bouguenec, and E. Denamur. 2004. A specific genetic background is required for acquisition and expression of virulence factors in Escherichia coli. Mol. Biol. Evol. 21:1085-1094. [DOI] [PubMed] [Google Scholar]
- 12.Escobar-Páramo, P., A. Sabbagh, P. Darlu, O. Pradillon, C. Vaury, G. Lecointre, and E. Denamur. 2004. Decreasing the effects of horizontal gene transfer on bacterial phylogeny: the Escherichia coli case study. Mol. Phylogenet. Evol. 30:243-250. [DOI] [PubMed] [Google Scholar]
- 13.Farrell, D., I. Morrissey, D. De Rubeis, M. Robbins, and D. Felmingham. 2003. A UK multicentre study of the antimicrobial susceptibility of bacterial pathogens causing urinary tract infection. J. Infect. 46:94-100. [DOI] [PubMed] [Google Scholar]
- 14.Gordon, D., and A. Cowling. 2003. The distribution and genetic structure of Escherichia coli in Australian vertebrates: host and geographic effects. Microbiology 149:3575-3586. [DOI] [PubMed] [Google Scholar]
- 15.Gu, Z., L. Hillier, and P. Kwok. 1998. Single nucleotide polymorphism hunting in cyberspace. Hum. Mutat. 12:221-225. [DOI] [PubMed] [Google Scholar]
- 16.Herzer, P., S. Inouye, M. Inouye, and T. Whittman. 1990. Phylogenic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolated of Escherichia coli. J. Bacteriol. 172:6175-6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson, J., P. Delavari, M. Kuskowski, A. L. Stell. 2001. Phylogenetic distribution of extraintestinal virulence-associated traits in Escherichia coli. J. Infect. Dis. 183:78-88. [DOI] [PubMed] [Google Scholar]
- 18.Lawrence, J., H. Ochman, and D. Hartl. 1991. Molecular and evolutionary relationships among enteric bacteria. J. Gen. Microbiol. 137:1911-1921. [DOI] [PubMed] [Google Scholar]
- 19.Lecointre, G., L. Rachdi, P. Darlu, and E. Denamur. 1998. Escherichia coli molecular phylogeny using incongruence length difference test. Mol. Biol. Evol. 15:1685-1695. [DOI] [PubMed] [Google Scholar]
- 20.Ochman, H., and R. Selander. 1984. Standard reference strains of Escherichia coli from natural populations. J. Bacteriol. 157:690-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Picard, B., J. Garcia, S. Gouriou, P. Duriez, N. Brahimi, E. Bingen, J. Elion, and E. Denamur. 1999. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect. Immun 67:546-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reid, S., C. Herbelin, A. Bumbaugh, R. Selander, and T. Whittam. 2000. Parallel evolution of virulence in pathogenic Escherichia coli. Nature 406:64-67. [DOI] [PubMed] [Google Scholar]
- 23.Schork, N., D. Fallin, and J. Lanchbury. 2000. Single nucleotide polymorphisms and the future of genetic epidemiology. Clin. Genet. 58:250-264. [DOI] [PubMed] [Google Scholar]
- 24.Selander, R., D. Caugant, and T. Whittnam. 1987. Genetic structure and variation in natural populations of Escherichia coli, p. 1625-1648. In F. C. Neidhardt (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology. American Society for Microbiology, Washington, DC.
- 25.Urwin, R., and M. Maiden. 2003. Multi-locus sequence typing: a tool for global epidemiology. Trends Microbiol. 11:479-487. [DOI] [PubMed] [Google Scholar]
- 26.Wang, D., J. Fan, C. J. Siao, A. Berno, P. Young, R. Sapolsky, G. Ghandour, N. Perkins, E. Winchester, J. Spencer, L. Kruglyak, L. Stein, L. Hsie, T. Topaloglou, E. Hubbell, E. Robinson, M. Mittmann, M. S. Morris, N. Shen, D. Kilburn, J. Rioux, C. Nusbaum, S. Rozen, T. J. Hudson, E. S. Lander, et al. 1998. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science 280:1077-1082. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.