Abstract
Because archaea are generally associated with extreme environments, detection of nonthermophilic members belonging to the archaeal division Crenarchaeota over the last decade was unexpected; they are surprisingly ubiquitous and abundant in nonextreme marine and terrestrial habitats. Metabolic characterization of these nonthermophilic crenarchaeotes has been impeded by their intractability toward isolation and growth in culture. From studies employing a combination of cultivation and molecular phylogenetic techniques (PCR-single-strand conformation polymorphism, sequence analysis of 16S rRNA genes, fluorescence in situ hybridization, and real-time PCR), we present evidence here that one of the two dominant phylotypes of Crenarchaeota that colonizes the roots of tomato plants grown in soil from a Wisconsin field is selectively enriched in mixed cultures amended with root extract. Clones recovered from enrichment cultures were found to group phylogenetically with sequences from clade C1b.A1. This work corroborates and extends our recent findings, indicating that the diversity of the crenarchaeal soil assemblage is influenced by the rhizosphere and that mesophilic soil crenarchaeotes are found associated with plant roots, and provides the first evidence for growth of nonthermophilic crenarchaeotes in culture.
The hallmark of the Archaea is their remarkable specialization; isolates have been recovered almost exclusively from extreme environments and specialized niches. Nevertheless, results from recent studies using molecular approaches to examine the phylogenetic breadth, ecological range, and abundance of members of the Archaea have challenged the assumption that these microorganisms are absent from, or only minor components of, nonextreme marine and terrestrial habitats (including mesophilic soils and rhizospheres). The accumulation of extensive sequence-based evidence by cultivation-independent approaches has made the broad ecological distribution and considerable abundance of archaea in these nonextreme habitats indisputable (9-11, 49). One intriguing group identified in this manner consists of low-temperature and mesophilic members of the division Crenarchaeota (termed “nonthermophilic”) (5) whose small subunit (SSU) rRNA gene sequences diverge deeply from those of cultured crenarchaeotes (3, 8, 15, 22, 51). Phenotypic properties of nonthermophilic crenarchaeotes are also presumed to be different from those of previously cultured members of this division isolated exclusively from thermophilic environments.
Their ubiquity, in combination with a high abundance in a variety of habitats (12, 27, 31, 38, 44, 45), suggests that they have major roles in one or more biosphere processes. However, to date, the physiological properties and functions in nature of these unforeseen archaea are largely the subject of speculation, in part because they have been recalcitrant toward isolation and growth in culture.
In an attempt to learn more about the ecophysiology of these organisms, we exploited our recent complementary discoveries that (i) mesophilic soil crenarchaeotes are found closely associated with plant roots (44) and (ii) the plant rhizosphere influences the diversity of the crenarchaeal assemblage in soil (47). A number of reports from other research groups are consistent with our findings and describe, for example, the recovery of SSU rRNA gene sequences of mesophilic crenarchaeotes from the rice rhizosphere (19), washed roots of maize (6), and mycorrhizospheres of pine seedlings (4). Here we extend our previous work by demonstrating that two phylogenetic types (phylotypes) of mesophilic crenarchaeotes from the monophyletic clade C1b (11) are predominantly associated with the tomato (Esculentum lycopersicon) rhizoplane of plants grown in soil collected from the West Madison Agricultural Research Station (referred to hereafter as WMAD soil) (3) and that the C1b.A1 phylotype (44, 46) is selectively enriched in culture amended with root extract as a putative nutrient source. Our results provide further evidence for a biologically relevant association of distinct members of the mesophilic soil Crenarchaeota with plant roots.
MATERIALS AND METHODS
Sample preparation and manipulation.
Soil collection and growth of tomatoes was carried out as previously described (44). Soil from the West Madison Agricultural Research station (Plano silt loam containing 61% sand, 23% silt, and 16% clay, with 1.7% organic matter, pH 7.0) (3) was collected between 2 and 8 cm below the surface using sterile implements. Inoculum was prepared by sonication for 30 s in a Branson 2200 Ultrasonic cleaner to displace attached microorganisms from gently and thoroughly rinsed roots of tomato cultivar M82a that were harvested after germination and growth of the plants in WMAD soil.
Preparation of tomato root extract.
Tomatoes were grown in a mixture of vermiculite and potting soil (1:3) and watered every 3 days with full-strength Hoagland's nutrient solution (2). Roots were harvested after 60 days by removing all of the soil mixture and rinsing thoroughly. Roots were weighed, frozen in liquid nitrogen, ground into a fine powder, and stored at −80°C. To prepare extract, powdered root material was measured into Milli-Q H2O and vortexed for 5 min. Coarse filtration was used to remove larger particles, followed by sterilization through a 0.22-μm filter.
Enrichment culture treatment and incubation.
In our enrichments for archaea, we used a combination of chemical and physical treatments and inhibitors to restrict the growth of bacteria. The inoculum was treated with lysozyme according to the method of Repaske (40). Freeze-thaw cycles were performed by three consecutive rounds of quick-freezing in a dry ice-ethanol bath for 5 min, followed by a 5-min thaw at 37°C. The inoculum (10% by vol) was incubated for growth in batch culture in a basal salts medium [5 mM KH2PO4, 5 mM Na2HPO4, 1 mM (NH4)SO4, 2 mM KCl, pH 7.0], with the addition of 1% Daniels' mineral elixir (7), 1% vitamin solution (43), 2.5 mM NaHSO3, 0.2× root extract (1× root extract is 25 g [wet root weight]/liter H2O), 100 mg/liter (each) carbenicillin and streptomycin, 200 mg/liter rifampin (in methanol), 6.7 mg/liter cephalothin, and 1.7 mg/liter clindamycin. Culture volumes were 1/10 to 1/5 of the incubating flask volume and were incubated on a rotary shaker at 200 rpm in low light.
DNA isolation and quantification.
Inoculum and culture samples were extracted using the FastDNA spin kit for soil (BIO101, MP Biomedicals, Irvine, CA) and further purified using Sepharose microspin columns as described previously (46). Plasmid DNA was isolated from cell culture using a standard alkaline lysis technique, with additional purification performed by phenol-chloroform extraction and concentration by precipitation with 0.1 volume of 3 M NaCl, pH 5.2, and 3 volumes of ethanol. Linearized plasmid DNA was quantified using the PicoGreen double-stranded DNA quantitation kit (Molecular Probes of Invitrogen, Eugene, OR) according to the manufacturer's protocol.
PCR, rRNA gene cloning, and sequencing.
Total enrichment DNA was isolated as previously described (44) or by a modified agarose plug method (41). PCR was performed in separate reactions with the SSU rRNA gene-specific primer sets, including the archaea-biased primers 133f, (44) and 915r (48) and the nonspecific cross-domain primer 1492r (29). PCR products were cloned by ligation using the pGEM-T vector system (Promega Corp., Madison, WI). Restriction enzymes used to generate amplified ribosomal DNA restriction analysis patterns were AluI, RsaI, and Sau3A1. Partial sequences were generated by bidirectional sequencing of cloned products using the vector primers SP6 and T7. Sequences were analyzed on an ABI Prism 377 DNA sequencer (University of Wisconsin Biotechnology Center) and tested for possible chimeric artifacts using the CHECK_CHIMERA (30) and Bellerophon (24) programs. Sequences were analyzed using BLAST (1) searches to determine the closest sequences available from the database. These sequence data have been submitted to the DDBJ/EMBL/GenBank databases (see “Nucleotide sequence accession number” below).
Phylogenetic analyses of sequence data.
