Abstract
A genome-wide search was performed to identify simple sequence repeat (SSR) loci among the available sequence databases from four strains of Xylella fastidiosa (strains causing Pierce's disease, citrus variegated chlorosis, almond leaf scorch, and oleander leaf scorch). Thirty-four SSR loci were selected for SSR primer design and were validated in PCR experiments. These multilocus SSR primers, distributed across the X. fastidiosa genome, clearly differentiated and clustered X. fastidiosa strains collected from grape, almond, citrus, and oleander. They are well suited for differentiating strains and studying X. fastidiosa epidemiology and population genetics.
Strains of Xylella fastidiosa cause economically important diseases that result in significant losses in several agricultural, horticultural, and landscape crops, including Pierce's disease (PD) of grapevines, almond leaf scorch (ALS) disease, citrus variegated chlorosis (CVC) disease, and oleander leaf scorch (OLS) disease (7, 11). This xylem-limited bacterium is transmitted by xylem-feeding insect vectors and colonizes the xylem, resulting in blockages that lead to desiccation of leaves, shoots, and fruits and, in some cases, death of the host plants (8, 10). The threat that X. fastidiosa poses to California agriculture was significantly increased by the recent introduction, establishment, and spread of Homalodisca coagulata, the glassy-winged sharpshooter (2). Currently, information regarding the population structure and genetic diversity, as well as the genetic, evolutionary, and epidemiological relationships among X. fastidiosa strains in agricultural populations, is unclear. Advances in the understanding of X. fastidiosa population structure and genetic diversity will greatly aid the development of effective pest-disease management strategies.
With the availability of the complete whole genome sequences of the CVC “9a5c” (2.67 Mbp) (12) and PD “Temecula” (2.52 Mbp) strains (14) and draft sequences of ALS “Dixon” (2.43 Mbp) and OLS “Ann-1” (2.67 Mbp) strains from GenBank (http://www.ncbi.nlm.nih.gov), identification of simple sequence repeat (SSR) loci is greatly facilitated. SSR markers, also known as microsatellites, are tandem repetitive DNA sequences with repeat motif lengths of 2 to 6 bp or more (13). In this study, we present multilocus SSR markers that were identified and designed from analyses of X. fastidiosa genome sequence databases.
A genome-wide search was performed to identify SSR loci with the Tandem Repeat Finder software, version 2.0 (1). The following criteria were used to identify and select SSR loci: (i) each locus has one copy per genome, and (ii) each SSR locus contains at least five or more repeat unit lengths. In silico pair-wise DNA sequence comparisons among selected loci were performed using sequence alignment to remove duplicate loci. Nonredundant SSR loci were then selected for primer design. For each SSR locus, BLAST analysis (http://www.ncbi.nlm.nih.gov/BLAST) was performed across the genomes of all four X. fastidiosa strains to select conserved regions that were 100 to 200 bp up- and downstream from the priming site locus, so that each designed primer would work for all X. fastidiosa strains. The Primer Premier 5 software (PremierBiosoft, Palo Alto, CA) was used for primer design with amplicon sizes ranging from 150 to 500 bp. Sequence specificity of each pair of primers was checked in silico by BLAST analysis against all available microbial sequence databases in GenBank to verify that the sequences are unique to X. fastidiosa. No significant match was found in any pair of primers (data not shown). This step is important to eliminate potential false-positive diagnoses. Thirty-four SSR primers were designed in this study (Table 1).
TABLE 1.
