Abstract
Toluene degradation in Pseudomonas putida KT2440 pWW0 plasmid is subjected to catabolite repression. Pu and PS1 promoters of the pWW0 TOL plasmid are down-regulated in vivo during exponential growth in rich medium. In cells growing on minimal medium, yeast extract (YE) addition mimics exponential-phase rich medium repression of these promoters. We have constructed and tested mutants in a series of global regulators described in Pseudomonas. We describe that a mutant in crc (catabolite repression control) partially relieves YE repression. Macroarray experiments show that crc transcription is strongly increased in the presence of YE, inversely correlated with TOL pathway expression. On the other hand, we have found that induced levels of expression from Pu and PS in the presence of YE are partially derepressed in a ptsN mutant of P. putida. PtsN but not Crc seems to directly interfere with XylR activation at target promoters. The effect of the double mutation in ptsN and crc is not the sum of the effects of each independent mutation and suggests that both regulators are elements of a common regulatory pathway. Basal expression levels from these promoters in the absence of inducer are still XylR dependent and are also repressed in the presence of yeast extract. Neither crc nor ptsN could relieve this repression.
The TOL plasmid catabolic pathway for the degradation of toluene and xylenes is a paradigm of specific and global regulation (5, 40, 41, 42). Expression of the catabolic operons involves the TOL plasmid-encoded XylR and XylS regulators, a set of sigma factors (σ70, σ54, σ32, and σ38), and DNA-bending proteins such as integration host factor and HU. Above this interplay of specific plasmid regulators and host transcriptional factors, catabolite repression plays a key role in the control of the expression of these pathways (5, 7, 9, 16, 17, 24, 30, 31, 41, 42).
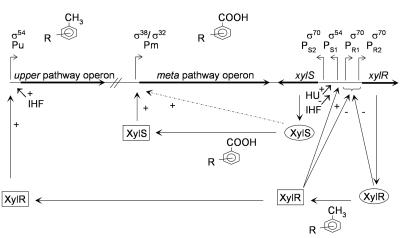
The current model of specific regulation of TOL plasmid expression is shown in Fig. 1 and can be summarized as follows: the so-called meta-regulatory loop operates when cells grow on toluates, whereas a more complex system, the cascade loop, operates when cells grow on xylenes and ensures that both the upper and the meta pathways are coordinately expressed. The master regulator involved in transcriptional control of the catabolic pathways in cells growing on xylenes is XylR. The xylR gene is transcribed from two σ70-dependent tandem promoters, PR1 and PR2. The cascade model is operational when cells grow in the presence of toluene or xylenes. In these conditions, XylR binds the aromatics and the inactive protein (XylRi) becomes activated (XylRa) to stimulate transcription from the upper pathway operon Pu promoter (1, 19, 35). This process requires σ54-containing RNA polymerase and the DNA-bending protein integration host factor (1, 37). In a similar pattern, XylRa also stimulates expression of the divergent xylS gene, normally transcribed at low levels by the constitutive PS2 promoter, by inducing transcription from a second σ54-dependent promoter, PS1 (18), a process that may be assisted by the chromatin-associated DNA-bending protein HU (36). In addition, translation of PS1 mRNA is more efficient than that of PS2 (20). As a consequence, in the presence of o-xylene, a nonmetabolizable XylR effector, the XylS protein is overproduced and transcription from the lower pathway promoter Pm is achieved even in the absence of meta pathway effectors.
FIG. 1.
TOL pathway regulatory network. The thick horizontal line depicts the TOL region including the upper and meta-cleavage pathways and the two regulatory genes XylR and XylS. Elliptical boxes indicate the inactive form of the regulatory proteins. Rectangular boxes indicate the active form of the regulatory proteins. Thin lines represent the connections between regulatory proteins and promoters, where a plus sign indicates transcription activation and a minus sign indicates inhibition of transcription. The dotted line indicates transcription activation of overproduced XylS in the absence of effector. The sigma factor(s) involved in transcription initiation is indicated above each promoter. Aromatic substrates of the pathways acting as effectors of the regulatory proteins are indicated. The regulatory circuits are explained in the text. R indicates possible substituent groups of the ring.
The PR and PS promoters are clustered in the 300-bp DNA region between the divergent xylR and xylS genes. The PS1 promoter shows the typical organization of σ54-dependent promoters. XylR upstream activator sequences (UASs) in PS1 overlap the two xylR tandem promoters so that XylR binding to its UASs to activate PS1 results in the repression of the two σ70-dependent PR promoters and consequently in its own synthesis (3, 29). The meta loop is operational when cells grow on benzoates. In these conditions, xylS is transcribed only from the constitutive, XylR-independent PS2 promoter. The basal levels of XylS protein are activated by a benzoate effector to promote transcription from Pm.
The expression of TOL plasmid operons is integrated into the overall metabolic control in Pseudomonas putida: both the upper pathway operon promoter Pu and the xylS PS1 promoter are subject to catabolite repression. As a consequence of the latter, expression of the meta-cleavage operon is also subject to moderate catabolite repression (16, 40). Early studies showed that P. putida (pWW0) cells did not express the TOL pathways during exponential growth in rich medium (24, 30), a phenomenon also referred to by Cases et al. as exponential silencing (7). These authors distinguished a second regulatory circuit based on their observation that specific carbon sources could reduce Pu activity to one-third (6). The rich medium exponential switch-off was overcome when spent Luria-Bertani (LB) medium was used instead (30). The TOL operons could be silenced in a similar way in cells growing on minimal medium with toluene if yeast extract (YE) was added (30).