SSU rRNA gene sequences were aligned in the ARB database (30a). Regions of ambiguous positional homology were removed from the data set, with additional manual corrections, where necessary, using the Seqlab editor within the GCG Wisconsin Package (Accelyrys). Phylogenetic analyses were performed using PAUP*, version 4.0b10 (Altivec; D. L. Swofford, Sinauer Associates, Sunderland, Mass.), and ARB. A data set of 39 sequences containing 1,080 homologous nucleotide positions was used to generate trees using the maximum-likelihood (ML), maximum-parsimony (MP), and evolutionary distance (ED) methods. To assess branch support, bootstrap resampling was performed in PAUP* with a total of 2,000 resamplings in all cases. MP trees were constructed using the default settings in PAUP*. ED analyses were conducted using the Kimura two-parameter and general time-reversible substitution model corrections with and without rate correction. Rate heterogeneities were corrected using a gamma distribution model (the shape parameter, α, was estimated to be 0.66 using a parsimony-based approximation in PAUP*). ML trees were constructed in ARB using the fastDNAml program (13, 35). Additional sequences with fewer than 1,080 homologous nucleotide positions were added to base trees using the parsimony addition function in ARB. All analyses were performed using multiple different out-group species.
Fluorescence in situ hybridization (FISH), cell counts, and microscopy.
Cell fixation, probe synthesis, hybridizations, microscopy, and image analysis were done as previously described (44), except that an Optronics “Magnafire” cooled charge-coupled device camera with motorized color wheel and RGB dichroic filters (resolution, 1,280 by 1,024 with 10 bits/pixel) was used in combination with Image-Pro Express, version 4.0, software. The following probes were hybridized simultaneously: Cren113a, Cren745a, Cren1209 (44), Bact338 (48), and Bact927 (17). DAPI (4′,6-diamidino-2-phenylindole) was used to stain for DNA. Cells were visualized using a BX-60 microscope (Olympus America) equipped for epifluorescence with an HBO 100-W mercury arc lamp and an Olympus UPlanF1 100× objective/1.3 NA, with a field area of 0.035 ± 0.002 mm2.
PCR-SSCP.
PCR-single-strand conformation polymorphism (PCR-SSCP) was conducted as described previously (46) using the crenarchaeote biased primer set 133FN6F/248R5P. Briefly, PCR mixtures were desalted with Sephadex G50 microspin columns packed in a UniFilter GF/B 800 microplate (Whatman, Inc., Ann Arbor, MI). Following purification, each PCR product was mixed (1:1, vol/vol) with SSCP loading buffer (60% deionized formamide, 25% ROX GeneFlo 625 standard [ChimerX, Milwaukee, WI], 7.5% 100 mM NaOH, 7.5% blue dextran EDTA [50 mg/ml blue dextran, 50 mM EDTA]). Samples were denatured at 94°C for 2 min and then immediately transferred onto ice. A 0.5-μl aliquot of each sample was spotted onto a membrane comb. Polyacrylamide SSCP gels and 1× MDE (BMA, Rockland, ME) were cast according to the manufacturer's instructions. The gel was loaded onto an ABI PRISM 377XL DNA Sequencer connected to an external cooling water bath to maintain the gel temperature at 23°C. The GS RUN 60W CHILLER module was selected to maintain constant wattage. PCR-SSCP data were analyzed using GeneScan software, version 3.1 (Applied Biosystems, Foster City, CA). Peak height cutoffs ranged from 5 to 50 relative fluorescence units depending on signal intensity. Samples were aligned to the internal size standard present in each lane.
Primer design and real-time PCR.
Real-time PCR was performed using the iCycler iQ thermal cycler with a 96- by 0.2-ml reaction module and iQ software (Bio-Rad Laboratories, Inc., Hercules, CA). In each experiment, three independent master mixes were prepared from each inoculum and culture sample and two independent master mixes for standard DNA were prepared and tested in duplicate within a single microtiter plate. Negative-control reactions (without added template) were also included in each experiment. Reactions were performed in a volume of 20 μl, using iQ SYBR green supermix (Bio-Rad Laboratories, Inc., Hercules, CA) and 0.2 μM concentrations of each primer and were optimized for magnesium concentration (2.5 mM MgCl2, final concentration, achieved by adding 0.5 mM EDTA) and annealing temperature. Reactions were performed in the iCycler (Bio-Rad Laboratories, Inc.) using the two-step amplification plus melting curve protocol as follows. Enzyme activation and well-factor determination were at 95°C for 3 min, followed by 40 cycles of 95°C for 10 s (denaturation) and 61.3°C for 45 s (annealing and elongation). The melt curve protocol began immediately after amplification and consisted of 1 min at 95°C, followed by 1 min at 55°C, and 80 10-s steps with a 0.5°C increase in temperature at each step. Threshold values for threshold cycle determination were generated automatically by the iCycler iQ software. Lack of variation in PCR products and the absence of primer dimers were ascertained from the melt curve profile of the PCR products. Slopes, regression coefficients, and PCR amplification efficiency curves were calculated using iCycler iQ software; efficiency (E) was calculated according to the equation E = 10(−1/slope) (39). Mean values and standard errors of mean values were calculated using Microsoft Excel, version 11.0.0.
Primer sets were designed using the Beacon Designer software program (PREMIER Biosoft International, Palo Alto, CA), the ARB database, and sequences recovered in our studies from cultures, roots, and soil. The predicted specificity of primers was examined by performing BLAST searches (1) against the GenBank database. The primer sequences designed for this study are as follows: 599F, 5′-GTA-GCC-GGT-TCT-ACA-AGT-C-3′; 669F, 5′-CGA-CGG-TGA-GGG-ATG-AAA-G-3′; 703R, 5′-ACT-GGT-GGT-CTT-CAA-TGG-ATC-3′; 886R, 5′-CCA-GGC-GGC-AAA-CTT-AAC-3′. The predicted bias of the 599F/703R primer set is toward C1b sequences (as is also the 133F/248R primer set), while the 669F/886R primer set is predicted to hybridize broadly to sequences across most crenarchaeal sequence groups recovered from nonthermophilic environments (i.e., C1, C2, and C3). Melting curve analysis (transition rate, 0.5°C s−1), together with separation of PCR DNA fragments performed by electrophoresis on 3% agarose gels, yielded a single product of the predicted size from both the enrichment culture and the plasmid standard DNA for each primer set used.
Nucleotide sequence accession numbers.
Sequence data have been submitted to the DDBJ/EMBL/GenBank databases under accession numbers AY487100 to AY487118.
RESULTS
PCR-SSCP analysis of the soil and rhizoplane microbial assemblage before and after incubation with root extract.
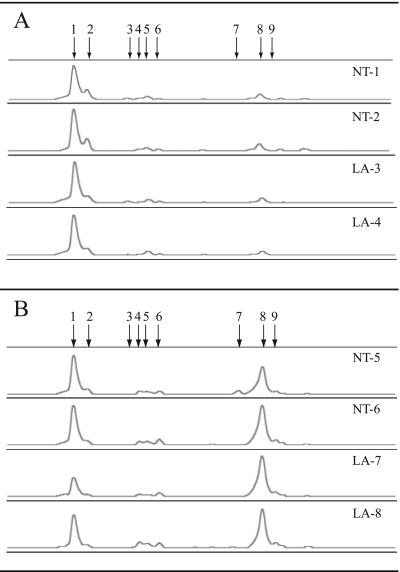
Using Crenarchaeota-biased primers (see Materials and Methods) and PCR-SSCP (20, 36), an electropherogram consisting of 9 different peaks was generated representing the crenarchaeal assemblage in WMAD bulk soil (free of any visible root material) and on the rhizoplane of tomato plants grown in WMAD soil (Fig. 1). The PCR-SSCP profiles from the WMAD soil samples were dominated by a single phylotype of the Crenarchaeota, represented by peak 1 (Fig. 1A), with a relative abundance based on peak area of >75% of the total peak area (Table 1). The rhizoplane PCR-SSCP profile (Fig. 1B) differed from the WMAD soil profile in that the phylotype represented by peak 8 was present in approximately equal, or greater, relative abundance to the phylotype represented by peak 1 (Table 1). Results from a different experiment examining the crenarchaeal assemblage on the tomato rhizoplane from plants grown in WMAD soil were similar; the relative abundance of peak 1 ranged from 28% to 39% of the total peak area (mean, 36%), while that of peak 8 ranged from 51% to 66% of total peak area (mean, 57%) (n = 6). Peaks representing other crenarchaeal phylotypes were detected at <5% relative abundance.