Characteristics of SSR primer sequences produced and used to study X. fastidiosa isolatesa
| Primer | Forward sequence | Reverse sequence | Type of repeat motif | No. of alleles | Amplicon size (bp) | ORF definition | Locus location in genome |
|---|---|---|---|---|---|---|---|
| OSSR-2 | TTGCTTCACCATTAGCCTTATC | GGCCGTACAGGACCGATC | (ATG)9 | 4 | 264 | Conserved hypothetical protein | 9403-9430 |
| OSSR-9 | TAGGAATCGTGTTCAAACTG | TTACTATCGGCAGCAGAC | (TTTCCGT)13 | 16 | 362 | Exopolyphosphatase | 24428-24519 |
| OSSR-12 | ACAGTCTGTGTCCGCAATTTG | CAGGCGCAGATAGCATTGATC | (AGAGGGTAT)9 | 7 | 360 | Conserved hypothetical protein | 71017-71098 |
| OSSR-14 | GGCGTAACGGAGGAAACG | ATGAACACCCGTACCTGG | (TGATCCATCCCTGTG)11 | 16 | 370 | copB | 35279-35444 |
| OSSR-16 | GCAAATAGCATGTACGAC | GTGTTGTGTATGTGTTGG | (CTGCTA)12 | 13 | 275 | NADH-ubiquinone oxidoreductase NQO12 subunit | 10784-11856 |
| OSSR-17 | AGTACAGCGAACAGGCATTG | AGCAACCAGGACGGGAAC | (TGCCTG)10 | 23 | 257 | Cell cycle protein | 25656-25716 |
| OSSR-19 | GCTGTGAACTTCCATCAATCC | GCAAGTAGGGGTAAATGTGAC | (CAGGATCA)10 | 11 | 274 | Adenylosuccinate lyase | 33208-33288 |
| OSSR-20 | ATCTGTGCGGCGGTTCTG | CACTTGCGGCGTAGATACTTC | (AGGATGCTA)20 | 14 | 154 | Competence-related protein | 1512703-1512883 |
| CSSR-4 | AACCCAATTCTTTTAATATGTG | TTGCAGCATTAGATATTTGAG | (TGCC)7 | 4 | 231 | Peptidyl-prolyl cis-trans isomerase | 1143270-1143298 |
| CSSR-6 | CGCACTGTCATCCATTTAATC | GCTGCTTCATCTAGACGTG | (GCTGTA)7 | 11 | 279 | Hypothetical protein | 98393-98435 |
| CSSR-7 | CACAGCGAACAGGCATTG | AGCAACCAAGACGGGAAC | (CTGTGC)14 | 14 | 279 | Alkaline phosphatase | 628252-628336 |
| CSSR-10 | GCAACCACAAAGCCGCAG | AGCACCTCTTAGCATCACTGG | (CAATGA)10 | 13 | 202 | tRNA/rRNA methylase | 1882243-1882303 |
| CSSR-12 | TAAGTCCATCACCGAGAAG | AAACGGATTTAGGAACACTC | (GAAGGCGTA)27 | 8 | 491 | Conserved hypothetical protein | 1220096-1220339 |
| CSSR-13 | CAATGTCACTCAGGTCAG | TTCTGGAATACATCAAATGC | (TGTTGGGG)10 | 14 | 305 | Hypothetical protein | 2639987-2640067 |
| CSSR-16 | CGATCAACCCATTCACTG | GCTCCTATTTGCATGATATTG | (GTGGTGGCA)6 | 5 | 199 | PilY1 | 30060-30114 |
| CSSR-17 | AGAAGTATTCGCTACGCTACG | GGTGATGATTCAGTTGGTGTTG | (CTGATGTG)9 | 13 | 199 | Hydroxyacylglutathione hydrolase | 2050614-2050686 |
| CSSR-18 | GTGCTTCCAGAAGTTGTG | GACTGTTCTCTTCGTTCAG | (GCCAA)12 | 9 | 304 | Hypothetical protein | 408996-409056 |