Definitive proof of catabolite repression was provided by Duetz et al. (17), who showed that o-xylene-induced expression of the TOL catabolic pathways did not occur in continuous cultures growing either at a high rate under nonlimiting conditions (i.e., excess of all nutrients) or at a low rate in cultures limited in N, P, or S, conditions which all result in an excess of carbon in the medium. However, when the culture was limited in C, the operon was expressed at a high level.
Catabolite repression in Pseudomonadaceae does not involve cyclic AMP (cAMP) as in Enterobacteriaceae (41). In fact, in P. putida and P. aeruginosa, cAMP levels are relatively constant regardless of the growth conditions (39, 44). Instead, catabolite repression seems to integrate different signals, a feature which increases the complexity of the system. Up to five different potential regulators have been related to catabolite repression in P. putida, namely, Crc (34, 41), Crp, called Vfr in P. aeruginosa (45, 49), CyoB (14, 38), RelA (25, 47), and the PTS system (8-10).
Crc is responsible for the catabolic repression of a number of functions in P. aeruginosa and P. putida, such as the expression of glucose-6-phosphate dehydrogenase and amidase activities and the branched-chain keto acid dehydrogenase (23). Recently, the alkane degradation pathway encoded by the OCT plasmid from P. putida GPo1 has also been shown to be under the control of Crc (51). Although the molecular mechanism underlying Crc activity is unknown, available data suggest that Crc is a component of a signal transduction pathway that modulates carbon metabolism in Pseudomonas.
P. aeruginosa vfr gene encodes a regulator homologous to Crp able to bind cAMP. Proteome analysis showed that the synthesis of at least 60 proteins is affected in a P. aeruginosa vfr mutant, confirming the role of this protein as a global regulator. However, vfr was not required for catabolite repression control in this strain (45). The cyoB gene codes for a subunit of one of the terminal oxidases in the branched respiratory system of Pseudomonas. It has been involved in catabolite repression of the phenol pathway of P. putida (38) and the alkane degradation pathway of P. putida GPo1 (previously P. oleovorans) (14). CyoB-deficient mutants partially escape from catabolite repression.
RelA and its counterpart SpoT are involved in the biosynthesis of (p)ppGpp. The level of these alarmones influences RNA polymerase activity at certain promoters and σ54 competitiveness for the RNA polymerase core. σ54 occupancy of RNA polymerase modulates the expression of the DmpR-dependent Po promoter that controls the phenol degradation operon in Pseudomonas sp. CF600 (25).
The ptsN gene product IIANtr shares characteristics with phosphotransferases of the PTS family. A knockout mutant in the ptsN gene relieves C source inhibition of the TOL plasmid upper pathway operon in cells growing exponentially in minimal medium with glucose or gluconate as a C source (9). However, expression from Pu in cells growing on rich medium in the exponential phase of growth was not affected (9).
In this study we used P. putida KT2440 and a series of knockout mutants in relA, cyoB, crp, crc, and ptsN to analyze expression from the TOL promoters. The approach we used reproduces rich medium repression under batch growth conditions by adding YE to cells growing exponentially on minimal medium. We also used macroarrays to simultaneously monitor variations in the level of expression of the potential global regulators and the expression of the TOL genes under repressive conditions. Our results show that repression of the TOL genes was concomitant with increase in the expression of the crc gene.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains and plasmids used are listed in Table 1. Escherichia coli strains were routinely grown at 30°C in liquid LB medium (43). P. putida strains were grown at 30°C in modified M9 minimal medium with glucose (25 mM) as the sole carbon source (30). When indicated, YE, which consists basically of a mixture of amino acids plus micrograms of vitamins, was added to a final concentration of 1%. This gave a final concentration of each amino acid in the culture medium ranging between 0.5 and 3 mM. When required, antibiotics were used at the following final concentrations (in micrograms per milliliter): kanamycin (Km), 50; tetracycline (Tc), 8; gentamicin (Gm), 100; chloramphenicol (Cm), 30; and streptomycin (Sm), 100.