FIG. 1.
PCR-SSCP electropherograms of independent soil (A) and rhizoplane (B) inoculum samples (also referred to as T0 samples). Abbreviations: NT, no treatment; LA, lysozyme and antibiotic treatment. For comparisons between samples, individual electropherograms were aligned on the basis of a common internal lane standard. The y axis represents relative fluorescence intensity, and the x axis represents the relative migration distance of the PCR-SSCP DNA fragment. Arrows indicate peaks representing different crenarchaeal phylotypes.
TABLE 1.
Results from PCR-SSCP analysis of WMAD soil (T0), rhizoplane (T0), and root extract enrichment cultures (30-day, 60-day)
| Time point (days) | WMAD soil samplea | Relative abundance (%) of peak:
|
Rhizo- plane samplea | Relative abundance (%) of peak:
|
||
|---|---|---|---|---|---|---|
| 1 | 8 | 1 | 8 | |||
| 0 | NT-1 | 75.9 | 11.4 | NT-5 | 47.1 | 37.0 |
| 0 | NT-2 | 81.4 | 13.6 | NT-6 | 41.0 | 44.8 |
| 0 | LA-3 | 79.4 | 9.7 | LA-7 | 24.6 | 65.9 |
| 0 | LA-4 | 81.0 | 8.8 | LA-8 | 42.8 | 57.2 |
| 30 | NT-9 | 36.4 | 40.6 | NT-13 | 12.1 | 81.2 |
| 30 | NT-10 | 85.2 | 17.4 | NT-14 | 0.0 | 88.8 |
| 30 | LA-11 | 49.0 | 16.0 | LA-15 | 5.1 | 93.2 |
| 30 | LA-12 | 45.5 | 17.3 | LA-16 | 6.6 | 93.4 |
| 60 | NT-17 | 29.1 | 59.1 | NT-21 | 5.4 | 87.9 |
| 60 | NT-18 | 31.4 | 53.1 | NT-22 | 7.7 | 82.8 |
| 60 | LA-19 | 12.2 | 82.2 | LA-23 | 2.1 | 97.9 |
| 60 | LA-20 | 20.4 | 9.1 | LA-24 | 2.0 | 91.9 |
NT, not treated; LA, lysozyme plus antibiotics.
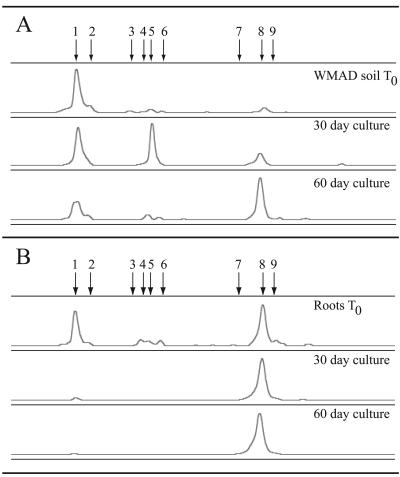
After 30 days of incubation under air at 24°C with root extract, the crenarchaeal assemblage detected in cultures that were inoculated with sonicate from the rhizoplane (details in Materials and Methods) consisted almost exclusively of the phylotype represented by peak 8 (Fig. 2B and Table 1). The relative abundance of the peak 8 phylotype was similar in cultures that had been incubated for an additional month (60 days total). The treatment group (either no treatment or treated with lysozyme and incubated with antibiotics), explained in greater detail below, did not show consistent effects, although there was a trend in the rhizoplane samples toward slightly higher relative abundance of the peak 8 phylotype in the lysozyme-treated cultures. Similar results were obtained from enrichment cultures in two other experiments: (i) 30-day cultures (n = 4), peak 1 ranged from 15% to 24% of total peak area (mean, 18%) and peak 8 ranged from 76% to 82% of total peak area (mean, 79%); (ii) 60-day cultures (n = 8), peak 1 ranged from undetectable to 5.4% of total peak area (mean, 2.4%) and peak 8 ranged from 87% to 100% of total peak area (mean, 94%).
FIG. 2.
Representative PCR-SSCP electropherograms of the crenarchaeal assemblage in soil (A) and rhizoplane (B) samples after incubation with root extract. For comparisons between samples, individual electropherograms were aligned on the basis of a common internal lane standard. The y axis represents relative fluorescence intensity, and the x axis represents the relative migration distance of the PCR-SSCP DNA fragment. Arrows indicate peaks representing different crenarchaeal phylotypes.
Upon incubation of WMAD soil inoculum in culture with root extract, the phylotype represented by peak 8 increased in relative abundance, while that represented by peak 1 decreased in relative abundance (Fig. 2A). This result was similar to that observed for cultures starting with root inoculum, in that a shift occurred in the crenarchaeal assemblage toward a higher percentage of the phylotype represented by peak 8. After 60 days of incubation in root extract culture, the relative abundance of the peak 8 phylotype was greater than that of the peak 1 phylotype in 3 of 4 samples (Table 1).
Phylogenetic analysis of root extract enrichment culture clones.
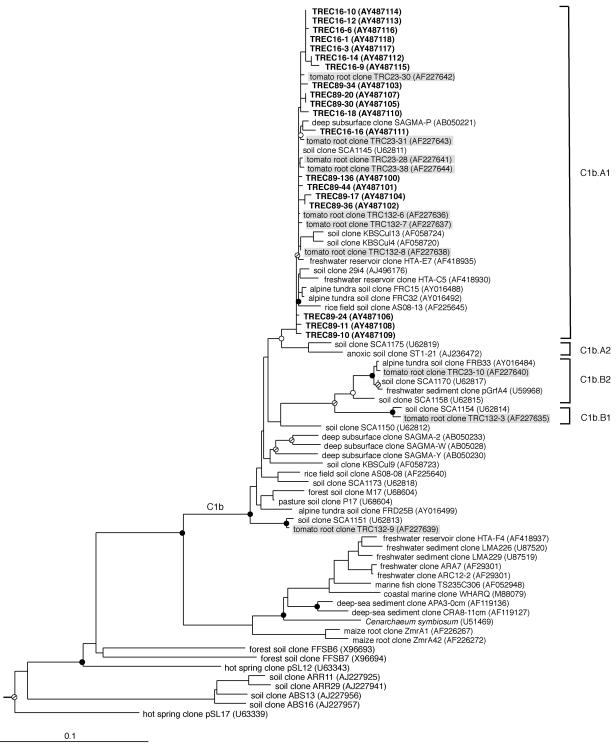
Crenarchaeota-biased primers were used in PCR to amplify and clone rRNA genes from root extract cultures in two experiments. Three unique restriction patterns were observed from amplified ribosomal DNA restriction analysis of 46 randomly chosen clones. Clones representative of the three patterns were chosen for sequence analysis, which revealed four identical and 15 nonidentical but highly similar sequences (≥98.4% identity over approximately 600 nucleotides [nt]). Longer regions of the SSU rRNA gene from five of the designated TREC (tomato root enrichment Crenarchaeota) clones were sequenced, and those sequences were 98.5% to 99.1% identical over >1,200 nt. Phylogenetic placement into clade C1b.A1 indicated that sequences from the crenarchaeotes in these cultures were most closely related to crenarchaeal sequences recovered most frequently by culture-independent methods directly from the tomato rhizoplane (44, 46) (Fig. 3); the near full-length TRC (tomato root Crenarchaeota) and TREC sequences share ≥98.9% sequence identity over approximately 1,200 nt positions. Consistent with their growth and function in mesophilic soil habitats, the range of G+C nucleotide content for SSU rRNA gene sequences from the crenarchaeotes in our enrichment cultures ranged from 54.5% to 55.7%. By comparison, the G+C nucleotide content for SSU rRNA gene sequences from other cultured crenarchaeotes, all thermophiles or hyperthermophiles, is in the range of 58% to 60% (16).