| CSSR-19 | TGCTGTGATTGGAGTTTTGC | TCAAACGAATCTGTCCATCAAG | (TGGTGAG)7 | 3 | 354 | Site-specific DNA-methyltransferase | 1717597-1717646 |
| CSSR-20 | GGTATCGCCTTTGGTTCTGG | GACAACCGACATCCTCATGG | (GTAGCA)8 | 15 | 247 | Phosphoglycolate phosphatase | 1155075-1155523 |
| ASSR-9 | GGTTGTCGGGCTCATTCC | TTGTCACAGCATCACTATTCTC | (CAAGTAC)11 | 16 | 259 | 50S ribosomal protein L13 | 35632-35709 |
| ASSR-11 | AGAGGCAACGCAGGAACAG | GTGAGTTATATCGGTGCAGCAG | (ACGCATC)10 | 9 | 260 | 50S ribosomal protein L20 | 10087-10157 |
| ASSR-12 | TGCTCATTGTGGCGAAGG | CGCAACGTGCATTCATCG | (GATTCAG)14 | 6 | 300 | Fructose-bisphosphate aldolase | 76999-77097 |
| ASSR-14 | TTGACTCAAGGAATAAAAC | GAAAAGAGTGTCAATACG | (CTGCGTGC)11 | 13 | 410 | Hypothetical protein | 61336-61424 |
| ASSR-16 | TTAATCAACAACGCTTATCC | TCGCAGTAGCCAGTATAC | (GCTCCGGTTCTA)26 | 12 | 497 | 1,4-β-Cellobiosidase | 803-1115 |
| ASSR-19 | CGCCGACTGTCTATGTGAC | TTCCTAGCAATGGCAATGTTG | (ACAACG)10 | 4 | 332 | Conserved hypothetical protion | 10957-11017 |
| ASSR-20 | TTACTATCGGCAGCAGACG | TGAAGCAATGGTGGATTTAGG | (ACAGAAA)10 | 16 | 302 | Exopolyphosphatase | 17324-17394 |
| GSSR-4 | GCGTTACTGGCGACAAAC | GCTCGTTCCTGACCTGTG | (ATCC)7 | 15 | 289 | Conserved hypothetical protein | 1729643-1729671 |
| GSSR-6 | TGTTCTCTTCGTTCAGCCAAGC | CGCAGCAGAGCAGCAGTG | (CTTGT)12 | 8 | 265 | None coding | 1941750-1941810 |
| GSSR-7 | ATCATGTCGTGTCGTTTC | CAATAAAGCACCGAATTAGC | (GGCAAC)24 | 17 | 385 | None coding | 19306-19850 |
| GSSR-12 | TTACGCTGATTGGCTGCATTG | GTCAAACACTGCCTATAGAGCG | (TATCTGT)20 | 10 | 471 | None coding | 52417-52557 |
| GSSR-14 | TTGATGTGCTTTTGCGGTAAG | GACAGGTCCTCTCATTGCG | (TCCCGTA)24 | 14 | 402 | None coding | 404211-404379 |
| GSSR-15 | CCGCAGAGTCCGTTGTAAC | AGCCGACGCACGGTATATC | (AGCCTGC)17 | 10 | 404 | Conserved hypothetical protein | 2229645-2229764 |
| GSSR-19 | GCCGATGCAGAACAAGAAC | TCAACTTCGCCACACCTG | (GAAAACAAG)19 | 13 | 388 | Phosphomannomutase | 153189-153360 |
| GSSR-20 | TGGATGGATAGATGATTCAGCC | CGATCAGTGGAGGATGTCTTG | (GAACCACTA)7 | 4 | 354 | None coding | 2203868-2203931 |
The O, C, A, and G prefixes in the primer names refer to the X. fastidiosa genome (oleander, citrus, almond, and grape strains, respectively) from which they were designed. The open reading frame (ORF) definition refers to the gene the SSR locus is in or close to.
