Abstract
We developed a novel method for the quantitative detection of the 16S rRNA of a specific bacterial species in the microbial community by using deoxyribozyme (DNAzyme), which possesses the catalytic function to cleave RNA in a sequence-specific manner. A mixture of heterogeneous 16S rRNA containing the target 16S rRNA was incubated with a species-specific DNAzyme. The cleaved target 16S rRNA was separated from the intact 16S rRNA by electrophoresis, and then their amounts were compared for the quantitative detection of target 16S rRNA. This method was used to determine the abundance of the 16S rRNA of a filamentous bacterium, Sphaerotilus natans, in activated sludge, which is a microbial mixture used in wastewater treatment systems. The result indicated that this DNAzyme-based approach would be applicable to actual microbial communities.
Microbes play essential roles in many environmental processes and are also utilized in environmental technology, including bioremediation, wastewater treatment, waste gas treatment, and solid waste treatment (1, 5, 16, 26). A variety of methods have been used to identify and quantify the microbes playing a key role in environmental processes and treatment systems (2, 6, 21, 22, 29). The quantification of 16S rRNA derived from such specific microbes is of considerable interest because the amount of 16S rRNA has been found to be positively correlated with the growth rate of the cell (3, 16, 17) and has been considered an indicator of bacterial activity (14). The majority of current approaches to the quantification of 16S rRNA utilize quantitative PCR after reverse transcription or dot blot hybridization. In quantitative PCR after reverse transcription, competitive PCR and real-time PCR are often used. Real-time PCR requires expensive instruments and reagents. Competitive PCR and dot blot hybridization are laborious and time-consuming. Rapid and cost-effective methods for the quantification of 16S rRNA should be developed to evaluate the activity of specific microbes in environments and treatment systems.
Activated sludge systems, used worldwide for wastewater treatment, utilize a complex microbial community (2, 26). One of the major operational problems of systems is the excessive growth of filamentous bacteria, which causes poor sludge settling (bulking) (10, 25, 26). The quantification of 16S rRNA derived from the filamentous bacteria causing bulking will be useful to estimate the growth activity of these organisms for the early detection of bulking. However, current methods for the quantification are either time-consuming or need expensive equipment and, therefore, a rapid and cost-effective method is required.
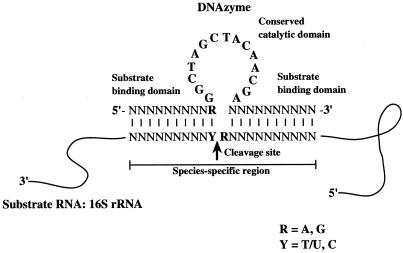
RNA can be cleaved by ribozyme and deoxyribozyme (DNAzyme) in a sequence-specific manner. DNAzyme has several advantageous features compared to ribozyme (11, 23). (i) DNAzyme is cheaper and easier to synthesize and more stable. (ii) DNAzyme is often more active (9). (iii) DNAzyme exhibits higher sequence specificity (15, 27). One model of a DNAzyme, denoted as the 10-23 DNAzyme (Fig. 1), is made of a single-stranded short DNA with a 15-deoxyribonucleotide catalytic domain, 5′-GGCTAGCTACAACGA-3′, flanked by two substrate-binding domains of approximately 10 deoxyribonucleotides each (19, 20). This DNAzyme has the potential to cleave any RNA at purine-pyrimidine junctions (Fig. 1). As in the case of oligonucleotide hybridization, DNAzyme achieves its target specificity by Watson-Crick interactions, which occur via two substrate-binding domains. A sequence-specific DNAzyme of this general structure would be applicable for the quantitative detection of a specific rRNA in a mixture of heterogeneous RNAs.
FIG. 1.
Secondary structure of the DNAzyme-RNA substrate complex. The cleavage site is designated by an arrow between R and Y.
In the present study, we explored the potential of using a DNAzyme to detect quantitatively the 16S rRNA of a specific bacterial species in a microbial community. Our strategy for quantitative detection consists of (i) RNA extraction from the microbial community, (ii) cleavage of the target sequence in the 16S rRNA with the DNAzyme, (iii) separation of the cleaved 16S rRNA from the intact 16S rRNA by gel electrophoresis, and (iv) quantitative detection of the target 16S rRNA by comparing the amount of cleaved 16S rRNA to that of intact 16S rRNA. We designed DNAzymes and optimized the reaction conditions using a synthesized 16S rRNA. Then we adopted one of the DNAzymes to determine the relative abundance of a filamentous bacterium, Sphaerotilus natans, in activated sludges.
