Abstract
Two genes, bshA and bshB, encoding bile salt hydrolase enzymes (EC 3.5.1.24) were identified in the genome sequence of Lactobacillus acidophilus NCFM. Targeted inactivation of these genes via chromosomal insertion of an integration vector demonstrated different substrate specificities for these two enzymes.
In humans and other mammals, primary bile salts are produced de novo in the liver from cholesterol (4). The steroid portion of the molecule is conjugated with an amide bond at the C-24 position to one of two amino acids, taurine or glycine. Following manufacture, conjugated bile salts are stored in the gall bladder and secreted via the bile duct into the small intestine. Here, these conjugates form spontaneous micelles that trap dietary cholesterol and fats, thus facilitating their absorption by the intestinal epithelium into the bloodstream (25). While more than 95% of bile salts enter the enterohepatic circulation in humans (17), up to 650 mg of bile salts per day elude absorption through the intestinal epithelium. Thus, high concentrations of these conjugates are present in the gastrointestinal tract.
Certain species of the indigenous microflora, including a number of lactobacilli and bifidobacteria, have evolved the ability to deconjugate bile salts. This action is dependent on the presence of an enzyme known as bile salt hydrolase (BSH; cholylglycine hydrolase; EC 3.5.1.24) that catalyzes the hydrolysis of glycine- and/or taurine-conjugated bile salts into the amino acid residue and the bile acid (13). Several theories have been proposed for the ecological significance of BSH activity in these organisms. These include their ability to use free amino acids as electron acceptors to obtain energy under anaerobic conditions (19, 35) and self-protection against the toxic effects of bile salts (32). However, studies on the impact of BSH-producing organisms in the colonized host have produced much conflicting evidence. Observations that a reduction in the levels of serum cholesterol is associated with the presence of BSH-producing organisms has led to increased interest in the possibility of their use in hypercholesterolemic individuals or to prevent elevated cholesterol levels in individuals with normal cholesterol status (8). Conversely, negative effects have also been reported including cases of contaminated small bowel syndrome, impaired lipid absorption, gallstone formation, and increased risk of colon cancer (25).
Lactobacillus acidophilus NCFM is a human isolate used commercially for over 25 years as a probiotic culture (31). The organism has the ability to survive in the gastrointestinal tract (31, 33), adhere to human epithelial cells in vitro (16), utilize fructooligosaccharides (3), modulate the host immune response, and prevent microbial gastroenteritis (36). Analysis of the NCFM genome sequence revealed the presence of two putative bile salt hydrolase genes (2). The bile-hydrolyzing capability associated with L. acidophilus NCFM had been previously identified by phenotypic screen (D.C. Walker, unpublished results). Due to the implications of the presence of bile salt hydrolase in several probiotic strains, this study was designed to further characterize this activity in NCFM through targeted gene inactivation.
Genomic analysis of the bsh loci.
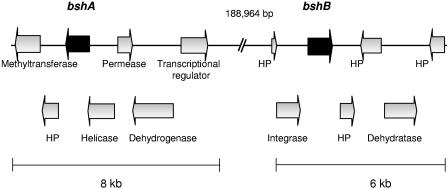
To identify the molecular foundations of the relationship between probiotic organisms and their hosts, genetic characterization of strains is essential (26). Thus, L. acidophilus NCFM was chosen for whole-genome sequencing. The 2.0-Mb genome sequence of L. acidophilus NCFM has recently been elucidated (2) and contains 1,864 predicted protein-encoding genes. Sequence analysis was performed using a combination of GCG (version 9.1; University of Wisconsin Genetics Computer Group), Clone Manager (version 6.0; SciEd Central) and GAMOLA (1). Protein homology searches were performed with the Basic Local Alignment Search Tool (PSI-BLAST), version 2.2, at the website of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov). Screening of early drafts of the genome sequence led to the identification of an open reading frame (ORF) whose predicted protein sequence showed significant similarity to BSH enzymes found in a number of other organisms (Table 1). These included Lactobacillus johnsonii (82%) (29), Listeria monocytogenes (65%) (9), and Bifidobacterium longum (54%) (34). This gene was designated bshA. Analysis of the genomic organization of the region surrounding bshA revealed the presence of a number of ORFs with homology to DNA helicases, transporters, and hypothetical proteins (Fig. 1). A promoter-type structure and ribosomal binding site preceding bshA were identified, as well as a putative terminator (ΔG = −24.7 kcal) downstream, suggesting that bshA is transcribed independently of the surrounding genes. Subsequent sequencing, leading to improved genome assemblies, resulted in the identification of another ORF, designated bshB, the putative gene product of which displayed 57% identity to that of bshA. Again, a ribosomal binding site and putative promoter were identified within the genome structure; a potential terminator (ΔG = −21.4 kcal) downstream also indicates that bshB is most likely to be monocistronic. Genes in the vicinity of bshB were predominantly similar to genes encoding hypothetical proteins of unknown function (Fig. 1). No other bsh homologues were found in the L. acidophilus NCFM genome. The GenBank accession number for the L. acidophilus NCFM genome sequence from which the bshA and bshB genes are derived is CP000033.