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Characteristics | Reference |
|---|---|---|
| Strains | ||
| P. putida KT2440 | mt-2 pWW0 cured | 2 |
| P. putida KT2440 (pWW0) | mt-2; ATCC 33015 | 50 |
| P. putida KT2440/102 | Kmr, P. putida KT2440 with a Km resistance cassette interrupting ORF102 | This study |
| P. putida KT2440/ptsN | Kmr, P. putida KT2440 with a Km resistance cassette interrupting the ptsN gene | This study |
| P. putida KT2440/284 | Kmr, P. putida KT2440 with a Km resistance cassette interrupting ORF284 | This study |
| P. putida KT2440/ptsO | Gmr, P. putida KT2440 with a Gm resistance cassette interrupting the ptsO gene | This study |
| P. putida KT2440/crc | Gmr, P. putida KT2440 with a Gm resistance cassette interrupting the crc gene | This study |
| P. putida KT2440/crp | Kmr, P. putida KT2440 with a Km resistance cassette interrupting the crp gene | This study |
| P. putida KT2440/cyoB | Tcr, P. putida KT2440 with a Tc resistance cassette interrupting the cyoB gene | F. Rojo |
| P. putida KT2440/aer | Kmr, P. putida KT2440 with a Km resistance cassette interrupting the aer gene | V. Shingler |
| P. putida KT2440/relA | Kmr, P. putida KT2440 with a Km resistance cassette interrupting the relA gene | V. Shingler |
| P. putida KT2440/spoT-relA | Kmr, Gmr, P. putida KT2440/relA with a Gm resistance cassette interrupting the spoT gene | V. Shingler |
| P. putida KT2440/crc-ptsN | Gmr, Kmr, P. putida KT2440/ptsN with a Gm resistance cassette interrupting the crc gene | This study |
| E. coli DH5α | F− Φ80d lacZΔM15Δ(lacZYA-argF) U169 recA1 endA1 hsdR17 R− M+supE44 thi1 gyrA relA1 | 21 |
| E. coli HB101 | Smr, hsd R− M+ pro leu thi recA | 43 |
| Plasmids | ||
| pGEM-T | PCR product cloning vector | Promega |
| pKNG101 | Smr, sucrose sensitive, ori RK2 | 26 |
| pKNG102 | Smr, Kmr (with a Km resistance cassette inserted in ORF102) | 9 |
| pKNG154 | Smr, Kmr (with a Km resistance cassette inserted in the ptsN gene) | 9 |
| pKNG284 | Smr, Kmr (with a Km resistance cassette inserted in ORF284) | 9 |
| pKNGptsO | Smr, Gmr (with a Gm resistance cassette inserted in the ptsO gene) | This study |
| pKNGcrc | Smr, Gmr (with a Gm resistance cassette inserted in the crc gene) | This study |
| pKNGcrp | Smr, Gmr (with a Km resistance cassette inserted in the crp gene) | This study |
| pRK600 | Cmr, oriColE1, mobRK2, traRK2 | 27 |
| pS10 | IncP1, Smr, xylR, transcriptional Pu::lacZ::tet fusion | This study |
| pWW0 | IncP9, mob+, tra+, 3MB+ | 50 |
Construction of mutants.
The P. putida KT2440 genome (http://tigrblast.tigr.org) was screened for the target genes, and the flanking region was identified. The different genes responsible for global regulation were cloned using PCR amplification of each region with specific oligonucleotides located approximately 1 kb upstream and downstream from the gene of interest. Every cloned gene was interrupted with an antibiotic resistance cassette and transferred to P. putida KT2440 by reverse genetics. To generate a crc mutant, we screened the genome for a sequence homologous to the crc gene of P. aeruginosa. Oligonucleotides 5′-GTAGCGTAGTGTGACTTGAAGGG-3′ and 5′-TGTACCGCGCTTCCTCAAAGGC-3′ located 1 kb upstream and downstream from the crc gene, respectively, were used to amplify a 2.8-kb fragment from the P. putida KT2440 chromosome. The fragment was first cloned in pGEM-T and sequenced to detect any unwanted mutation. A gentamicin cassette was introduced at the unique SmaI site, thus interrupting the crc open reading frame (ORF) in the 225th codon. The knockout gene was then cloned in the suicide delivery plasmid pKNG101 between the SmaI and SpeI sites. The resulting plasmid was transferred to P. putida KT2440 in a triparental mating where transfer functions were provided in trans by E. coli HB101 (pRK600). P. putida mutant strains bearing the knockout crc gene were directly selected as streptomycin-sensitive, gentamicin-resistant, sucrose-resistant colonies. Accuracy of the double recombination event was confirmed by PCR and Southern blot analysis.
The knockout cyoB mutant where the cyoB gene was interrupted by a tetracycline resistance cassette was obtained from F. Rojo. Delivery plasmids containing a knockout mutant of ORF284, ptsN, and ORF102 were obtained from V. de Lorenzo and were transferred to P. putida as described above. Mutants were selected as kanamycin-resistant, sucrose-resistant, streptomycin-sensitive strains and checked by PCR and Southern blot analysis. To obtain a ptsO mutant, oligonucleotides located ca. 1 kb upstream (5′-GCCCACCTTGAACTTCTGCG-3′) and downstream (5′-GTCCGGAATACATCGGTGCC-3′) of the ptsO gene were designed and a 2.3-kb fragment was amplified from P. putida KT2440 chromosome. The fragment was first cloned in pGEM-T and sequenced to detect any unwanted mutation. A Gm cassette was introduced in the unique SmaI site, thus interrupting the ptsO ORF. The knockout gene was then cloned in the suicide delivery plasmid pKNG101 between the SspI and SpeI sites. The resulting plasmid was transferred to P. putida KT2440 in a triparental mating as described above.
To obtain a crp mutant, oligonucleotides located 1 kb upstream (5′-GGCTCACCGTTCAGTTGGG-3′) and downstream (5′-GGATACGCCGCTGGTGGG-3′) from the crp gene were used to amplify a 2.75-kb fragment from the P. putida KT2440 chromosome. The fragment was first cloned in pGEM-T and sequenced. A kanamycin cassette was introduced in the unique NruI site, thus interrupting the crp ORF in the 116th codon. The knockout gene was then cloned in the suicide delivery plasmid pKNG101 and transferred to P. putida KT2440 as described above, and mutants were selected as kanamycin-resistant, sucrose-resistant, streptomycin-sensitive strains and checked by PCR and Southern blot analysis.
In addition to the previously described mutants, aer, relA, and relA/spoT P. putida KT2440 mutant strains were obtained from V. Shingler. Finally, the TOL plasmid pWW0 was transferred to each mutant strain by conjugation. To obtain the crc-ptsN double mutant, the above approach was used, except that the recipient strain was P. putida KT2440 containing the ptsN mutation.