Design and construction of DNAzyme.
The sequences of the substrate-binding domain in the DNAzyme were designed by using the same sequence as the species-specific probes for fluorescence in situ hybridization for Escherichia coli (18), Pseudomonas putida (4), and S. natans (25). The complementary sequences for the cleavage site which consisted of purine-pyrimidine (RY) were selected at the center of the sequences, and the pyrimidine nucleotide (Y) was replaced with the 15-nucleotide (nt) catalytic domain. The substrate-binding domain was extended to both sides to possess 12 nt each. The name, sequence, and target gene of each DNAzyme are given in Table 1.
TABLE 1.
Sequence of DNAzyme
| DNAzyme | Sequence (5′-3′)a | 16S rRNA target siteb | Specificity | Reference |
|---|---|---|---|---|
| DzECO-24 | CATCTCTGAAAAggctagctacaacgaTTCCGTGGATGT | 994-1018 | Escherichia coli | 18 |
| DzPPU-24 | GCCAGTTTTGGAggctagctacaacgaGCAGTTCCCAGG | 623-647 | Pseudomonas putida | 4 |
| DzSNA-24 | CGCGGAATTCCAggctagctacaacgaCCCCCTCTACCG | 659-683 | Sphaerotilus natans | 25 |
The conserved 15-base motif of the 10-23 DNAzyme's catalytic domain is denoted in lowercase.
E. coli numbering.
Species-specific cleavage of 16S rRNA by DNAzyme.
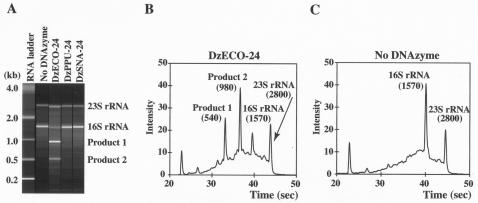
E. coli K-12 strain MG1655, purchased from the American Type Culture Collection (ATCC), was grown in Luria-Bertani (LB) medium at 37°C. Total RNA was extracted from 10 ml of the bacterial culture with FastRNA Pro Blue and FastPrep Instrument (Qbiogene, Inc., Irvine, CA). The extracted RNA (300 ng) was mixed with DNAzyme DzECO-24, DzPPU-24, or DzSNA-24 (15 μmol/liter). Then, 3 μl of Tris-HCl (pH 8.0) at 500 mmol/liter was added. The mixture was made up to 27.5 μl with nuclease-free water and preheated at 90°C for 1 min. Reactions were initiated by the addition of 2.5 μl of MgCl2 at 300 mmol/liter, and the mixture was incubated at 37°C. After 1 h, the reaction was terminated by cooling on ice and by adding an equal volume of 100 mmol of EDTA/liter, and the RNA in the reaction mixture was purified with the RNeasy MinElute Cleanup Kit (QIAGEN, Hilden, Germany). The purified RNA was analyzed by using an Agilent 2100 bioanalyzer (Agilent Technologies, Inc., Palo Alto, CA), which utilizes capillary electrophoresis on a microchip device, the RNA Nano LabChip. As a result, cleaved RNA fragments were detected when DNAzyme DzECO-24 specific for E. coli was used (Fig. 2A). The two cleaved fragments were ca. 540 and 980 nt long, suggesting that the E. coli 16S rRNA was cleaved at the target site by DzECO-24 (Fig. 2B). On the other hand, no cleaved products were observed when the extract was incubated with DNAzymes DzPPU-24 and DzSNA-24 (Fig. 2A), which were specific for P. putida and S. natans, respectively, or when incubated without DNAzymes (Fig. 2C). In all electrophoretograms, relatively high backgrounds were observed in the presence (Fig. 2B) or absence (Fig. 2C) of DNAzymes. The backgrounds might be derived from the random hydrolysis of RNA during preheating and incubation.
FIG. 2.