TABLE 1.
Comparison of amino acid sequences of some bacterial BSH enzymes
| Bacterial strain/peptide | Accession no. | BshA
|
BshB
|
Reference(s)a | ||
|---|---|---|---|---|---|---|
| % Identity | % Similarity | % Identity | % Similarity | |||
| NCFM BshA | CP000033 | 100 | 100 | 57 | 57 | 2 |
| L. johnsonii NCC533 (1147) | NC_005362 | 69 | 82 | 55 | 72 | 29 |
| Lactobacillus gasseri ADH | AAK07837 | 65 | 77 | 56 | 73 | 30 |
| L. gasseri | ZP_00046418 | 65 | 77 | 57 | 73 | DS* |
| L. johnsonii NCC533 (1412) | NC_005362 | 61 | 79 | 57 | 76 | 29 |
| L. johnsonii 100-100 alpha | AAG22541 | 61 | 79 | 57 | 76 | 10, 11 |
| Enterococcus faecium FAIR-E 345 | AY260046 | 50 | 67 | 57 | 73 | DS* |
| L. monocytogenes EGD-e | NP_465591 | 47 | 65 | 56 | 74 | 9 |
| E. faecalis V583 | NC_004668 | 49 | 68 | 55 | 73 | 28 |
| Lactobacillus plantarum WCFS1 | NC_004567 | 46 | 65 | 52 | 71 | 23 |
| L. plantarum 80 | Q06115 | 46 | 64 | 52 | 71 | 7 |
| Clostridium perfringens | NP_561625 | 37 | 56 | 38 | 56 | 5 |
| B. longum SBT2928 | AAF67801 | 35 | 54 | 37 | 54 | 34 |
| L. johnsonii 100-100 beta | AAC34381 | 35 | 52 | 33 | 54 | 10, 24 |
| Bifidobacterium bifidum ATCC11863 | AY506536 | 34 | 55 | 37 | 55 | 22 |
| L. acidophilus KS-13 | AAD03709 | 34 | 51 | 32 | 53 | DS* |
*, direct submission to the National Center for Biotechnology Information.
FIG. 1.
Schematic representation of the genetic organization of the bsh loci of L. acidophilus NCFM. Arrows indicate ORFs and the directions of transcription; bsh genes are represented by solid black arrows. Annotations of flanking genes are also indicated. HP denotes a hypothetical protein of unknown function. This representation is not drawn to scale.
Construction of bsh mutants.