Sampling and isolation of RNA.
Cells grown overnight in M9 minimal medium with glucose as the carbon source were diluted to a turbidity of 0.2 at 660 nm in the same medium. When the cultures reached a turbidity of 0.7 (in the exponential growth phase), they were divided into four fractions, which were supplemented with o-xylene (a nonmetabolizable inducer of the Pu and PS1 promoters in the TOL plasmid pWW0), 1% (wt/vol) yeast extract as repressor agent, or both. The fourth fraction was left unsupplemented as a control. Cultures were incubated at 30°C for 30 min, and 10-ml samples were harvested by centrifugation at 4°C in disposable plastic tubes precooled in liquid N2 and were kept at −80°C until use. Total RNA was extracted with the phenol-guanidine thiocyanate mixture Tri Reagent LS (Molecular Research Center, Inc.) according to the manufacturer's instructions, except that the initial lysis step was carried out at 60°C. The relative levels of each specific messenger were estimated by reverse primer extension analysis of equal amounts of total RNA, using the following oligonucleotides: 5′-GGCCAGCGTCACAGACTCCAGGCG-3′ for Pu-dependent transcripts, 5′-GAGACTGCATAGGGCTCGGCGTGG-3′ for PS, and 5′-ACGGATCTGGCTGCTAAGGTCTTGC-3′ for PR transcripts. Primer extension analysis of 10 to 20 μg total RNA samples was carried out as described previously (31) using the 32P-end-labeled oligonucleotides described above. Samples were run in urea sequencing gels, and gels were exposed to a phosphorimaging screen (Fuji Photo Film Co. Ltd.) for 5 to 12 h. Phosphorimaging screens were scanned with a phosphorimaging instrument (Molecular imager FX; Bio-Rad). Data were quantified with Quantity One software (Bio-Rad).
Construction of the macroarray and cDNA labeling.
The DNA arrays used in the hybridization experiments were produced by Newbiotechnic (Seville, Spain). Each DNA array consisted of a positively charged nylon membrane on which each PCR-amplified, ORF-specific DNA fragment was printed in duplicate with a robotic technique. For radioactive cDNA labeling, we used 20 μg total RNA in 12 μl diethyl pyrocarbonate-treated H2O containing 250 ng random hexamer oligonucleotides (Amersham). Samples were heated to 70°C for 10 min and chilled on ice. Probe synthesis was carried out at 42°C for 2 h in a 50-μl reaction volume containing 50 mM Tris-HCl (pH 8.5), 30 mM KCl, 8 mM MgCl2, 1 mM dithiothreitol, 0.5 mM dATP, 0.5 mM dTTP, 0.5 mM dGTP, 0.05 mM dCTP, 100 μCi [α-32P]dCTP at 3000 Ci/mmol (Amersham), 40 U RNAsin (Promega), and 200 U Superscript II RNase H− reverse transcriptase (Invitrogen LifeTechnologies). Unincorporated nucleotides were removed in Micro Bio-Spin chromatography columns (Bio-Rad Laboratories), and samples were treated with RNase H (U.S. Biochemicals) for 20 min at 37°C. Prior to hybridization, high density arrays were prewetted in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.5% sodium dodecyl sulfate (SDS) and prehybridized for 2 h at 64°C in roller bottles containing 20 ml 1× hybridization buffer (0.5% blocking reagent [Roche], 5× SSC, 0.1% N-lauroylsarcosine, 0.02% SDS, 0.1 mg/ml sheared salmon sperm DNA). Prehybridization buffer was removed and replaced with 1× hybridization buffer containing 4 × 107 cpm/ml cDNA probe, and hybridization continued at 64°C for 40 h. After hybridization, arrays were washed twice at room temperature and once at 65°C in 2× SSC with 0.5% SDS, followed by one wash at 65°C in 0.1× SSC with 0.5% SDS. Arrays were then sealed in thin polypropylene bags to avoid drying and exposed to a phosphorimaging screen (Fuji Photo Film Co. Ltd.) for 12 h. Phosphorimaging screens were scanned with a phosphorimaging instrument (Molecular imager FX; Bio-Rad).
β-Galactosidase activity assay.
The wild-type P. putida cells and a series of isogenic mutants deficient in the synthesis of Aer, RelA, and SpoT proteins were transformed with pS10, a Kms derivative of the low-copy-number plasmid pJB3 (4) bearing the xylR gene and a transcriptional Pu::′lacZ::tet fusion preceded by a Smr cassette to prevent read-through transcription from vector promoters. Cells grown overnight on M9 minimal medium with glucose as the carbon source were diluted to an optical density at 660 nm of 0.5 in the same medium. After 30 min at 30°C with shaking, cultures were divided into four fractions: one was kept as a control, and to the other three, we added o-xylene, 1% (wt/vol) yeast extract, or both. After 90 min, culture samples were analyzed for β-galactosidase activity with the standard colorimetric assay described by Miller (32). At least two independent assays with duplicate samples were done in each case.
RESULTS AND DISCUSSION
Expression of the TOL plasmid Pu and PS promoters.