Analysis of RNA fragments using the Agilent 2100 bioanalyzer. (A) A gel-like electropherogram image shows the results of the incubation of total RNA extracted from E. coli with three kinds of sequence-specific DNAzymes. (B and C) An electrophoretogram exhibits the results with DNAzyme DzECO-24 (B) and without DNAzymes (C). The numbers in parentheses are bases of the RNA fragment.
The same experiments were repeated with the total RNA extracted from P. putida KT2440 and S. natans ATCC 13338T. P. putida KT2440, donated by K. N. Timmis (GBF-National Research Center for Biotechnology, Braunschweig, Germany), was grown in LB medium at 30°C. S. natans ATCC 13338T, purchased from ATCC, was grown in 0.1% nutrient broth (NB; Becton Dickinson, Franklin Lakes, NJ) at 30°C. The total RNA was extracted from 10 ml of the culture of P. putida KT2440 and S. natans ATCC 13338T and incubated with DzECO-24, DzPPU-24, or DzSNA-24. The cleavage was observed only by DzPPU-24 and DzSNA-24, respectively (data not shown). These results showed the species-specific cleavage by the three DNAzymes.
Effect of various factors on the DNAzyme reaction.
The synthesized 16S rRNA used in DNAzyme experiments as RNA substrates were prepared according to the method of Juzumiene and Wollenzein (7) as follows. Almost-complete DNAs coding for 16S rRNA were amplified from the purified genomic DNA of E. coli MG1655, P. putida KT2440, and S. natans ATCC 13338T by PCR with the primer 8f-T7 (5′-TAATACGACTCACTATAGGGAGAGTTTGATYMTGGCTCAG-3′; Y = C or T, M = A or C) (28) containing the T7 promoter at the 5′ end (shown by an underline), and the primer 1492r (5′-GGYTACCTTGTTACGACTT-3′; Y = C or T) (28), and then the amplified products were transcribed in vitro to RNA.
The influence of the length of the substrate-binding domain of each DNAzyme was examined by using DNAzymes with arms of various lengths (16 nt [8+8, 5′+3′] to 24 nt [12+12, 5′+3′]). The synthesized 16S rRNA (30 nmol/liter) and a DNAzyme (15 μmol/liter) were mixed, and the cleavage reaction was carried out at 37°C for 1 h. The proportion of 16S rRNA cleaved by the DNAzyme was determined from the relative ratio of the areas of peaks in the electropherogram as follows: R = [P1+P2]/[P1+P2+P16S] (13, 24), where R was the cleavage ratio, P1 and P2 were the peak areas of the cleaved products, and P16S was the peak area of intact 16S rRNA (Fig. 2B). The signal intensities of individual peaks in the electropherograms were determined with the Bio Sizing software associated with the Agilent 2100 Bioanalyzer, which collected the peak area above the background for the calculation. As a result, the cleavage ratios with DNAzymes DzECO with 16-, 18-, 20-, 22-, and 24-nt-long arms were 17, 66, 73, 79, and 81%, respectively. Those with DNAzymes DzSNA with 16-, 18-, 20-, 22-, and 24-nt-long arms were 20, 24, 32, 75, and 82%, respectively. In DNAzyme DzPPU, those with 16- to 22-nt-long arms exhibited no activity, but that with 24-nt-long arm showed a cleavage ratio of 71%. Thus, the DNAzymes possessing a longer arm showed a higher ratio of cleavage. In general, a longer binding domain has less sequence specificity. Santro and Joyce (20) reported that a DNAzyme with a 7+7-nt-long arm exhibited 20- to 50-fold more sensitivity to single base mismatches than that with a 8+8-nt-long arm. The shorter arm length is preferable to attain higher sequence specificity. Therefore, we selected the DNAzymes with the shortest arm which still achieve a cleavage ratio of more than 50%. In subsequent experiments, we used DNAzymes DzECO-18, DzPPU-24, and DzSNA-22.