To evaluate the polymorphisms detected by the designed SSR primers, 43 X. fastidiosa strains isolated from four crops were used (22 from grape, 10 from citrus, 6 from almond, and 5 from oleander) (Table 2). PD strains of X. fastidiosa were isolated from infected grape stems and cultured on periwinkle wilt medium plates at 28°C for 7 to 10 days until colonies developed (4). Isolated colonies were confirmed as X. fastidiosa by enzyme-linked immunosorbent assay (11) and with the X. fastidiosa-specific PCR primers RST 33 and RST 31 (9). Bacterial DNA was extracted using the hexadecyltrimethylammonium bromide method (15). DNA samples of X. fastidiosa CVC strains were kindly provided by E. G. D. Lemos (Universidade Estadual Paulista, Jaboticabal, SP, Brazil). Almond and oleander X. fastidiosa strains were kindly provided by Alexander H. Purcell (University of California, Berkeley). For SSR PCR assays, PCR mixtures consisted of 20-μl volumes containing 1.0 mM MgCl2, 0.2 mM each deoxynucleoside triphosphate, and 0.5 U AmpliTaq Gold polymerase in 2 μl of 10× reaction buffer (Applied Biosystems, Foster City, CA), 10 pmol SSR primer with either 2 μl of genomic DNA (10 ng/μl) or 2 μl of bacterial cell suspension (2 × 105 CFU/ml). The PCR tests were conducted in a model ABI 9700 thermal cycler with the following temperature profile: the initial denaturation step was 95°C for 6 min, followed by 30 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 1 min, and a final extension at 72°C for 7 min. SSR products were mixed with sample loading dye (10 mM NaOH, 95% formamide, 0.05% bromophenol blue, and 0.05% xylene cyanol) at a 1:2 ratio. A 2-μl aliquot of this mixture was resolved in a 5% polyacrylamide gel. The gel was run in TBE buffer (89 mM Tris, 89 mM boric acid, and 2.0 mM EDTA; pH 8.3) at a constant 100 W for 2.5 to 4.5 h depending on amplicon size (150 bp to 500 bp). The gels were visualized by silver staining (Promega Biosciences Inc., San Luis Obispo, CA).
TABLE 2.
Identification, host plant, and collection location of the 43 X. fastidiosa isolates tested
| Strain name | Location | Host of origin |
|---|---|---|
| PD-1, PD-2, PD-3, PD-4, PD-8, PD-9, PD-10, PD-11, PD-13, PD-14 | Kern, Calif. | Grape |
| PD-5, PD-6, PD-7, PD-22a | Riverside (Temecula), Calif. | Grape |
| PD-15, PD-16, PD-17, PD-18, PD-19, PD-20, PD-21 | Napa, Calif. | Grape |
| PD-12 | Baja, Calif. | Grape |
| CVC-1, CVC-2, CVC-3, CVC-4, CVC-5, CVC-6, CVC-7, CVC-8, CVC-9, CVC-10a | São Paulo, Brazil | Citrus |
| ALS-1 | Tulare, Calif. | Almond |
| ALS-2 | Contra Costa, Calif. | Almond |
| ALS-3, ALS-4, ALS-5 | San Joaquin, Calif. | Almond |
| ALS-6a | Solano, Calif. | Almond |
| OLS-1, OLS-2, OLS-3, OLS-4,a OLS-5 | Riverside, Calif. | Oleander |
Labels in bold are the strains whose genomes have been sequenced.
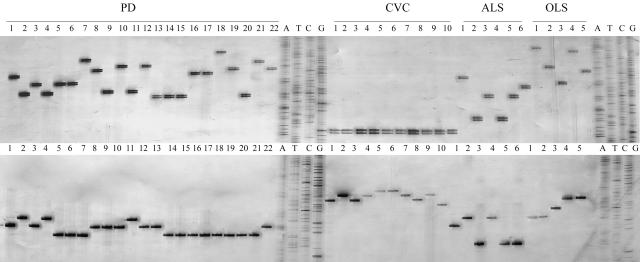
The 34 SSR primers presented here are capable of differentiating X. fastidiosa strains regardless of host origin. Figure 1 presents examples of the CSSR-6 and OSSR-9 primers, which detected 43 X. fastidiosa isolates. The average level of polymorphism among 34 SSR primers used against the 43 isolates was 11.3 alleles per locus, which is strong evidence of the ability of these markers to distinguish genetically similar isolates. The 34 SSR primers were divided into three groups based on the number of polymorphisms they resolved: (i) high, detected 15 or more alleles, (ii) intermediate, detected 5 to 14 alleles, and (iii) low, detected less than 5 alleles (Table 1). Fidelity of these SSR alleles was verified by sequence validation (data not presented).