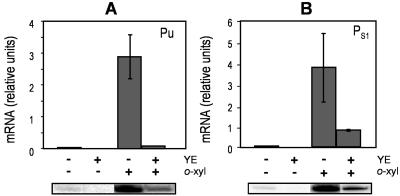
As a first step in identifying the global regulators involved in catabolite repression of the TOL plasmid promoters, we analyzed the o-xylene induction of TOL catabolic promoters in batch cultures in the presence and in the absence of 1% (wt/vol) YE. o-Xylene is a nonmetabolizable inducer able to activate XylR, which promotes transcription from both σ54-dependent promoters Pu and PS1, while the addition of YE reproduces rich medium repression (30). P. putida (pWW0) cells were grown on M9 minimal medium with glucose as the carbon source. When the culture reached a turbidity of 0.7, it was split into four fractions: one was kept as a control, one was supplemented with 1% YE, and o-xylene in the gas phase with or without 1% YE was added to the other two fractions. After 30 min of incubation at 30°C, mRNA was extracted and expression levels from the TOL promoters were determined by primer extension analysis. As expected, Pu and PS1 were strongly induced when o-xylene was added (Fig. 2). However, o-xylene-induced Pu and PS1 expression in the presence of YE was about 80 to 90% lower than that in its absence (Fig. 2A and B). High-level expression from Pu and PS1 was strictly dependent on the presence of the inducer o-xylene. Overall, these results confirm previous findings that the two σ54-dependent TOL plasmid promoters are the targets of catabolite repression control.
FIG. 2.
Yeast-extract-mediated repression of mRNA expression from Pu (A) and PS1 (B) promoters in P. putida KT2440 (pWW0). Cells grown overnight in M9 minimal medium with glucose as the carbon source were diluted to an optical density at 660 nm of 0.2. When the cultures reached exponential growth, they were divided into four fractions, three of which were supplemented with o-xylene (o-xyl), 1% (wt/vol) yeast extract, or both. The fourth fraction was left unsupplemented as a control. Cultures were incubated at 30°C for 30 min, and samples were collected for primer extension mRNA analysis. cDNA bands corresponding to each promoter (134 nucleotides for Pu and 206 for PS1 [bottom]) were quantified and compared.
The same assays were repeated with the whole series of P. putida mutants in genes that could potentially be involved in catabolite repression, as well as in mutants in the open reading frames adjacent to ptsN. Our results showed that the pattern described above for the wild type was almost identical in the mutants ORF102 and ORF284 of the rpoN gene cluster and for crp. In the cyoB mutant, only a moderate relief of YE-dependent Pu and PS1 repression was observed (not shown). These results ruled out the involvement of these proteins in TOL pathway catabolite repression. By using pS10, we measured expression from Pu in the presence of o-xylene and 1% (wt/vol) YE in isogenic P. putida backgrounds lacking the Aer protein, an aerotaxis and energy sensor thought to monitor the redox state of the electron transport chain in E. coli (22), and RelA- and SpoT-deficient backgrounds. The RelA and SpoT proteins are involved in the synthesis of (p)ppGpp, which influences RNA polymerase activity (11), and they are known to respond to amino acid and nutrient limitations. Although the inhibition caused by YE points toward (p)ppGpp playing a role in this repression, this does not seem the case. In all these mutants, o-xylene-induced β-galactosidase levels were similar to levels in the wild type, therefore indicating that these genes were not involved in catabolite repression of the Pu promoter either. This is in accordance with previous results comparing the (p)ppGpp effect on the activity of the two analogous systems DmpR/Po and XylR/Pu (46). Despite the clear contribution of this alarmone to upregulate the DmpR-dependent Po transcription, it appeared to play a minor role in XylR-dependent Pu transcription (46), which is consistent with our findings.
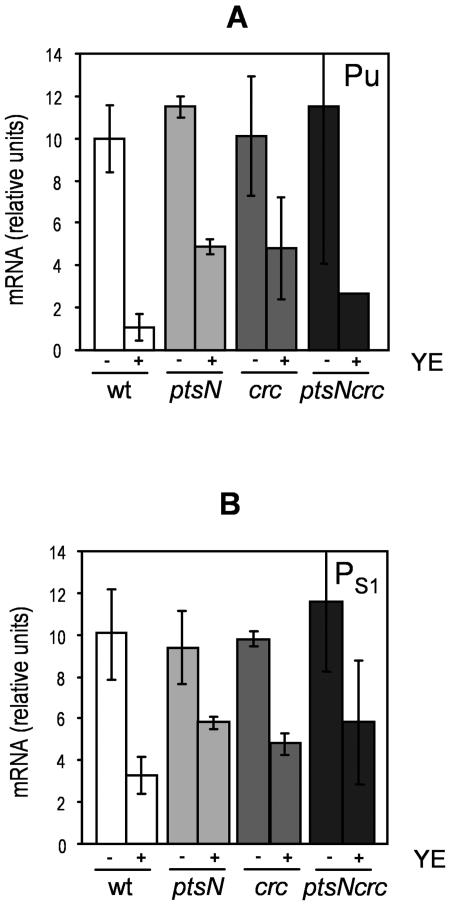
In contrast, pronounced derepression was observed in the ptsN and crc mutants (Fig. 3). The level of repression from Pu and PS1 in the ptsN- and crc- deficient mutants was only about 50%. PtsN repression of Pu (9) and PS1 (15) had been described before, but this is the first report of Crc-mediated repression of Pu and PS1. To test the potential synergistic effects of these two factors on catabolite repression, we constructed a ptsN-crc double mutant and tested the expression from Pu and PS1. The results revealed that the degree of derepression was not increased with respect to that observed in the crc or ptsN single mutant, which yielded the highest level of derepression.
FIG. 3.