FIG. 3.
Inferred phylogenetic (ML) tree of archaeal SSU rRNA gene sequences cloned from rhizoplane enrichment cultures. The habitat of each environmental sequence and sequences from this study is indicated before the clone name. Tomato root enrichment culture clones are shown in boldface type. Clone number prefixes 16 and 89 designate different experiments. Tomato root clones from direct extraction of tomato rhizoplane DNA are shown on a shaded background. GenBank accession numbers are listed parenthetically. Branch points supported by bootstrap values of >90% in all MP and ED methods are indicated by filled circles. Circles with a slash indicate bootstrap support values of >70%, open circles indicate bootstrap support values of >50%, and branches without circles were not resolved (bootstrap support values of <50%). Bar, 0.1 change per nucleotide.
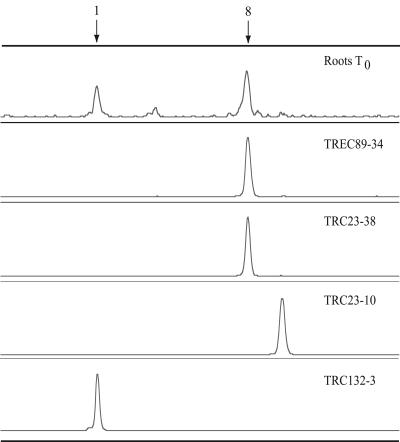
PCR-SSCP analysis of crenarchaeal clones.
Two of the TREC clones recovered from enrichments, TREC89-17 and TREC89-34, were selected for PCR-SSCP analysis. The PCR-SSCP profiles of those clones contained a peak that comigrated with peak 8 from total tomato rhizoplane DNA (representative profile shown by TREC89-34 in Fig. 4). TRC23-30 and TRC23-38, two clones that were recovered directly from the rhizoplane (44) and that clustered with the TREC clones within clade C1b.A1, were also analyzed by PCR-SSCP, and each revealed a similar peak (representative profile shown by TRC23-38 in Fig. 4). Two additional TRC clones, TRC23-10 and TRC132-3, and a clone from WMAD soil, MWS38 (46), chosen in part because they did not place within clade C1b.A1 (clones TRC132-3 and MWS38 clustered together in clade C1b.B1, while clone TRC23-10 placed in clade Clb.B2) (46), were analyzed and found to generate peaks migrating at different positions from peak 8 on PCR-SSCP electropherograms. The peaks generated by clones TRC132-3 and MWS38 comigrated instead with peak 1 from WMAD soil and root samples (representative profile shown by TRC132-3 in Fig. 4). Although the phylogenetic relationships of the clones described here were predictive of their PCR-SSCP profiles, this is not always the case (46).
FIG. 4.
PCR-SSCP electropherograms of a crenarchaeal rhizoplane sample and clones recovered from the rhizoplane and root extract enrichment culture. For comparisons between samples, individual electropherograms were aligned on the basis of a common internal lane standard. The y axis represents relative fluorescence intensity, and the x axis represents the relative migration distance of the PCR-SSCP DNA fragment. Arrows indicate peaks corresponding to the two most abundant crenarchaeal phylotypes detected on the rhizoplane.
Crenarchaeal cell quantification in inoculum samples subjected to different treatments.
We quantified the number of crenarchaeotes relative to total microorganisms present in root extract enrichment cultures. The inoculum was prepared in a manner similar to that for the PCR-SSCP experiments, by sonication from gently rinsed tomato roots that were harvested from plants grown in WMAD soil in the growth chamber. Cell numbers were estimated with FISH probes designed to selectively detect mesophilic Crenarchaeota or organisms within the domain Bacteria. Crenarchaeal cell numbers in untreated inoculum samples from three experiments ranged from 1.2% to 2.1% of total probe-positive cells (mean, 1.8%) (Table 2). These values are similar to those from our previous study in which we quantified crenarchaeal cells attached directly to rinsed, nonsenescent tomato roots (44).
TABLE 2.
Inoculum cell counts
| Inoculum samplea | Mean no. of cellsb± SEM (% of total cells)
|
||
|---|---|---|---|
| Crenarchaeotes | Bacteria | Eukaryotes | |
| A1 | 1.8 ± 0.5 (1.9) | 87.7 ± 4.2 (97.2) | 0.7 ± 0.3 (0.7) |
| A2 | 1.5 ± 0.5 (1.8) | 78.3 ± 5.2 (97.7) | 0.3 ± 0.3 (0.4) |
| B1 | 1.2 ± 0.3 (1.2) | 94.0 ± 5.5 (97.9) | 0.8 ± 0.5 (0.9) |
| B2 | 0.8 ± 0.2 (1.7) | 48.7 ± 5.4 (98.3) | < 0.2 (< 0.1) |
| C1 | 2.0 ± 0.5 (3.0) | 68.0 ± 4.0 (95.8) | 1.0 ± 0.4 (1.4) |
| C2 | 1.7 ± 0.5 (1.4) | 124 ± 8.3 (98.5) | 0.2 ± 0.2 (0.1) |
A, B, and C indicate different experiments; 1 and 2 indicate different subsamples.
Mean values were calculated from counts of 6 microscopic fields per sample.
In addition to the use of root extract as a growth substrate, treatments were chosen to select against bacteria that were present in the starting inoculum. The inoculum was treated with lysozyme or lysozyme combined with freeze-thaw cycles before incubation for growth. Lysozyme hydrolyzes glycosidic linkages of peptidoglycan (which is present in cell walls of bacteria but not in those of archaea) and freeze-thaw treatment should enhance lysis of weakened cell walls. Both lysozyme (mean, 4.6%) and lysozyme combined with freeze-thaw (mean, 3.4%) treatments had a significant effect (an approximate twofold increase, P < 0.05) on the proportion of crenarchaeotes compared to total cells in inoculum samples prior to growth. This suggests that the treatments lysed a portion of bacterial cells in the inoculum.
Persistence of crenarchaeotes in root extract enrichment cultures.
As an additional selection, cultures were incubated with antibiotics that target bacteria but not archaea. We monitored the presence of crenarchaeotes in root extract cultures by PCR of SSU rRNA gene sequences with Crenarchaeota-biased primer sets. Results indicated that, while crenarchaeotes were initially present in all inoculated samples, over time, Crenarchaeota-specific PCR products were detected only in cultures subjected to treatments with particular combinations of antibiotics (Table 3). While various antibiotics and eukaryotic cell growth inhibitors were evaluated initially, in the experiments described below, a combination of carbenicillin and streptomycin was used.
TABLE 3.