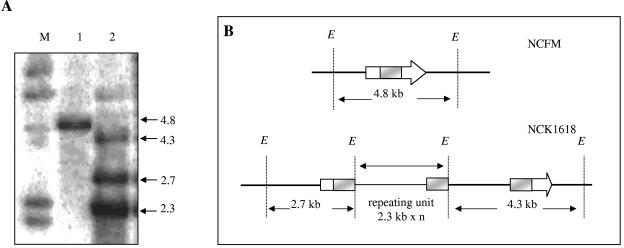
The directed integration system described previously by Russell and Klaenhammer (30) was used to inactivate first bshA and then bshB in the genome of L. acidophilus NCFM, creating two separate mutants. Primers were designed to amplify a 588-bp internal region of bshA (forward primer, 5′-AAA GTC GAC GAA AAG GGG CTT GGT A-3′; reverse primer, 5′-AAG AAT TCC CAT CAG GTT GTT CTA C-3′). The underlined restriction sites were used to clone the amplified product into the Ori+ RepA− integration plasmid, pORI28. The resultant plasmid, pTRK734, was transformed into L. acidophilus NCFM containing pTRK669, a temperature-sensitive helper plasmid that provides repA in trans for the replication of pORI28. A temperature increase from 37°C to 42°C resulted in the integration of pTRK734 into the NCFM genome, concurrent with the loss of pTRK669 and its associated Cmr phenotype at the nonpermissive temperature. To confirm the integration of pTRK734 at the correct genome locus, Southern hybridizations were performed using the 588-bp fragment, labeled with nonradioactive digoxigenin (DIG) (Roche Diagnostics Corporation, Indianapolis, IN), as a probe. The bshA probe hybridized to a 4.8-kb EcoRI fragment in the wild type (Fig. 2A). In the mutant NCFMΔbshA (or NCK1618), this band was absent, and due to the presence of a single EcoRI site in the integration vector sequence, junction fragments of approximately 4.3 kb and 2.7 kb were observed (Fig. 2A). This genome structure is indicative of the occurrence of a single crossover homologous recombination event (Fig. 2B). An additional band of 2.3 kb was detected in the mutant (Fig. 2); this fragment corresponds to the integration vector, pTRK734, which had amplified and was present in more than one copy. In the case of bshB, a 618-bp internal fragment was amplified (forward primer, 5′-AGG ATC CAG TTA GTT CCA TCA GAA TA-3′; reverse primer, 5′-TAT AAG CTT GGT ATG GCC GGA CTC AAC-3′) and cloned into pORI28 to create pTRK735 and a similar strategy adopted for inactivation. The integration event was checked and verified in a manner similar to that shown in Fig. 2 for the bshA mutant. The bshB mutant was designated NCFMΔbshB (or NCK1619).
FIG. 2.
(A) Southern hybridization analysis of L. acidophilus NCFM and NCK1618. Chromosomal DNA was digested with EcoRI (lanes 1 and 2) and hybridized with the DIG-labeled 588-bp internal bshA fragment as a probe. Lanes: M, DIG-labeled λ-HindIII marker (Roche Molecular Biochemicals); 1, NCFM; 2, NCK1618. DNA sizes are indicated in kilobases. (B) Schematic representation of the relevant regions of the NCFM and NCFMΔbshA (NCK1618) chromosomes. Heavy lines represent chromosomal DNA; thin lines represent plasmid DNA. bshA is represented by the arrow, and the shaded box is the internal fragment corresponding to the hybridization probe. EcoRI sites (E) are indicated.
Determination of bile tolerance.
Bile tolerance is an important criterion in the selection of probiotic strains. Previous reports have suggested that a concentration of 0.3% oxgall (Sigma Chemical Co., St. Louis, MO) closely approximates the bile levels found in the gastrointestinal tract (14, 15). To evaluate their ability to grow in the presence of bile, the parent, NCFMΔbshA, and NCFMΔbshB were screened for growth on MRS plates or in MRS broth supplemented with concentrations of oxgall up to 0.5%. Typically, Lactobacillus spp. do not grow at levels higher than 0.3% (20, 21). When incubated under anaerobic conditions, the parent was capable of growth in up to 0.25% oxgall, as were both NCFMΔbshA and NCFMΔbshB (data not shown). At concentrations of 0.3% and higher, no growth was observed for any strain, either in broth or on plates. This indicated that inactivation of one or other of the bile salt hydrolase enzymes of L. acidophilus NCFM did not affect its ability to grow in the presence of bile.
Detection of BSH activity.