One factor affecting the efficiency of the cleavage reaction is the ratio of DNAzyme to RNA substrate. A fixed amount (30 nmol/liter) of synthesized E. coli 16S rRNA was incubated at 37°C for 1 h with DzECO-18 at a DNAzyme/substrate molar ratio of 1:1 to 600:1. As a result, the cleavage ratio increased on increasing the ratio of DNAzyme to substrate and plateaued at a value of 400. DNAzymes DzPPU-24 and DzSNA-22 with their substrate gave similar results. Consequently, a DNAzyme/substrate molar ratio of 500:1 (15 μmol/liter of DNAzyme and 30 nmol/liter of synthesized 16S rRNA) was used in the subsequent experiments.
To determine the optimal time period for the cleavage reaction, each DNAzyme was incubated for 80 min with their corresponding synthesized 16S rRNA. The cleavage ratio increased with incubation time for all DNAzymes. DzPPU-24 exhibited an abrupt increase in cleavage ratio in the initial 5 min and then showed a slight increase until 10 min. DNAzymes DzECO-18 and DzSNA-22 showed a significant increase until 60 min. These disparities of cleavage rate may be derived from the differential accessibility of each DNAzyme toward target site on 16S rRNA because of the inherent higher structure of each 16S rRNA molecule. A shorter incubation is preferable for saving time and, moreover, a long incubation may bring about the degradation of RNA due to its unstable structure. The appropriate incubation periods for DzECO-18, DzPPU-24, and DzSNA-22 were determined to be 60, 10, and 60 min, respectively, and these times were used in the subsequent experiments.
To determine the effect of one-base mismatch and temperature, we prepared various DNAzyme variants that possessed one base mismatch in the binding domains and carried out the experiments at 37, 45, and 50°C. We found that the perfectly matched DNAzyme had a higher ratio of cleavage than any of the mismatched DNAzymes (Table 2) at all temperatures. Incubation at 50°C produced the highest level of sequence specificity. In the experiments on P. putida, only the perfectly matched DzPPU-24 cleaved the target 16S rRNA at 50°C. In the experiments on E. coli and S. natans, two variants which had one base mismatch at the 3′ end or 5′ end [“(a)” and “(e)” of DzECO-18 and DzSNA-22] showed little cleavage activity even at 50°C. The ratio of cleavage by these DzSNA-22 variants was 20%, while that by the perfectly matching DNAzyme was 85%. The mismatch located proximal to the cleavage site [variant “(c)” in Table 2] resulted in the ablation of cleavage activity even at 37°C, presumably due to greater distortion of the active site in the catalytic domain in DNAzyme caused by the one base mismatch. Incubation at 55 and 60°C was also examined and resulted in a decrease in the ratio of cleavage and enhancement of the random hydrolysis of RNA (date not shown). Therefore, the appropriate reaction temperature for sequence-specific cleavage was determined to be 50°C. The cleavage ratios of DNAzymes DzECO-18, DzPPU-24, and DzSNA-22 at 50°C were 61, 53, and 85%, respectively. In the subsequent experiments, the abundance of target 16S rRNA in the total 16S rRNA mixture was calculated by using these cleavage ratios as follows: A = R/CDz, where A was the abundance of target rRNA, and CDz was the cleavage ratio of each DNAzyme. In the present study, abundance is shown as the average value ± the standard deviation for three independent experiments.
TABLE 2.