Yeast-extract-mediated repression of mRNA expression from Pu (A) and PS1 (B) promoters in mutants P. putida KT2440/ptsN (pWW0), P. putida KT2440/crc (pWW0), and P. putida KT2440/ptsNcrc (pWW0) compared to the control strain P. putida KT2440 (pWW0). Cell growth, sampling, and analysis done were as described in the legend to Fig. 2.
In the wild-type strain, basal levels of Pu and PS1 promoters in the absence of inducer were low but measurable and were XylR dependent (results not shown). In the presence of YE, these levels were repressed to 10% of their initial value. We measured YE effect on basal expression levels from both promoters in the ptsN and crc mutants. Neither ptsN nor crc mutants were able to relieve the strong repression of these uninduced levels (not shown). This suggests that the effect of YE is partially mediated by PtsN or Crc only in the presence of effector.
PtsN but not Crc directly interferes with XylR activation at target promoters.
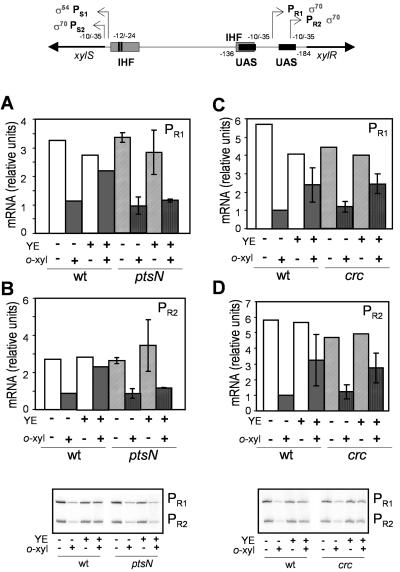
Expression from both promoters of the xylR gene is known to be repressed in the presence of an effector for XylR, which was attributed to the stronger binding of activated XylR to its target UASs in the −140 region with respect to the +1 region of PS1. In fact, the UASs overlap Eσ70 binding sites at PR1 and PR2 (3, 29) (Fig. 4). In vitro analysis of PR promoter expression has previously shown that Eσ70-dependent transcription from PR1 and PR2 decreases markedly in the presence of a truncated version of XylR which mimics the effector activated conformation. This was apparently a consequence of the binding and multimerization of the regulator at the UASs (3). Similar conclusions were obtained in vivo by comparing PR1 and PR2 promoter activity in a wild type and a xylR-deleted mutant of TOL plasmid. A strong derepression (more than fivefold) of both promoters was observed in the absence of XylR, and consistently, this expression, which was the highest observed for xylR in vivo in any condition, was independent of the presence of an aromatic effector (29). Based on these previous observations, we can assume that XylR occupancy of the UASs can be inferred from the activity of these two PR promoters. XylR binding to its UASs is influenced by a number of factors. Besides the presence of effector, which increases XylR affinity for its targets (1, 3, 29), the presence and activity of Eσ54 seems to influence UAS occupancy by allowing XylR release after each transcription cycle (29). The results in Fig. 4 show that the presence of 1% (wt/vol) YE partially relieved o-xylene-dependent PR repression in the wild-type strain. Therefore, the presence of YE could be influencing any of the above-mentioned processes to produce a lower occupancy of UASs by XylR, as reflected by the higher activity of PR1 and PR2. This lower binding of XylR correlates with the low level of PS1 (and hence of Pu) expression in the presence of YE (Fig. 2). However, in the ptsN mutant, PR promoter expression with o-xylene was not affected by the presence of YE (Fig. 4A and B). This indicates that PtsN was in part responsible for YE effect on XylR binding to its UASs and suggests that PtsN exerts its influence directly at the level of the transcriptional machinery at the TOL σ54-dependent promoters, somehow interfering with o-xylene-XylR interaction to bind its UASs or with the Eσ54-binding and activation mechanism.
FIG. 4.
Expression from PR1 (A and C) and PR2 (B and D) promoters in mutants P. putida KT2440/ptsN (pWW0) and P. putida KT2440/crc (pWW0) compared to the control strain P. putida KT2440 (pWW0) in the presence (+) or absence (−) of effector (o-xylene [o-xyl]) and repressor agent (yeast extract). Cell growth, sampling, and analysis were done as described in the legend to Fig. 2, except that oligonucleotides complementary to xylR were used. cDNA bands corresponding to each PR promoter (208 n for PR1 and 180 n for PR2 [bottom]) were quantified and compared. IHF, integration host factor; wt, wild type.
The behavior of the crc mutant was different from that of ptsN, namely, XylR expression in this mutant in the presence of YE was similar to its expression in the wild type, i.e., YE partially relieved the o-xylene-dependent repression of PR1 and PR2 (Fig. 4C and D). Therefore, Crc does not seem to influence XylR UAS occupancy in the presence of YE. Then Crc effect on PS1 expression (and hence Pu expression) is not likely to be exerted directly at the level of the transcription machinery, which would probably influence UAS occupancy. The molecular mechanism behind Crc activity is unknown. Sequence similarities suggest a relation with endo- and exonucleases, which would point out to processes such as messenger stability. However, such a function would produce an additive phenotype in a double mutant, ptsN-crc, which was not observed. A general role of Crc generating a metabolic signal is more plausible. Recently, Velázquez et al. (48) suggested that catabolites of the Entner-Doudoroff pathway were responsible for C-source repression of Pu. The authors observed a degree of relief of glucose repression of the Pu promoter in a crc knockout mutant, connecting this result with the observation in a crc mutant of P. aeruginosa of an increase in Entner-Doudoroff pathway activity, detected as a higher level of glucose-6-phosphate dehydrogenase (12). Although this effect on the Entner-Doudoroff pathway has not been confirmed in P. putida, we cannot rule out this possibility that it is responsible for the crc effect on YE repression. However, this scenario is hardly conceived when YE, containing essentially a mixture of amino acids, is added as a repressing agent.