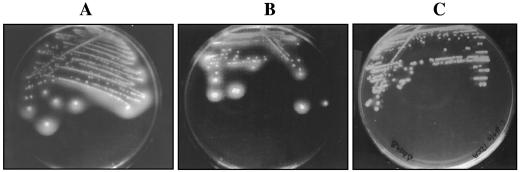
A direct plate assay for detection of BSH activity was employed to compare the NCFMΔbshA and NCFMΔbshB mutants with the wild-type strain. A number of glycine- and taurine-conjugated bile salts were selected for the assays; these are listed in Table 2. By the inclusion of bile salts in agar medium, BSH-positive strains can be identified by halos of precipitated free bile acids surrounding the colonies due to hydrolysis and acidification of the medium. The assay used in this study is a modified version of that developed by Dashkevicz and Feighner (6); in our assays, the upper concentration of bile salts used was reduced from 0.5% to 0.2% due to the inability of L. acidophilus NCFM to grow at 0.5%. In the cases of taurochenodeoxycholic acid (TCDCA) and glycochenodeoxycholic acid (GCDCA), this concentration was reduced further to 0.02 to 0.05%. For the wild-type strain, hydrolysis of taurodeoxycholic acid (TDCA) (Table 2 and Fig. 3A) and taurocholic acid (TCA) (Table 2) under anaerobic conditions resulted in significant amounts of deoxycholic acid precipitating around active colonies. Similar levels of precipitation were observed surrounding colonies of NCFMΔbshA when plated on TDCA (Table 2 and Fig. 3B) and TCA (Table 2), indicating that this mutant had retained the ability to hydrolyze these deoxy-conjugated compounds. Furthermore, NCFMΔbshA retained the ability to hydrolyze the glycine-conjugated compounds such as glycodeoxycholic acid and glycocholic acid. Hydrolysis of TCDCA and GCDCA by L. acidophilus NCFM was observed as a cloudiness in the agar rather than as distinct zones of precipitation observed for the other bile salts. However, while both the parent and mutant grew, albeit slowly, on 0.05% TCDCA and GCDCA, NCFMΔbshA appeared to have lost the ability to hydrolyze both salts (Table 2), as indicated by the lack of precipitation in the agar. Interestingly, when both TCDCA and GCDCA concentrations in the agar were further reduced to 0.02%, NCFMΔbshA showed some signs of BSH activity (Table 2). The creation of a ΔbshB mutant allowed the activities of BshB to be assessed in a similar manner. Comparisons with the parent strain demonstrated that while NCFMΔbshB displayed growth on and precipitation of all glycoconjugates tested, this mutant was capable of growth but incapable of precipitation of the tauroconjugated bile salts, TCA, TDCA, and TCDCA (Table 2 and Fig. 3C).
TABLE 2.
Analysis of L. acidophilus NCFM and bsh mutants on different bile saltsa
| Bile salts (%)b | Value (%) for indicated strain
|
|||||
|---|---|---|---|---|---|---|
| NCFM
|
NCFMΔbshA
|
NCFMΔbshB
|
||||
| Growth | Precipitation | Growth | Precipitation | Growth | Precipitation | |
| TDCA (0.2) | 100 | 100 | 100 | 100 | 100 | 0 |
| TCA (0.2) | 100 | 60 | 100 | 60 | 100 | 0 |
| TCDCA (0.05) | 100 | 100 | 100 | 0 | 5 | 0 |
| TCDCA (0.02) | 100 | 100 | 100 | 5 | 5 | 0 |
| GDCA (0.2) | 60 | 60 | 60 | 60 | 60 | 60 |
| GCA (0.2) | 100 | 100 | 100 | 100 | 100 | 100 |
| GCDCA (0.05) | 30 | 30 | 30 | 0 | 30 | 30 |
| GCDCA (0.02) | 30 | 30 | 30 | 5 | 30 | 30 |
Numeric values represent an arbitrary scoring system; levels of growth and precipitation are scored relative to NCFM on 0.2% TDCA, which is given a score of 100%.
GDCA, glycodeoxycholic acid; GCA, glycocholic acid.
FIG. 3.
Plate assays for the detection of BSH activity. (A) L. acidophilus NCFM; (B) NCFMΔbshA (NCK1618); (C) NCFMΔbshB (NCK1619) on MRS agar containing 0.2% TDCA. In the assays shown, precipitation in the agar is indicative of BSH activity specific for TDCA.
Thus, to summarize, while NCFMΔbshA was capable of the hydrolysis of some tauro- and glycoconjugated bile salts, this mutant had a reduced ability to hydrolyze TCDCA and GCDCA, bile salts containing chenodeoxycholic acid as the steroid moiety. Conversely, inactivation of bshB revealed that the BshB enzyme encoded by this gene appears to exhibit substrate specificity dictated by the amino acid conjugated to the bile salt. This conclusion was made based on the inability of NCFMΔbshB to hydrolyze any bile salt conjugated to taurine. Previous studies have demonstrated that the specificity of the catalytic activities of BSH enzymes may be influenced either by the amino acid in the conjugate or by other side chains in the steroid moiety (13, 18, 27). Our results suggest that BshA activity is dictated by the steroid nucleus of the conjugated bile salt, while the specificity of BshB is determined by the presence of taurine in the bile salt structure.