FIG. 1.
Examples of SSR markers produced with primers OSSR-9 (top) and CSSR-6 (bottom) among 43 X. fastidiosa isolates separated by 5% polyacrylamide gel. The A, T, C, and G lanes are molecular size markers.
The genome-wide search across the sequences of all four crop-associated strains found the most abundant motif repeats ranged between 6 and 9 bp. Coletta-Filho et al. (3) reported that there are no mono- or direpeats in the X. fastidiosa CVC 9a5c strain. Based on our results, this is also the case for the three other X. fastidiosa strains used in this study. These results are in contrast to other gram-negative bacteria, such as Escherichia coli. For example, in E. coli (strain K-12), 19,200 mono- and 7,575 direpeats with repeat units equal to or greater than six were identified (5), and hexa- or longer repeats were rare in the E. coli genome. The evolutionary and adaptive implications of the various classes of repeat motifs among bacteria are not known. SSR allele sizes were determined relative to a known sequencing molecular size marker with a precision of ±1 bp.
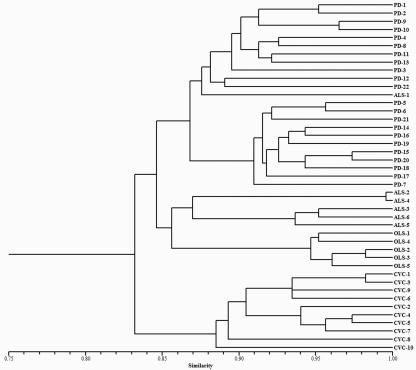
Data were colleted based on the presence-or-absence binary scoring method. The binary data set was converted into a similarity matrix. Unweighted paired-group method using arithmetic averages cluster analysis with simple matching coefficient of resemblance was performed with NTSYSpc, version 2.01 (Exeter Software, Setauket, NY). Cluster analysis of genetic distances divided the 43 isolates into four major clusters (Fig. 2). Each subcluster clearly defined the crop-associated isolates (grape, citrus, almond, and oleander). The exception was ALS-1, which was closely linked with PD strains. Hendson et al. (6) reported that this ALS-1 strain (an almond leaf scorch strain from Tulare County) was tightly clustered with PD strains when randomly amplified polymorphic DNA analysis and contour-clamped homogeneous electric field electrophoresis DNA marker systems were used for cluster analyses (6). It was suggested that some of the X. fastidiosa strains collected from almond may cause either PD or ALS under natural conditions. Cluster analysis also showed that the 10 CVC strains were more distantly related to the rest of the strains and their groupings. Within the 22 PD strains, SSR markers were able to group the Kern County strains as separate from Napa County strains, except for the PD-14 strain, which was grouped with Napa's strains, while strains isolated in Temecula in Riverside County were mixed between these two groups.
FIG. 2.
Dendrogram of genetic similarity among 43 X. fastidiosa isolates based on unweighted paired-group method using arithmetic averages cluster analysis of data from 34 SSR loci.
The 34 X. fastidiosa SSR markers presented here provide a powerful tool for many applications. For example, these markers can be used for differentiating X. fastidiosa strains both within and among crop associations as demonstrated in this study. They can also be used to study the population structure and genetic diversity of X. fastidiosa strains and aid in epidemiological and strain virulence studies. This marker system will be easy to adapt to a multiplex PCR process. When this multiplex format is combined with a fluorescence-based automated sequencing analyzer, it will provide an accurate and high-throughput platform for large-scale pathogen detection.