Expression of the crc gene inversely correlates with levels of catabolite repression from Pu and PS1.
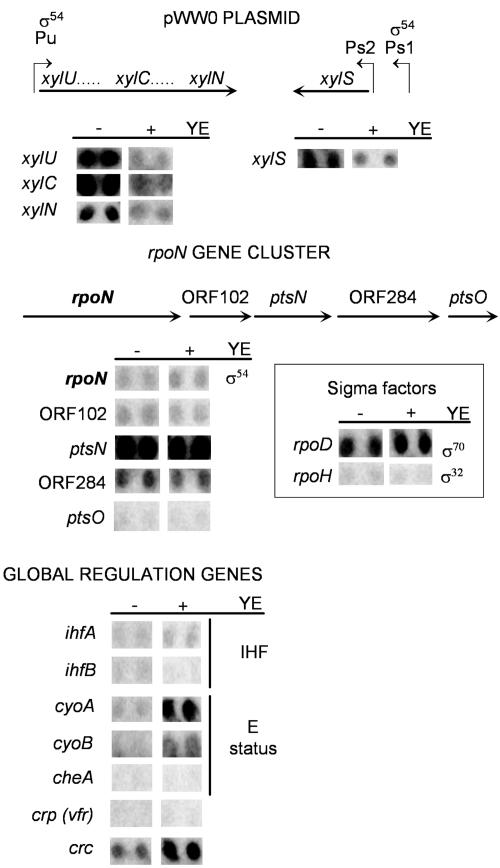
We set up a macroarray to monitor the mRNA level of rpoN and ihf, whose gene products are directly involved in the transcription of Pu and PS1, together with genes of the TOL upper pathway and xylS. We also monitored expression of the genes in the rpoN gene cluster (orf102, ptsN, orf284, and ptsO) and genes potentially involved in catabolite repression, such as crp, crc, and cyoB. Total RNA was isolated from o-xylene-induced P. putida KT2440 (pWW0) cells growing in the presence and in the absence of YE, and its derived cDNA was radioactively labeled and hybridized to the macroarray membrane as described in the Materials and Methods section. Figure 5 shows that expression of xylS and of the Pu-dependent catabolic genes was repressed in the presence of YE. Interestingly, these results also revealed that, in general, YE did not affect the level of expression of the genes directly involved in the expression of Pu. This was also the case for the genes in the rpoN cluster, although ptsO expression levels were below the detection limits and no conclusion could be drawn out for this gene. This was also the case for crp, ihfB, and cheA. However, we observed a marked increase in the expression of crc, cyoA, and cyoB transcripts. Interestingly, crc expression increase correlated with a decrease in the expression from Pu and PS1. The increase in the mRNA of the two subunits of the terminal oxidase cyoA and cyoB, which are encoded by the same operon, can be related to an increase in respiratory rates, in agreement with the observation made in E. coli in response to the addition of Casamino Acids (33). The terminal oxidase levels have been shown to play a minor role in the YE repression of TOL catabolite promoters.
FIG. 5.
Effect of the presence of yeast extract on the mRNA levels of genes involved in the control of the expression in TOL plasmid catabolic operons. Genes in the DNA macroarray used to determine the mRNA levels are displayed in the panels. P. putida KT2440 (pWW0) cells were grown on M9 minimal medium with glucose as the carbon source. When the cultures reached exponential growth, they were divided into two fractions, both of them supplemented with o-xylene in the gas phase, and to one of them, 1% (wt/vol) yeast extract was added. Cultures were incubated at 30°C for 30 min, and samples were collected for mRNA isolation. Radioactively labeled cDNA was synthesized and hybridized to the DNA macroarray as described in Materials and Methods. IHF, integration host factor.
Concluding remarks.
Several functions have been described in Pseudomonadaceae as responsible for the catabolite repression of different genes and pathways. Among these genes, cyoB, crc, and ptsN have been directly related to the global regulation of different operons encoding enzymes for the metabolism of hydrocarbon (6, 14, 46, 51). However, except for (p)ppGpp in the DmpR-activated Po promoter of the phenol degradation pathway (28), the molecular mechanism underlying each process remains to be elucidated.
PtsN, the IIANtr phosphotransferase present in the rpoN gene cluster, has been shown to mediate the glucose-dependent repression of Pu transcription, although the phenomenon known as exponential silencing, i.e., the repression of transcription in the early-exponential phase during growth in LB medium, was unaltered in this mutant. Using primer extension analysis to directly track the transcription process, we found that Pu and PS1 expression in the presence of YE is derepressed in a ptsN mutant. The addition of YE seems to interfere with XylR binding at its UASs, and the absence of PtsN prevents this interference. We envisage that PtsN may function by affecting XylR activity (46) or at least by modulating the overall mechanism of XylR activation of both σ54-dependent promoters.
Crc has been implicated in carbon source regulation of the alk operon for alkane degradation in P. putida GPo1 (51), although in this pathway, cyoB, which appears to “sense” the energy status of the cell, is the main player in catabolite repression (13). We have shown here that crc transcription is markedly increased in the presence of YE, which inversely correlates with TOL pathway expression. A crc mutant was partially derepressed in the presence of YE. However, this derepression does not seem to interfere with XylR binding at its UASs.