Persistence of crenarchaeotes in root extract enrichment cultures
| Culture designation | Treatmenta | Antibioticsb | PCR product detectede on day after inoculation:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 8 | 12 | 45 | 115 | 166 | 178 | 197 | 208c | 238 | 378 | |||
| 143d | None | None | − | − | − | − | − | − | − | |||
| 144 | None | None | + | + | + | − | − | − | − | |||
| 145 | L | None | + | + | + | + | − | − | − | |||
| 146 | L | S, C | + | + | + | + | — | + | + | + | + | + |
| 147 | L | S, C, R | + | + | − | − | − | − | − | |||
| 148 | L | S, C, Cl, Ce | + | + | + | + | + | + | + | + | + | + |
| 149 | FTL | None | + | + | + | + | − | − | − | |||
| 150 | FTL | S, C | + | + | + | + | + | + | + | + | + | + |
| 151 | FTL | S, C, R | + | + | − | − | − | − | − | |||
| 152 | FTL | S, C, Cl, Ce | + | + | + | + | + | + | + | + | + | + |
L, lysozyme; FTL, freeze-thaw lysozyme (details in Materials and Methods).
C, carbenicillin; Ce, cephalothin; Cl, clindamycin; R, rifampin; S, streptomycin.
After 208 days, samples were taken only from cultures that were positive for crenarchaeotes.
Uninoculated control.
—, not sampled; +, PCR product was detected; −, no PCR product was detected.
Crenarchaeal cell quantification in root extract enrichment culture.
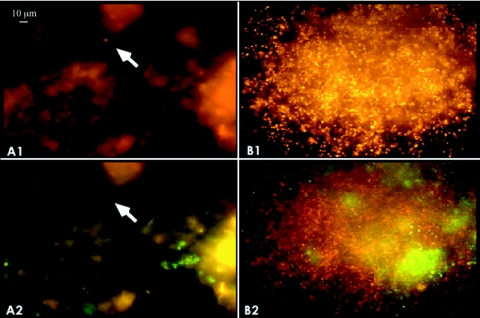
Cell counts were performed on crenarchaeotes by FISH and microscopy after incubation in root extract enrichment culture. The results suggested that enrichment of crenarchaeotes occurred in incubations with root extract and antibiotics (Table 4). Consistent with results from the persistence studies described above, no increase in the ratio of crenarchaeal cells to the total was observed in the absence of antibiotics. Enrichment was significantly enhanced by treatment with lysozyme (P < 0.05). Treatments combining antibiotics with lysozyme and freeze-thaw cycles did not result in significantly different enrichments from treatments with antibiotics alone. Taken together, these conditions led to enrichments with an average 10-fold increase in the relative abundance of crenarchaeotes in the inoculum, with some individual cultures enriched as much as 40% for crenarchaeotes. Microscopic analysis of enrichment cultures using FISH and probes specific for mesophilic crenarchaeotes revealed the formation of microcolonies with large numbers of cells (Fig. 5), which were never observed in inoculum samples. Enrichment of crenarchaeotes was not observed in parallel experiments in which cultures were amended with a variety of organic compounds other than root extract (e.g., complex substrates, such as yeast extract, tryptone, or filtered culture medium from root extract enrichments, and defined substrates, such as bicarbonate, monosaccharides, organic acids, and amino acids) (data not shown). Crenarchaeotes were also never prevalent in dilution cultures when dilutions greater than 10−4 were performed (data not shown).
TABLE 4.
Enrichment of crenarchaeotes in root extract culturese
| Treatmenta | Antibioticsb | Meanc % crenarchaeotad |
|---|---|---|
| None | − | 1.6 A |
| L, FTL | − | 2.6 A |
| None | + | 16.7 B |
| L | + | 22.3 C |
| FTL | + | 20.2 B, C |
L, lysozyme; FTL, freeze-thaw and lysozyme (details in Materials and Methods).
Antibiotic additions (+, added; −, not added) are described in Materials and Methods.
Means were calculated from counts of 6 microscopic fields per sample.
Mean values with different letters (A, B, and C) differ significantly (P < 0.05).
Analysis of variance was performed using the general linear model, and mean values were compared using Duncan's multiple-range test (42). Degrees of freedom: model, 13; error, 250; total, 263; pooled standard error, 9.82. The average time of incubation for cultures in each treatment was 22 to 24 weeks.
FIG. 5.
Fluorescence micrographs of crenarchaeotes from enrichment cultures. Representative color micrographs showing inoculum from the rhizoplane before (A) and after (B) incubation in root extract enrichment culture and indicating crenarchaeotes (panels labeled 1, orange) and bacteria (panels labeled 2, green). Crenarchaeal cells can also be seen in panel B2 (red) with the filter set used to visualize bacteria. The arrow indicates the location of a crenarchael cell doublet.
Quantification of crenarchaeal growth by real-time PCR.
We used real-time PCR with primers biased toward the SSU rRNA gene sequences of mesophilic soil crenarchaeotes in SYBR green I assays to quantify growth of these organisms in root extract cultures. Results from these experiments were similar using two different primer sets (Table 5). After 60 days, there was a four- to fivefold increase in the SSU rRNA gene copy number in cultures that were incubated with root extract but without antibiotics. In cultures that were treated with lysozyme and incubated with both root extract and antibiotics; however, there was a 10-fold increase in SSU rRNA gene copy number after 30 days and a 60- to 70-fold increase after 60 days. Since the crenarchaeal assemblage in inoculum samples consisted almost entirely of the phylotypes represented by PCR-SSCP peaks 1 and 8 in approximately equal abundance, there is an additional twofold increase in abundance for the phylotype represented by peak 8, as it was detected almost exclusively in our cultures following incubation.
TABLE 5.
Results from real-time PCR analysis of crenarchaeaotes in rhizoplane inoculum and root extract enrichment culture
| Primer pair | Treatment and templatea | Enrichment culture/ inoculum ratio ± SEM |
|---|---|---|
| None (untreated) | ||
| 599F/703R | Inoculum | 6.6 ± 0.8 |
| 599F/703R | 30-day culture | 3.6 ± 0.5 |
| 599F/703R | 60-day culture | 33.3 ± 3.6 |
| 669F/886R | Inoculum | 3.3 ± 0.3 |
| 669F/886R | 30-day culture | 2.3 ± 0.4 |
| 669F/886R | 60-day culture | 12.2 ± 0.8 |
| LAb | ||
| 599F/703R | Inoculum | 1.8 ± 0.2 |
| 599F/703R | 30-day culture | 16.9 ± 3.4 |
| 599F/703R | 60-day culture | 129.0 ± 5.9 |
| 669F/886R | Inoculum | 1.8 ± 0.1 |
| 669F/886R | 30-day culture | 17.9 ± 2.7 |
| 669F/886R | 60-day culture | 107.0 ± 5.4 |
The results given in each row represent the combined average results from PCR assays of two individually prepared cultures, with the exception of inoculum samples, in which results from one of the pair of individually prepared inoculum samples are used as the basis for comparison to the other samples and the second is compared to the first.
LA, treatment with lysozyme and antibiotics.
Two experiments were performed, with both primer sets, to test for the possibility that a PCR inhibition factor (present in T0 [inoculum] samples and subsequently degraded upon incubation in culture) resulted in an apparent increase in the number of 16S rRNA gene copies. In the first, 1 μl of 60-day culture DNA from LA-23 was mixed with 1 μl (each) T0 DNA from NT-1, NT-2, LA-3, and LA-4 in separate reactions. In the second, 20 fg plasmid standard DNA was mixed with 1 μl (each) T0 DNA from NT-1, NT-2, LA-3, and LA-4 in separate reactions. There was no evidence for inhibition in either test (data not shown).