Effect of one base mismatch on the DNAzyme cleavage reaction
| Name | Sequence (5′-3′)a | Mean ratio of cleaved target 16S rRNA (%) ± SD at:
|
||
|---|---|---|---|---|
| 37°C | 45°C | 50°C | ||
| DzECO-18 | CTCTGAAAA<>TTCCGTGGA | 66.2 ± 2.1 | 75.4 ± 2.8 | 61.3 ± 2.9 |
| DzECO-18-(a) | G--------<>--------- | 18.4 ± 1.0 | 12.0 ± 0.4 | 8.0 ± 0.1 |
| DzECO-18-(b) | ----C----<>--------- | NAb | NA | NA |
| DzECO-18-(c) | --------T<>--------- | NA | NA | NA |
| DzECO-18-(d) | ---------<>----C---- | 8.7 ± 0.3 | NA | NA |
| DzECO-18-(e) | ---------<>--------T | 36.1 ± 2.2 | 19.4 ± 0.4 | 10.1 ± 0.2 |
| DzPPU-24 | GCCAGTTTTGGA<>GCAGTTCCCAGG | 82.3 ± 3.1 | 61.9 ± 3.0 | 53.0 ± 1.6 |
| DzPPU-24-(a) | C-----------<>------------ | NA | NA | NA |
| DzPPU-24-(b) | -----A------<>------------ | NA | NA | NA |
| DzPPU-24-(c) | -----------T<>------------ | NA | NA | NA |
| DzPPU-24-(d) | ------------<>------G----- | 11.5 ± 1.1 | NA | NA |
| DzPPU-24-(e) | ------------<>-----------C | 25.8 ± 1.3 | 5.0 ± 0.1 | NA |
| DzSNA-22 | GCGGAATTCCA<>CCCCCTCTACC | 82.7 ± 5.0 | 92.2 ± 4.6 | 85.0 ± 3.9 |
| DzSNA-22-(a) | C----------<>----------- | 49.8 ± 2.1 | 40.0 ± 0.4 | 20.2 ± 0.7 |
| DzSNA-22-(b) | ---C-------<>----------- | 7.5 ± 0.2 | 5.1 ± 0.2 | NA |
| DzSNA-22-(c) | ----------T<>----------- | NA | NA | NA |
| DzSNA-22-(d) | -----------<>------G---- | 6.1 ± 0.2 | NA | NA |
| DzSNA-22-(e) | -----------<>----------G | 34.3 ± 0.3 | 29.6 ± 0.2 | 20.1 ± 0.6 |
<>, the sequence of 15-nt conserved catalytic domain; -, Identical to the matched DNAzyme sequence.
NA, no activity.
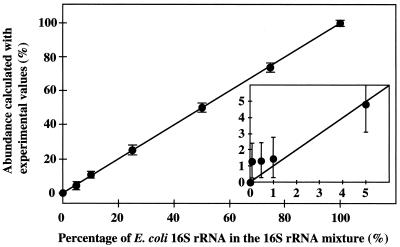
To evaluate the accuracy of the quantification of abundance of target 16S rRNA, the mixtures of synthesized 16S rRNA of E. coli and P. putida (totally 1 μg; 0, 0.1, 0.5, 1, 5, 10, 25, 50, 75, and 100% of E. coli 16S rRNA) were incubated with DzECO-18 at 50°C for 60 min. When the content of target 16S rRNA was 5% or more, the calculated values were in good agreement with the theoretical values (Fig. 3). Although the detection of target 16S rRNA was feasible at 1%, the accuracy was low. Thus, the lower limit for quantification was 5% of the total 16S rRNA. These results demonstrated that the DNAzyme would be useful for the quantification of a specific 16S rRNA in heterogeneous rRNA samples.
FIG. 3.
Accuracy of the quantification of abundance of target 16S rRNA. The synthesized E. coli 16S rRNA used as a target was mixed with the synthesized P. putida 16S rRNA and incubated with DzECO-18.
Determination of the relative abundance of the 16S rRNA of filamentous bacteria in activated sludge.
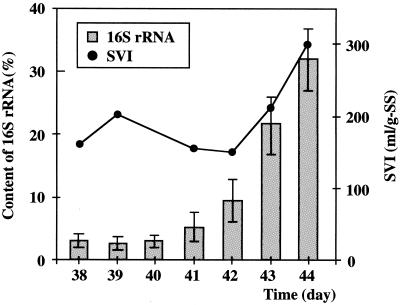
The DNAzyme method was adopted to determine the relative abundance of the 16S rRNA of filamentous bacteria in wastewater treatment systems using activated sludge. The laboratory-scale activated sludge reactor used in the present study was constructed and operated by Liu et al. as described previously (12). It consisted of two tanks; an aeration tank (2.36 liters) connected to a settling tank (0.84 liters) with a bottom flow passage. Activated sludge obtained from a sewage treatment plant was inoculated into the reactor. Air was supplied into the aeration tank at a flow rate of 1.2 to 1.5 liters/min. Synthetic wastewater containing glucose (0.375 g/liter), ammonium sulfate (0.035 g/liter), and potassium dihydrogen phosphate (0.007 g/liter) was continuously added via a peristaltic pump at a flow rate of 0.25 liters/h. The retention time of the activated sludge was adjusted to 20 days by discharging sludge. The sludge volume index (SVI), which is the parameter used to characterize the settlement of the sludge, was determined through a 30-min zone-settling test in a 100-ml cylinder. The reactor was continuously operated at a constant temperature of 25°C. We did not expect a sludge bulking in these conditions, but a bulking occurred with the excess growth of filamentous bacteria. The SVI increased to the bulking level (>200) on the 43rd day, and the sludge began to flow out on the 45th day. Sphaerotilus-like filamentous bacteria occupied ca. 60% of total filaments that extended out of the flocs.