To our knowledge, this is the first report of the presence of two BSH enzymes with different substrate specificities in a single strain of Lactobacillus. Although BSH production is not an essential attribute for organisms that colonize the gastrointestinal tract, its importance in colonizing species of Lactobacillus is highlighted by the high level of conservation between the various enzymes indicated in Table 1. However, after a comparison of both predicted protein sequences to the deduced amino acid sequences of other BSH enzymes, it was interesting that they both share a higher level of identity to enzymes from other Lactobacillus species than to each other. BshA shares 69% identity with the enzyme from L. johnsonii NCC533 (29), while BshB is more similar (57% identity; 76% similarity) to that of BSH alpha from L. johnsonii 100-100 (11). No such identity was observed at the DNA level. However, the G+C contents of both genes are similar to those of other genes in the L. acidophilus NCFM genome. Therefore, it is possible that bshA and bshB may have originated from different sources through horizontal gene transfer from a closely related genome, but we have no tangible evidence for this at the present time. Another possibility is that these genes may have arisen from a duplication event, following which the genes have subsequently diverged to encode enzymes with different substrate specificities. However, since the proteins share only 57% identity, this scenario is unlikely.
While the evolution of BSH enzymes with different activities in L. acidophilus NCFM is striking, the significance of BSH activity in lactobacilli is still far from understood. Since the free bile acids appear to be more inhibitory than the conjugated bile salts themselves, it is most likely not a detoxification mechanism for these organisms. While some positive effects of the consumption of BSH-producing probiotics have been documented, the effects are often transient and unsustainable. Generally, human studies have yielded mixed results with no clear-cut reduction in cholesterol observed due to probiotic consumption (12). It seems that the debate will continue as to whether or not this enzyme activity is a desirable property in probiotic bacteria.
Acknowledgments
We thank Mike Russell for his technical assistance and Eric Altermann for his help with sequence analysis. Raul Cano provided DNA sequence information prior to publication.
This research was funded by the North Carolina Dairy Foundation, Danisco, Inc. (Madison, WA), the Southeast Dairy Foods Research Center, and Dairy Management, Inc., the California Dairy Research Foundation, and The Environmental Biotechnology Institute.
REFERENCES
- 1.Altermann, E., and T. R. Klaenhammer. 2003. GAMOLA: a new local solution for sequence annotation and analyzing draft and finished genomes. OMICS 7:161-169. [DOI] [PubMed] [Google Scholar]
- 2.Altermann, E., W. M. Russell, M. A. Azcarate-Peril, R. Barrangou, B. L. Buck, O. McAuliffe, N. Souther, A. Dobson, T. Duong, M. Callanan, S. Lick, A. Hamrick, R. Cano, and T. R. Klaenhammer. 2005. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc. Natl. Acad. Sci. USA 102:3609-3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrangou, R., E. Altermann, R. Hutkins, R. Cano, and T. R. Klaenhammer. 2003. Functional and comparative genomic analyses of an operon involved in fructooligosaccharide utilization by Lactobacillus acidophilus. Proc. Natl. Acad. Sci. USA 100:8957-8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bortolini, O., A. Medici, and S. Poli. 1997. Biotransformations on steroid nucleus of bile acids. Steroids 62:567-577. [DOI] [PubMed] [Google Scholar]
- 5.Coleman, J. P., and L. L. Hudson. 1995. Cloning and characterization of a conjugated bile acid hydrolase gene from Clostridium perfringens. Appl. Environ. Microbiol. 61:2514-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dashkevicz, M. P., and S. D. Feighner. 1989. Development of a differential medium for bile salt hydrolase active Lactobacillus spp. Appl. Environ. Microbiol. 55:11-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeBoever, P., and W. Verstraete. 1999. Bile salt deconjugation by Lactobacillus plantarum 80 and its implications for bacterial toxicity. J. Appl. Microbiol. 87:345-352. [DOI] [PubMed] [Google Scholar]
- 8.DeSmet, I., L. Van Hoorde, M. De Saeyer, M. Vande Woestyne, and W. Verstraete. 1994. In vitro study of bile salt hydrolase (BSH) activity of BSH isogenic Lactobacillus plantarum 80 strains and estimation of cholesterol lowering through enhanced BSH activity. Microb. Ecol. Health Dis. 7:315-329. [Google Scholar]