Experiments were also performed using primer set 133F/248R (46), which was used to generate the PCR-SSCP profiles, and the 771F/957R primer set designed and used in real-time PCR in a study conducted by Ochsenreiter et al. (34). Compared to the 599F/703R and 669F/886R primer sets, however, the 113F/248R and the 771F/957R primer sets performed poorly in our experiments, with regard to PCR amplification efficiency (14, 37). Even so, the same general trend in relative increase of SSU rRNA gene copy number was obtained with 133F/248R as that reported in Table 5 (data not shown). Studies with the 771F/957R primer set were not pursued further.
Based on a calibration curve (32, 37) generated in real-time PCR with purified plasmid DNA from clone TRC23-30 (44) and the 599F/703R primer set, we estimated the copy number of crenarchaeote SSU rRNA genes in 60-day cultures to be 4.1 × 106 gene copies per ml of culture. The slope of the plasmid standard curve was −3.51, the correlation coefficient was >0.99, and the PCR amplification efficiency for the standard DNA was 1.926. The slope of the curve based on a fivefold dilution series of culture DNA (from no dilution to 1.6 × 10−3) was −3.50, the correlation coefficient was >0.95, and the PCR amplification efficiency for the culture DNA was 1.932. The reproducibility of the data at higher dilutions was poor, corresponding to a limit of detection of approximately 25 copies of the target gene in these experiments. This number is similar to detection limits obtained by others (21, 28). The estimated doubling time for crenarchaeotes in root extract cultures was approximately 8 days, with about 8 generations in 60 days.
DISCUSSION
Plants are known to deposit up to 20% of their total photosynthate into the rhizosphere, allowing microorganisms there to achieve much greater abundance and activity than they can in habitats of lower nutrient availability, such as bulk soil (reviewed in references 53 and 54). Microscopic and molecular phylogenetic studies of archaea in soil (3, 46), in the rhizosphere (47), and on the rhizoplane (44) led us to the first description of the colonization of plant surfaces by mesophilic crenarchaeotes and the influence of plant roots on the diversity of the crenarchaeal assemblage in soil. It was our prediction that the root environment selects for microorganisms that are actively metabolizing root exudates and/or other root materials and that crenarchaeotes associated with the root surface would therefore be more amenable to growth in culture than those residing in bulk soil. Consistent with this hypothesis, we demonstrated growth of one of the two dominant crenarchaeal phylotypes found associated with the tomato rhizoplane, represented by PCR-SSCP peak 8, in enrichment culture amended with root extract. These results provide additional support for a biological role for soil crenarchaeotes in rhizosphere microbial ecosystems.
Colonization of tomato roots by mesophilic soil crenarchaeotes was previously described (44) and was based on (i) the recruitment of these microorganisms to roots from soil, (ii) consistent recovery of crenarchaeal sequences and cells from roots, (iii) a tight association with roots, i.e., cells remained attached even after thorough rinsing, and (iv) broad distribution of cells and microcolonies on roots. Although the tomato rhizoplane is not exclusively colonized by the peak 8 phylotype, the work presented here suggests that this particular phylotype comprises a dominant, if not the dominant, crenarchaeal population on tomato roots grown in WMAD soil. Detection of this phylotype in WMAD soil itself, however, appears to be more variable. For example, although it was detected at very low levels in the WMAD soil sampled in experiments presented here, the peak 8 phylotype has been noted in greater relative abundance in PCR-SSCP profiles of WMAD soil generated in other research (46). From our sampling thus far, the phylotype represented by peak 1 appears to be the dominant member of the crenarchaeal assemblage in WMAD soil (in addition to results presented in this paper, another study by Sliwinski and Goodman [46] demonstrated that the relative abundance of the peak 1 phylotype was higher than that of other phylotypes, ranging between 43% and 53% of the total). The fact that the peak 8 phylotype consistently colonizes the rhizoplane in high relative abundance compared to other phylotypes, yet is detected variably in WMAD soil, suggests that its growth requirements are met more consistently on the rhizoplane than in bulk (WMAD) soil habitats. More investigation is required to define the parameters that influence the distribution and abundance of this and other crenarchaeal phylotypes in soil.
We have demonstrated the association of mesophilic crenarchaeotes with plant roots using several approaches. Previous studies described the direct extraction of crenarchaeal DNA from the rhizoplane (44) and rhizosphere soil (47) and visualization of crenarchaeote cells directly on the root surface using phylogenetic probes. We have now provided additional evidence for association by a specific phylotype of soil crenarchaeotes with plant roots. The peak 8 phylotype is one of two dominant phylotypes recovered directly from roots and the only phylotype recovered from cultures after 60 days of incubation with root extract. In two different studies, the majority of clones recovered directly from the rhizoplane and all of the clones recovered from root extract enrichment cultures placed within clade C1b.A1; sequences from the recovered clones comigrated on PCR-SSCP gels with peak 8 from total DNA extracted directly from the rhizoplane. Furthermore, four lines of evidence, taken together, provide strong support for growth of the peak 8 phylotype when it is incubated in root extract enrichment culture: (i) persistence for >1 year in culture, (ii) the increase in proportion of crenarchaeotes to total cells, (iii) the increase in SSU rRNA gene copy number of crenarchaeotes per unit volume of culture, and (iv) the formation of microcolonies containing large numbers of crenarchaeal cells. In addition, the fact that three different primer sets, each designed to be specific for the SSU rRNA genes of mesophilic soil crenarchaeotes and one also used in PCR-SSCP analyses, yielded similar results in real-time PCR assays strengthens the conclusion that growth of these microorganisms occurred in root extract enrichment cultures. Although we achieved enrichment in cultures amended with root extract, we were not as successful using a variety of common organic compounds, including many known to be present in plant exudates. These results may suggest that the actual substrates supporting growth are unusual exudate compounds produced either by the plant roots or by associated bacteria. Identification of the actual substrates for growth will be critical for growing the crenarchaeotes in axenic culture.
Crenarchaeal enrichment was dependent upon the presence of antibiotics in the medium, although there was also a small (four- to fivefold) increase in crenarchaeal 16S rRNA gene copies in cultures incubated without antibiotics. The antibiotics used in this study were chosen because they are known to inhibit bacteria but not archaea. It was surprising, therefore, that the addition of rifampin to the cultures resulted in the loss of the crenarchaeal PCR signal sooner than it occurred in the absence of antibiotics. While it is currently unknown whether the inhibition was due to a direct or an indirect effect, RNA polymerases from other archaea are not targets for the antibiotic. On the other hand, sensitivity to rifampin has been observed in a number of different archaea, including some crenarchaeotes, at concentrations similar to that used in this study (23, 52). A detergent-like machanism of cell lysis has been suggested for the inhibitory effect of rifampin on Halobacterium halobium (55). Possible indirect effects of rifampin on enrichment of crenarchaeotes include the inhibition of an obligate bacterial partner; involvement by the crenarchaeotes in a symbiotic partnership(s) with other microbial species has not yet been explored. Consistent with this idea is the failure of serial dilutions to lead to enrichment. Another possible explanation for the effect of rifampin could be that certain bacteria present in the enrichments were able to metabolize the small amount of methanol used as a solvent that was introduced into the culture along with the antibiotic and, by doing so, were able to overgrow the crenarchaeotes. Bacteria that were present and growing in the enrichments may have developed resistance to rifampin (and to other antibiotics used), or, alternatively, diffusion of the antibiotics may have been impeded by extracellular polysaccharide that was produced in the cultures.