We extracted total RNA from 10 ml of activated sludge with the Fast RNA Pro Blue Kit and FastPrep Instrument (Qbiogene, Inc.) and dissolved final RNA pellets in 50 μl of nuclease-free water. Then we mixed 10 μl of RNA sample with 15 μmol/liter of DNAzyme DzSNA-22 and incubated the mixture at 50°C for 60 min. As a result, two products of expected size were obtained by electrophoresis, whereas no product was observed without the DNAzyme. The amount of target 16S rRNA was found to begin increasing at the 41st day and reached 32.1% at the 44th day (Fig. 4). DzSNA-22 was designed to specifically detect S. natans, but the sequence comparison made with Ribosomal Database Project II showed that it would detect some nonfilamentous bacteria, Aquabacterium citratiphilum, A. commune, Comamonas acidovorans, and Rhodoferax fermentas, as well as filamentous bacteria, Leptothrix discophora, and L. mobilis. In addition, DzSNA-22 will likely cleave a one base mismatched target though the cleavage efficiency is low (Table 2). Sequence comparison showed that Acidvorax avenae, A. konjaci, Aquaspirillun metamorphum, C. testosteroni, C. terrigena, Delfita acidovorans, Hydrogenophaga flava, H. palleronii, and Rubrivivax gelatinosus, and Variovorax paradoxus possessed one base mismatch in 16S rRNA at the third position from the 5′ end of the DNAzyme-binding site. There were no other one base mismatched rRNA sequences in the database except for a few 16S rRNA gene clone samples. Leptothrix spp. have a morphology very similar to that of S. natans and may cause bulking, so measuring the amount of 16S rRNA in both species is useful for estimating the bulking tendency of activated sludge. The other species possessing the sequences that matched perfectly or had one base mismatch are considered to have no relationship with sludge bulking. There are no reports of their detection in activated sludge in significant amounts, and so their levels are considered low to have any significant influence on the estimation of bulking tendency.
FIG. 4.
Changes in the SVI and the percentage of the target 16S rRNA in the activated sludge. The target 16S rRNA content was measured with DzSNA-22.
Our results showed an increase in the target 16S rRNA from the 41st day. On the other hand, the increase in SVI was observed on the 43rd day, and the occurrence of bulking was obvious at the 44th day. Wastewater treatment was impossible on the 45th day due to the flowing out of activated sludge. Our results demonstrate that the measurement of 16S rRNA using DNAzyme in the bacteria causing bulking can provide an early warning in activated sludge systems. The early warning is important to prevent bulking.
In the present study, the experimental results indicated that the DNAzyme method would be applicable to actual microbial complexes. The quantification of the rRNA of a specific bacterial species is useful to predict its activity because the bacterial growth rate and cellular rRNA concentration are positively correlated (3, 16, 17). Furthermore, the high level of rRNA in cells makes possible direct quantification without amplification by PCR. One can therefore avoid the various problems caused by PCR amplification such as PCR bias and artifacts (8), which provide incorrect information in PCR-dependent DNA analyses.
In the DNAzyme method, a standard curve or internal standard was not required, and the content of the target rRNA was calculated from the amount of cleaved RNA and intact RNA in a single experiment (Fig. 2). These in turn saved the time and money. The DNAzyme method required less than 4 h and comprised the extraction of RNA using a standard commercial kit (2 h), incubation for the DNAzyme reaction (1 h), and separation of the reaction products by electrophoresis and their detection (1 h). Recently, Ueno et al. reported a method to quantitative detection a specific rRNA sequence that uses the sequence-specific cleavage of rRNA with an oligonucleotide and an enzyme, RNase H (24). This approach also provides a rapid and simple means for the quantitative detection of specific rRNA sequences. It requires an oligonucleotide and RNase H, whereas the DNAzyme method requires an oligonucleotide only. Therefore, the DNAzyme method is more cost-effective. Moreover, a DNAzyme is more stable than RNase H, and the solution of DNAzyme can be preserved at room temperature, while the solution of RNase H should be preserved at −20°C. Thus, a DNAzyme is easier to handle than RNase H.