- 9.Dussurget, O., D. Cabanes, P. Dehoux, M. Lecuit, the European Listeria Genome Consortium, C. Buchrieser, P. Glaser, and P. Cossart. 2002. Listeria monocytogenes bile salt hydrolase is a PrfA-regulated virulence factor involved in the intestinal hepatic phases of listeriosis. Mol. Microbiol. 45:1095-1106. [DOI] [PubMed] [Google Scholar]
- 10.Elkins, C. A., and D. C. Savage. 1998. Identification of genes encoding conjugated bile salt hydrolase and transport in Lactobacillus johnsonii 100-100. J. Bacteriol. 180:4344-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elkins, C. A., S. A. Moser, and D. C. Savage. 2001. Genes encoding bile salt hydrolases and conjugated bile salt transporters in Lactobacillus johnsonii 100-100 and other Lactobacillus species. Microbiology 147:3404-3412. [DOI] [PubMed] [Google Scholar]
- 12.Gardiner, G. E., R. P. Ross, P. M. Kelly, and C. Stanton. 2002. Microbiology of therapeutic milks, p. 431-478. In R. K. Robinson (ed.), Dairy microbiology handbook—the microbiology of milk and milk products. Wiley and Sons, Inc., New York, N.Y.
- 13.Gilliland, S. E., and M. L. Speck. 1977. Deconjugation of bile acids by intestinal lactobacilli. Appl. Environ. Microbiol. 33:15-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilliland, S. E., T. E. Stanley, and L. J. Bush. 1984. Importance of bile tolerance of Lactobacillus acidophilus used as a dietary adjunct. J. Dairy Sci. 67:3045-3051. [DOI] [PubMed] [Google Scholar]
- 15.Goldin, B. R., and S. L. Gorbach. 1992. Probiotics for humans, p. 355-376. In R. Fuller (ed.), Probiotics, the scientific basis. Chapman and Hall, London, United Kingdom.
- 16.Greene, J. D., and T. R. Klaenhammer. 1994. Factors involved in adherence to lactobacilli in human Caco-2 cells. Appl. Environ. Microbiol. 60:4487-4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hofmann, A. F. 1989. Concepts of biliary secretion. Dig. Dis. Sci. 34:16S-20S. [DOI] [PubMed] [Google Scholar]
- 18.Huijghebaert, S. M., and A. F. Hoffman. 1986. Influence of the amino acid moiety on deconjugation of bile acid amidates by cholylglycine hydrolase or human fecal cultures. J. Lipid Res. 27:742-752. [PubMed] [Google Scholar]
- 19.Huijghebaert, S. M., J. A. Mertens, and H. J. Eyssen. 1982. Isolation of a bile salt sulfatase-producing Clostridium strain from rat intestinal microflora. Appl. Environ. Microbiol. 43:185-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hyronimus, B., C. le Marrec, A. Hadj Sassi, and A. Deschamps. 2000. Acid and bile tolerance of spore-forming lactic acid bacteria. Int. J. Food Microbiol. 61:193-197. [DOI] [PubMed] [Google Scholar]
- 21.Jacobsen, C. N., V. Rosenfeldt Nielsen, A. E. Hayford, P. L. Moller, K. F. Michaelsen, A. Paerrgaard, B. Sandstrom, M. Tvede, and M. Jacobsen. 1999. Screening of probiotic activities of 47 strains of Lactobacillus spp. by in vitro techniques and evaluation of the colonization ability of 5 selected strains in humans. Appl. Environ. Microbiol. 65:4949-4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, G. B., C. M. Miyamoto, E. A. Meighen, and B. H. Lee. 2004. Cloning and characterisation of the bile salt hydrolase gene from Bifidobacterium bifidum strains. Appl. Environ. Microbiol. 70:5603-5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kleerebezem, M., J. Boekhorst, R. van Kranenburg, D. Molenaar, O. P. Kuipers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. W. Fiers, W. Stiekema, R. M. Lankhorst, P. A. Bron, S. M. Hoffer, M. N. Groot, R. Kerkhoven, M. de Vries, B. Ursing, W. M. de Vos, and R. J. Siezen. 2004. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 100:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lundeen, S. G., and D. C. Savage. 1992. Multiple forms of bile salt hydrolase from Lactobacillus sp. strain 100-100. J. Bacteriol. 174:7217-7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marteau, P., M. F. Gerhardt, A. E. MyaraBouvier, F. Trivin, and J. C. Rambaud. 1995. Metabolism of bile salts by alimentary bacteria during transit in the human small intestine. Microb. Ecol. Health Dis. 8:151-157. [Google Scholar]
- 26.McAuliffe, O. E., and T. R. Klaenhammer. 2002. Genomic perspectives on probiotics and the gastrointestinal microflora. In G. W. Tannock (ed.), Probiotics and prebiotics: where are we going? Caister Academic Press, Norfolk, United Kingdom.