Our research represents a significant advance in the study of the mesophilic Crenarchaeota, first, because it reveals a biological relationship between members of the C1b.A1 clade and plants and, second, because neither isolation nor growth of these organisms in culture has been reported in the decade or so since nonthermophilic crenarchaeotes were initially discovered in mesophilic soils (3, 51) or their planktonic counterparts were discovered in marine waters (8, 15). Our result of a modest increase (102) in SSU rRNA gene copy number over an 8-week incubation period is in line with results from a number of other studies that have attempted enrichments and isolations of archaea and bacteria identified first by molecular phylogenetic methods (that had not been previously detected from culture-dependent work). Examples include isolates from the bacterial divisions Acidobacteria, Verrucomicrobia, Gemmatimonadetes, Actinobacteria, and Proteobacteria that were obtained only after extended incubations (25, 26, 50) and enrichments of anaerobic methane-oxidizing archaea from marine sediments, with apparent 2- to 100-fold (or greater) increases in SSU rRNA gene copies after 24 weeks of incubation (although the latter data are somewhat inconclusive because no minimum detection level was established for the target gene in sediment cores prior to incubation (18).
Based on evidence from thermophilic and hyperthermophilic crenarchaeotes that have been isolated in pure culture and contain a single rRNA operon in each of their genomes, it is not unreasonable to make the assumption that individual genomes of mesophilic crenarchaeotes also contain only one rRNA operon. Using this assumption and comparing results from real-time PCR of enrichment culture DNA with plasmid standard DNA, we estimate that our enrichment cultures contained 4.1 × 106 crenarchaeal cells per ml after 60 days of incubation and, by extrapolation, ∼104 crenarchaeal cells per ml in the rhizoplane sonicates used for the inoculum. Another group investigated crenarchaeal populations in the rhizosphere of the grass Festuca ovina s.l. using real-time PCR (34) and found a relatively low abundance of crenarchaeal to bacterial 16S rRNA gene copies in the rhizosphere versus bulk soil. Based on the numerous differences that exist between the two studies (e.g., soil type, plant species, growth chamber versus field study, and units of measurement), direct comparisons between them are not valid. It is worth noting, however, that studies by Sliwinski and Goodman, and others, examining crenarchaeal assemblages in a variety of soils (46) and in the rhizosphere of diverse plant species (33, 47) suggests that the abundances of crenarchaeal populations differ in different soil and rhizosphere habitats.
In earlier work (44), we found a 10-fold increase in the abundance of crenarchaeotes on senescent roots over nonsenescent roots of tomato. One intriguing possibility raised by that result and by our studies here is the idea that plants that are stressed, for example, as they might be when grown in unfertilized soil within a growth chamber, harbor larger populations of crenarchaeotes on their roots than their unstressed counterparts. This possibility and other factors potentially affecting the abundance, distribution, and diversity of crenarchaeotes in soil and rhizosphere habitats are the subjects of ongoing investigations in our laboratories.
Acknowledgments
We thank John Helgeson for the generous use of his Olympus BX-60 microscope, Ena Urbach and Brian Manske for advice with phylogenetic analyses, Doreen Gillespie for discussions, and Gary Roberts and Peter Zuber for comments on the manuscript.
A National Science Foundation grant (no. MCB-0136441) and contributions from the McKnight Foundation supported this work.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Baker, A. 1981. Accumulators and excluders—strategies in the response of plants to heavy metals. J. Plant Nutr. 3:643-654. [Google Scholar]
- 3.Bintrim, S. B., T. J. Donohue, J. Handelsman, G. P. Roberts, and R. M. Goodman. 1997. Molecular phylogeny of Archaea in soil. Proc. Natl. Acad. Sci. USA 94:277-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bomberg, M., G. Jurgens, A. Saano, R. Sen, and S. Timonen. 2003. Nested PCR detection of archaea in defined compartments of pine mycorrhizospheres developed in boreal forest humus microcosms. FEMS Microbiol. Ecol. 43:163-171. [DOI] [PubMed] [Google Scholar]
- 5.Buckley, D. H., J. R. Graber, and T. M. Schmidt. 1998. Phylogenetic analysis of nonthermophilic members of the kingdom Crenarchaeota and their diversity and abundance in soils. Appl. Environ. Microbiol. 64:4333-4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chelius, M. K., and E. W. Triplett. 2001. The diversity of archaea and bacteria in association with the roots of Zea mays L. Microb. Ecol. 41:252-263. [DOI] [PubMed] [Google Scholar]
- 7.Daniels, L., N. Belay, and B. S. Rajagopal. 1986. Assimilatory reduction of sulfate and sulfite by methanogenic bacteria. Appl. Environ. Microbiol. 51:703-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeLong, E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 89:5585-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeLong, E. F. 1998. Archaeal means and extremes. Science 280:542-543. [DOI] [PubMed] [Google Scholar]
- 10.Delong, E. F. 1998. Everything in moderation: archaea as ‘non-extremophiles’. Curr. Opin. Genet. Dev. 8:649-654. [DOI] [PubMed] [Google Scholar]
- 11.DeLong, E. F., and N. R. Pace. 2001. Environmental diversity of bacteria and archaea. Syst. Biol. 50:470-478. [PubMed] [Google Scholar]
- 12.DeLong, E. F., K. Y. Wu, B. B. Prezelin, and R. V. M. Jovine. 1994. High abundance of archaea in Antarctic marine picoplankton. Nature 371:695-697. [DOI] [PubMed] [Google Scholar]
- 13.Felsenstein, J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17:368-376. [DOI] [PubMed] [Google Scholar]
- 14.Ferré, F. 1992. Quantitative or semi-quantitative PCR: reality versus myth. PCR Methods Appl. 2:1-9. [DOI] [PubMed] [Google Scholar]
- 15.Fuhrman, J. A., K. McCallum, and A. A. Davis. 1992. Novel major archaebacterial group from marine plankton. Nature 356:148-149. [DOI] [PubMed] [Google Scholar]
- 16.Galtier, N., N. Tourasse, and M. Gouy. 1999. Relationships between genomic G+C content, RNA secondary structures, and optimal growth temperature in prokaryotes. Science 283:220-221. [DOI] [PubMed] [Google Scholar]
- 17.Giovannoni, S. J., T. B. Britschgi, C. L. Moyer, and K. G. Field. 1990. Genetic diversity in Sargasso Sea bacterioplankton. Nature 345:60-63. [DOI] [PubMed] [Google Scholar]
- 18.Girguis, P. R., V. J. Orphan, S. J. Hallam, and E. F. Delong. 2003. Growth and methane oxidation rates of anaerobic methanotrophic archaea in a continuous-flow bioreactor. Appl. Environ. Microbiol. 69:5472-5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Groβkopf, R., S. Stubner, and W. Liesack. 1998. Novel euryarchaeotal lineages detected on rice roots and in the anoxic bulk soil of flooded rice microcosms. Appl. Environ. Microbiol. 64:4983-4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayashi, K. 1991. PCR-SSCP: a simple and sensitive method for detection of mutations in the genomic DNA. PCR Methods Appl. 1:34-38. [DOI] [PubMed] [Google Scholar]
- 21.Hein, I., A. Lehner, P. Rieck, K. Klein, E. Brandl, and M. Wagner. 2001. Comparison of different approaches to quantify Staphylococcus aureus cells by real-time quantitative PCR and application of this technique for examination of cheese. Appl. Environ. Microbiol. 67:3122-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hershberger, K. L., S. M. Barns, A. L. Reysenbach, S. C. Dawson, and N. R. Pace. 1996. Wide diversity of Crenarchaeota. Nature 384:420. [DOI] [PubMed] [Google Scholar]
- 23.Huber, R., J. K. Kristjansson, and K. O. Stetter. 1987. Pyrobaculum gen. nov., a new genus of neutrophilic, rod-shaped archaebacteria from continental solfataras growing optimally at 100°C. Arch. Microbiol. 149:95-101. [Google Scholar]
- 24.Huber, T., G. Faulkner, and P. Hugenholtz. 2004. Bellerophon; a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317-2319. [DOI] [PubMed] [Google Scholar]
- 25.Janssen, P. H., P. S. Yates, B. E. Grinton, P. M. Taylor, and M. Sait. 2002. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl. Environ. Microbiol. 68:2391-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joseph, S. J., P. Hugenholtz, P. Sangwan, C. A. Osborne, and P. H. Janssen. 2003. Laboratory cultivation of widespread and previously uncultured soil bacteria. Appl. Environ. Microbiol. 69:7210-7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karner, M. B., E. F. DeLong, and D. M. Karl. 2001. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409:507-510. [DOI] [PubMed] [Google Scholar]
- 28.Kolb, S., C. Knief, S. Stubner and R. Conrad. 2003. Quantitative detection of methanotrophs in soil by novel pmoA-targeted real-time PCR assays. Appl. Environ. Microbiol. 69:2423-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, New York, N.Y.