In summary, the DNAzyme method for the quantitative detection of specific RNA is rapid, cost-effective, and easy to perform. This method will be useful for measuring the activity of a specified bacterial species in the actively growing community used for an environmental biotechnology such as wastewater treatment. It may also have applications in clinical microbiology to detect the activity of pathogenic bacteria.
REFERENCES
- 1.Abraham, W.-R., B. Nogales, P. N. Golyshin, D. H. Piper, and K. N. Timmis. 2002. Polychlorinated biphenyl-degrading microbial communities in soils and sediments. Curr. Opin. Microbiol. 5:246-253. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R. I., H. Lemmer, and M. Wagner. 1998. Monitoring the community structure of wastewater treatment plants: a comparison of old and new techniques. FEMS Microbiol. Ecol. 25:205-215. [Google Scholar]
- 3.DeLong, E. F., G. S. Wickham, and N. R. Pace. 1989. Phylogenetic strains: ribosomal RNA-based probes for the identification of single microbial cells. Science 243:1360-1363. [DOI] [PubMed] [Google Scholar]
- 4.DuTeau, N. M., J. D. Rogers, C. T. Bartholomay, and K. N. Reardon. 1998. Species-specific oligonucleotides for enumeration of Pseudomonas putida F1, Burkholderia sp. strain JS150, and Bacillus subtilis ATCC 7003 in biodegradation experiments. Appl. Environ. Microbiol. 64:4994-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Furukawa, K., H. Suenaga, and M. Goto. 2004. Biphenyl dioxygenases: functional versatilities and directed evolution. J. Bacteriol. 186:5189-5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hellyer, T. J., L. E. DesJardin, G. L. Hehman, M. D. Cave, and K. D. Eisenach. 1999. Quantitative analysis of mRNA as a marker for viability of Mycobacterium tuberculosis. J. Clin. Microbiol. 37:290-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Juzumiene, D. I., and P. Wollenzein. 2001. Arrangement of the central pseudoknot region of 16S rRNA in the 30S ribosomal subunit determined by site-directed 4-thiouridine crosslinking. RNA 7:71-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanagawa, T. 2003. Bias and artifacts in multitemplate polymerase chain reaction (PCR). J. Biosci. Bioeng. 96:317-323. [DOI] [PubMed] [Google Scholar]
- 9.Kurreck, J., B. Bieber, R. Jahnel, and V. A. Erdmann. 2002. Comparative study of DNAzymes and ribozymes against the same full-length messenger RNA of the vanilloid receptor subtype I. J. Biol. Chem. 277:7099-7107. [DOI] [PubMed] [Google Scholar]
- 10.Layton, A. C., P. N. Karanth, C. A. Lajoie, A. J. Meyers, I. R. Gregory, R. D. Stapleton, D. E. Taylor, and G. S. Sayler. 2000. Quantification of Hyphomicrobium populations in activated sludge from an industrial wastewater treatment system as determined by 16S rRNA analysis. Appl. Environ. Microbiol. 63:1167-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li, Y., and R. R. Breaker. 1999. Deoxyribozymes: new players in the ancient game of biocatalysis. Curr. Opin. Struct. Biol. 9:315-323. [DOI] [PubMed] [Google Scholar]
- 12.Liu, R., S. Aruga, M. Kawaharasaki, S. Okunuki, and T. Kanagawa. 2002. Induced filamentous bulking under laboratory conditions, p 315-322. IWA Conference on Environmental Biotechnology: Biotechnology Applications for Treatment and Utilization of Industrial Wastes. International Water Association, London, United Kingdom.