- 27.Moser, S. A., and D. C. Savage. 2001. Bile salt hydrolase activity and resistance to toxicity of conjugated bile salts are unrelated properties in lactobacilli. Appl. Environ. Microbiol. 67:3476-3480. [DOI] [PMC free article] [PubMed]
- 28.Paulsen, I., L. Banerjei, G. S. A. Myers, K. E. Nelson, R. Seshadri, T. D. Read, D. E. Fouts, J. A. Eisen, S. R. Gill, J. F. Heidelberg, H. Tettelin, R. J. Dodson, L. Umayam, L. Brinkac, M. Beanan, S. Daugherty, R. T. DeBoy, S. Durkin, J. Kolonay, R. Madupu, W. Nelson, J. Vamathevan, B. Tran, J. Upton, T. Hansen, J. Shetty, H. Khouri, T. Utterback, D. Radune, K. A. Ketchum, B. A. Dougherty, and C. M. Fraser. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071-2074. [DOI] [PubMed] [Google Scholar]
- 29.Pridmore, R. D., B. Berger, F. Desiere, D. Vilanova, C. Barretto, A. C. Pittet, M. C. Zwahlen, M. Rouvet, E. Altermann, R. Barrangou, B. Mollet, A. Mercenier, T. R. Klaenhammer, F. Arigoni, and M. A. Schell. 2004. The genome sequence of the probiotic intestinal bacterium Lactobacillus johnsonii NCC 533. Proc. Natl. Acad. Sci. USA 101:2512-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Russell, W. M., and T. R. Klaenhammer. 2001. Identification and cloning of gusA, encoding a new β-glucuronidase from Lactobacillus gasseri ADH. Appl. Environ. Microbiol. 67:1253-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanders, M. E., and T. R. Klaenhammer. 2001. Invited review: the scientific basis of Lactobacillus acidophilus NCFM functionality as a probiotic. J. Dairy Sci. 84:319-331. [DOI] [PubMed] [Google Scholar]
- 32.Savage, D. C. 1992. Gastrointestinal microbial ecology: possible modes of action of direct-fed microbials in animal production—a review of the literature, p. 11-81. In Direct-fed microbials in animal production. National Feed Ingredients Association, West Des Moines, Iowa.
- 33.Sui, J., S. Leighton, F. Busta, and L. Brady. 2002. 16S ribosomal DNA analysis of the faecal lactobacilli composition of human subjects consuming a probiotic strain Lactobacillus acidophilus NCFM. J. Appl. Microbiol. 92:907-912. [DOI] [PubMed] [Google Scholar]
- 34.Tanaka, H., H. Hashiba, J. Kok, and I. Mierau. 2000. Bile salt hydrolase of Bifidobacterium longum—biochemical and genetic characterization. Appl. Environ. Microbiol. 66:2502-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Eldere, J., J. Robben, G. De Pauw, R. Merckx, and H. Eyssen. 1988. Isolation and identification of intestinal steroid-desulfating bacteria from rats and humans. Appl. Environ. Microbiol. 54:2112-2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Varcoe, J. J., G. Krejcarek, F. Busta, and L. Brady. 2003. Prophylactic feeding of Lactobacillus acidophilus NCFM to mice attenuates overt colonic hyperplasia. J. Food Prot. 66:457-465. [DOI] [PubMed] [Google Scholar]