- 30.Larsen, N., G. J. Olsen, B. L. Maidak, M. J. McCaughey, R. Overbeek, T. J. Maeke, T. L. Marsh, and C. R. Woese. 1993. The ribosomal database project. Nucleic Acids Res. 21:3021-3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30a.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, St. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermannl, R. Jost, A. König, T. Liss, R. Lübmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Massana, R., L. T. Taylor, A. E. Murray, K. Y. Wu, W. H. Jeffrey, and E. F. DeLong. 1998. Vertical distribution and phylogenetic characterization of marine planktonic Archaea in the Santa Barbara channel. Limnol. Oceanogr. 43:607-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morrison, T., J. J. Weis, and C. T. Wittwer. 1998. Quantification of low-copy transcripts by continuous SYBR Green I monitoring during amplification. Biotechniques 24:954-962. [PubMed] [Google Scholar]
- 33.Nicol, G. W., L. A. Glover, and J. I. Prosser. 2003. The impact of grassland management on archaeal community structure in upland pasture rhizophere soil. Environ. Microbiol. 5:152-162. [DOI] [PubMed] [Google Scholar]
- 34.Ochsenreiter, T., D. Selezi, A. Quaiser, L. Bonch-Osmolovskaya, and C. Schleper. 2004. Diversity and abundance of Crenarchaeota in terrestrial habitats studied by 16S RNA surveys and real time PCR. Environ. Microbiol. 5:787-797. [DOI] [PubMed] [Google Scholar]
- 35.Olsen, G. J., H. Matsuda, R. Hagstrom, and R. Overbeek. 1994. fastDNAmL: a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Comput. Appl. Biosci. 10:41-48. [DOI] [PubMed] [Google Scholar]
- 36.Orita, M., H. Iwahana, H. Kanazawa, K. Hayashi, and T. Sekiya. 1989. Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc. Natl. Acad. Sci. USA 86:2766-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pfaffl, M. W. 2001. Development and validation of an externally standardised quantitative insulin like growth factor-1 (IGF-1) RT-PCR using LightCycler SYBR Green I technology, p. 281-291. In S. Meuer, C. Wittwer, and K. Nakagawara (ed.), Rapid cycle real-time PCR, methods and applications. Springer Press, Heidelberg, Germany.
- 38.Preston, C. M., K. Y. Wu, T. F. Molinski, and E. F. DeLong. 1996. A psychrophilic crenarchaeon inhabits a marine sponge: Crenarchaeum symbiosum gen. nov., sp. nov. Proc. Natl. Acad. Sci. USA 93:6241-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rasmussen, R. 2001. Quantification on the LightCycler, p. 21-34. In S. Meuer, C. Wittwer, and K. Nakagawara (ed.), Rapid cycle real-time PCR, methods and applications. Springer Press, Heidelberg, Germany.
- 40.Repaske, R. 1956. Lysis of gram-negative bacteria by lysozyme. Biochim. Biophys. Acta 22:189-191. [DOI] [PubMed] [Google Scholar]
- 41.Rondon, M. R., S. J. Raffel, R. M. Goodman, and J. Handelsman. 1999. Toward functional genomics in bacteria: analysis of gene expression in Escherichia coli from a bacterial artificial chromosome library of Bacillus cereus. Proc. Natl. Acad. Sci. USA 96:6451-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.SAS Institute. 1985. SAS user's guide, version 5. SAS Institute, Cary, N.C.
- 43.Schaefer, D. M., C. L. Davis, and M. P. Bryant. 1980. Ammonia saturation constants for predominant species of rumen bacteria. J. Dairy Sci. 63:1248-1263. [DOI] [PubMed] [Google Scholar]
- 44.Simon, H. M., J. A. Dodsworth, and R. M. Goodman. 2000. Crenarchaeota colonize terrestrial plant roots. Environ. Microbiol. 2:506-515. [DOI] [PubMed] [Google Scholar]
- 45.Sinninghe Damsté, J. S., W. I. C. Rijpstra, E. C. Hopmans, F. G. Prahl, S. G. Wakeham, and S. Schouten. 2002. Distribution of membrane lipids of planktonic Crenarchaeota in the Arabian Sea. Appl. Environ. Microbiol. 68:2997-3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sliwinski, M. K., and R. M. Goodman. 2004. Spatial heterogeneity of crenarchaeal assemblages within mesophilic soil ecosystems as revealed by PCR-single-stranded conformation polymorphism profiling. Appl. Environ. Microbiol. 70:1811-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sliwinski, M. K., and R. M. Goodman. 2004. Comparison of crenarchaeal consortia inhabiting the rhizosphere of diverse terrestrial plants with those in bulk soil in native environments. Appl. Environ. Microbiol. 70:1821-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stahl, D. A., and R. I. Amann. 1991. Development and application of nucleic acid probes in bacterial systematics, p. 205-248. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, New York, N.Y.
- 49.Stein, J. L., and M. I. Simon. 1996. Archaeal ubiquity. Proc. Natl. Acad. Sci. USA. 93:6228-6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stevenson, B. S., S. A. Eichorst, J. T. Wertz, T. M. Schmidt, and J. A. Breznak. 2004. New strategies for cultivation and detection of previously uncultured microbes. Appl. Environ. Microbiol. 70:4748-4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ueda, T., Y. Suga, and T. Matsuguchi,. 1995. Molecular phylogenetic analysis of a soil microbial community in a soybean field. Eur. J. Soil Sci. 46:415-421. [Google Scholar]
- 52.Watrin, L., E. Corre, and D. Prieur. 1996. In vivo susceptibility of sulfothermophilic archaea to antimicrobial agents. J. Mar. Biotechnol. 4:215-219. [Google Scholar]
- 53.Whipps, J. M. 1990. Carbon economy, p. 59-97. In J. M. Lynch (ed.), The rhizosphere. John Wiley and Sons, Chichester, United Kingdom.
- 54.Whipps, J. M., and J. M. Lynch. 1985. Energy losses by the plant in rhizodeposition. Annu. Proc. Phytochem. Soc. 26:59-71. [Google Scholar]
- 55.Zillig, W., K. O. Stetter, W. Schulz, and D. Janekovic. 1980. Comparative studies of structure and function of DNA-dependent RNA polymerases from eubacteria and archaebacteria, p. 159-178. In P. Mildner and B. Rieds (ed.), Enzyme regulation and mechanism of action. Pergamon Press, Oxford, United Kingdom.