- 13.Lu, C.-Y., D.-J. Tso, T. Yang, Y.-J. Jong, and Y.-H. Wei. 2002. Detection of DNA mutations associated with mitochondrial disease by Agilent 2100 bioanalyzer. Clin. Chem. Acta 318:97-105. [DOI] [PubMed] [Google Scholar]
- 14.McKillip, J. L., L. A. Jaykus, and M. Drake. 1998. rRNA stability in heat-killed and UV-irradiated enterotoxigenic Staphylococcus aureus and Escherichia coli O157:H7. Appl. Environ. Microbiol. 64:4264-4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nelson, J. W., and I. Tinoco. 1982. Comparison of the kinetics of ribo-, deoxyribo-, and hybrid oligonucleotide double-strand formation by temperature-jump kinetics. Biochemistry 21:5289-5295. [DOI] [PubMed] [Google Scholar]
- 16.Nogales, B., E. R. B. Moore, E. Llobet-Brossa, R. Rossello-More, R., Amann, and K. N. Timmis. 2001. Combined use of 16S ribosomal DNA and 16S rRNA to study the bacterial community of polychlorinated biphenyl-polluted soil. Appl. Environ. Microbiol. 67:1874-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poulsen, L., G. Ballard, and D. A. Stahl. 1993. Use of rRNA fluorescence in situ hybridization for measuring the activity of single cells in young and established biofilms. Appl. Environ. Microbiol. 29:1354-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Regnault, B., S. M.-Delautre, M. L.-Collin, M. Lefévre, and P. A. D. Grimont. 2000. Oligonucleotide probe for the visualization of Escherichia coli/Escherichia fergusonii cells by in situ hybridization: specificity and potential applications. Res. Microbiol. 151:521-533. [DOI] [PubMed] [Google Scholar]
- 19.Santro, S. W., and G. F. Joyce. 1997. A general purpose RNA-cleaving DNA enzyme. Proc. Natl. Acad. Sci. USA 94:4262-4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Santro, S. W., and G. F. Joyce. 1998. Mechanism and utility of an RNA-cleaving DNA enzyme. Biochemistry 37:13330-13342. [DOI] [PubMed] [Google Scholar]
- 21.Sayler, G. S., M. S. Shields, E. T. Tedford, A. Breen, and S. W. Hooper. 1985. Application of DNA-DNA colony hybridization to the detection of catabolic genotypes in environmental samples. Appl. Environ. Microbiol. 49:1295-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stahl, D. A., B. Flesher, H. R. Mansfield, and L. Montgomery. 1988. Use of phylogenetically based hybridization probes for studies of nominal microbial ecology. Appl. Environ. Microbiol. 54:1079-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun, L. Q., M. J. Cairns, E. G. Saravolac, A. Baker, and W. L. Gerlach. 2000. Catalytic nucleic acids: from lab to applications. Pharmacol. Rev. 52:325-347. [PubMed] [Google Scholar]
- 24.Ueno, Y., Y. Sekiguchi, A. Sunaga, H. Yoshida, and Y. Kamagata. 2004. Sequence-specific cleavage of small-subunit (SSU) rRNA with oligonucleotides and ribonuclease H: a rapid and simple approach to the SSU rRNA-based quantitative detection of microorganisms. Appl. Environ. Microbiol. 70:3650-3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wagner, M., R. Amann, P. Kampfer, B. Assmus, A. Hartmann, P. Hutzler, N. Springer, and K. H. Schleifer. 1994. Identification and in situ detection of gram-negative filamentous bacteria in activated sludge. Syst. Appl. Microbiol. 17:405-417. [Google Scholar]
- 26.Wagner, M., and A. Loy. 2002. Bacterial community composition and function in sewage treatment systems. Curr. Opin. Biotechnol. 13:218-227. [DOI] [PubMed] [Google Scholar]
- 27.Warashina, M., T. Kuwabara, Y. Nakamatsu, and K. Taira. 1999. Extremely high and specific activity of DNA enzymes in cells with a Philadelphia chromosome. Chem. Biol. 6:237-250. [DOI] [PubMed] [Google Scholar]
- 28.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson, M. S., C. Bakerman, and E. L. Madsen. 1999. In situ, real-time catabolic gene expression: extraction and characterization of naphthalene dioxygenase mRNA transcripts from groundwater. Appl. Environ. Microbiol. 65:80-87. [DOI] [PMC free article] [PubMed] [Google Scholar]