Acknowledgments
We gratefully acknowledge funding from the California Department of Food and Agriculture's Pierce's Disease Board.
We also thank Elena Lemos for providing the CVC strain DNA samples and Alexander H. Purcell for providing almond and oleander X. fastidiosa strains.
REFERENCES
- 1.Benson, G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blua, M. J., P. A. Phillips, and R. A. Redak. 1999. A new sharpshooter threatens both crops and ornamentals. Calif. Agric. 53:22-25. [Google Scholar]
- 3.Coletta-Filho, D. H., M. A. Takita, A. A. de Souza, C. I. Aguilar-Vildoso, and M. A. Machado. 2001. Differentiation of strains of Xylella fastidiosa by a variable number of tandem repeat analysis. Appl. Environ. Microbiol. 67:4091-4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis, M. J., W. J. French, and N. W. Schaad. 1981. Axenic culture of the bacteria associated with phony disease of peach and plum leaf scald. Curr. Microbiol. 6:309-314. [Google Scholar]
- 5.Gur-Arie, R., C. J. Cohen, Y. Eitan, L. Shelef, E. M. Hallerman, and Y. Kashi. 2000. Simple sequence repeats in Escherichia coli: abundance, distribution, composition, and polymorphism. Genome Res. 10:62-71. [PMC free article] [PubMed] [Google Scholar]
- 6.Hendson, M., A. H. Purcell, D. Chen, C. Smart, M. Guilhabert, and B. Kirkpatrick. 2001. Genetic diversity of Pierce's disease strains and other pathotypes of Xylella fastidiosa. Appl. Environ. Microbiol. 67:895-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hopkins, D. L. 1989. Xylella fastidious: xylem-limited bacterial pathogen of plants. Annu. Rev. Phytopathol. 27:271-290. [DOI] [PubMed] [Google Scholar]
- 8.Krivanek, A. F., and M. A. Walker. 2005. Vitis resistance to Pierce's disease is characterized by differential Xylella fastidiosa populations in stems and leaves. Phytopathology 95:44-52. [DOI] [PubMed] [Google Scholar]
- 9.Minsavage, G. V., C. M. Thompson, D. L. Hopkins, R. M. V. B. C. Leite, and R. E. Stall. 1994. Development of a polymerase chain reaction protocol for detection of Xylella fastidiosa in plant tissue. Phytopathology 84:456-461. [Google Scholar]
- 10.Purcell, A. H., and D. L. Hopkins. 1996. Fastidious xylem-limited bacterial plant pathogens. Annu. Rev. Phytopathol. 34:131-151. [DOI] [PubMed] [Google Scholar]
- 11.Purcell, A. H., S. R. Saunders, M. Hendson, M. E. Grebus, and M. J. Henry. 1999. Causal role of Xylella fastidiosa in oleander leaf scorch disease. Phytopathology 89:53-58. [DOI] [PubMed] [Google Scholar]
- 12.Simpson, A. J. G., F. C. Reinach, P. Arruda, F. A. Abreu, M. Acencio, et al. 2000. The genome sequence of the plant pathogen Xylella fastidiosa. Nature 406:151-157. [DOI] [PubMed] [Google Scholar]
- 13.van Belkum, A., S. Scherer, L. Van Alphen, and H. Verbrugh. 1998. Short-sequence DNA repeats in prokaryotic genomes. Microbiol. Mol. Biol. Rev. 62:275-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Sluys, M. A., M. C. de Oliveira, C. B. Monteiro-Vitorello, C. Y. Miyaki, L. R. Furlan, et al. 2003. Comparative analyses of the complete genome sequences of Pierce's disease and citrus variegated chlorosis strains of Xylella fastidiosa. J. Bacteriol. 185:1018-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson, K. 1987. Preparation of genomic DNA from bacteria, p. 2.4.1-2.4.2. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. Greene Publishing Associates and Wiley Interscience, New York, N.Y.