Our data show that the effect of the double mutation in ptsN and crc is not the sum of the effects of each independent mutation. A genetic interpretation of this result suggests that both regulators are elements of a common regulatory pathway. However, on the basis of current knowledge on the phosphotranferase mechanism in the PTS systems and the observations of XylR binding in both mutants, it is difficult to envisage such a scenario. The precise function and mechanism of Crc in the cell are still unknown, but current evidence points to a very different mechanism. The only plausible explanation for our findings at this time appears to be that Crc acts by sensing the presence of YE in an early step of the regulatory process.
Acknowledgments
We thank Patricia Marín for skillful technical assistance, V. Shingler, F. Rojo, and V. de Lorenzo for generous gifts of strains and plasmids, and Manuel Rey from Newbiotechnic, Seville, Spain, for imprinting the macroarrays and technical advice. We thank M. M. Fandila for secretarial assistance.
This work was supported by EC grant Biocarte (CLK-2002-01923) and MCYT grant BMC2001-0515.
REFERENCES
- 1.Abril, M. A., M. Buck, and J. L. Ramos. 1991. Activation of the Pseudomonas TOL plasmid upper pathway operon. Identification of binding sites for the positive regulator XylR and for integration host factor protein. J. Biol. Chem. 266:15832-15838. [PubMed] [Google Scholar]
- 2.Bagdasarian, M., R. Lurz, B. Ruckert, F. C. Franklin, M. M. Bagdasarian, J. Frey, and K. N. Timmis. 1981. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene 16:237-247. [DOI] [PubMed] [Google Scholar]
- 3.Bertoni, G., S. Marqués, and V. de Lorenzo. 1998. Activation of the toluene-responsive regulator XylR causes a transcriptional switch between sigma54 and sigma70 promoters at the divergent PR/PS region of the TOL plasmid. Mol. Microbiol. 27:651-659. [DOI] [PubMed] [Google Scholar]
- 4.Blatny, J. M., T. Brautaset, H. C. Winther-Larsen, K. Haugan, and S. Valla. 1997. Construction and use of a versatile set of broad-host-range cloning and expression vectors based on the RK2 replicon. Appl. Environ. Microbiol. 63:370-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cases, I., and V. de Lorenzo. 2001. The black cat/white cat principle of signal integration in bacterial promoters. EMBO J. 20:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cases, I., and V. de Lorenzo. 1998. Expression systems and physiological control of promoter activity in bacteria. Curr. Opin. Microbiol. 1:303-310. [DOI] [PubMed] [Google Scholar]
- 7.Cases, I., V. de Lorenzo, and J. Pérez-Martín. 1996. Involvement of sigma factor σ54 in exponential silencing of the Pseudomonas putida TOL plasmid Pu promoter. Mol. Microbiol. 19:7-17. [DOI] [PubMed] [Google Scholar]
- 8.Cases, I., J.-A. López, J.-P. Albar, and V. de Lorenzo. 2001. Evidence of multiple regulatory functions for the PtsN (IIANtr) protein of Pseudomonas putida. J. Bacteriol. 183:1032-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cases, I., J. Pérez-Martín, and V. de Lorenzo. 1999. The IIANtr (PtsN) protein of Pseudomonas putida mediates the C source inhibition of the sigma54-dependent Pu promoter of the TOL plasmid. J. Biol. Chem. 274:15562-15568. [DOI] [PubMed] [Google Scholar]
- 10.Cases, I., F. Velázquez, and V. de Lorenzo. 2001. Role of ptsO in carbon-mediated inhibition of the Pu promoter belonging to the pWW0 Pseudomonas putida plasmid. J. Bacteriol. 183:5128-5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cashel, M., D. Gentry, V. J. Hernandez, and D. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt, R. Curtis III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, D.C.
- 12.Collier, D. N., C. Spence, M. J. Cox, and P. V. Phibbs. 2001. Isolation and phenotypic characterization of Pseudomonas aeruginosa pseudorevertants containing suppressors of the catabolite repression control-defective crc-10 allele. FEMS Microbiol. Lett. 196:87-92. [DOI] [PubMed] [Google Scholar]
- 13.Dinamarca, M. A., I. Aranda-Olmedo, A. Puyet, and F. Rojo. 2003. Expression of the Pseudomonas putida OCT plasmid alkane degradation pathway is modulated by two different global control signals: evidence from continuous cultures. J. Bacteriol. 185:4772-4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dinamarca, M. A., A. Ruíz-Manzano, and F. Rojo. 2002. Inactivation of cytochrome o ubiquinol oxidase relieves catabolic repression of the Pseudomonas putida GPo1 alkane degradation pathway. J. Bacteriol. 184:3785-3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du, Y., A. Holtel, J. Reizer, and M. H. Saier, Jr. 1996. Sigma54-dependent transcription of the Pseudomonas putida xylS operon is influenced by the IIANtr protein of the phosphotransferase system in Escherichia coli. Res. Microbiol. 147:129-132. [DOI] [PubMed] [Google Scholar]
- 16.Duetz, W. A., S. Marqués, C. de Jong, J. L. Ramos, and J. G. van Andel. 1994. Inducibility of the TOL catabolic pathway in Pseudomonas putida (pWW0) growing on succinate in continuous culture: evidence of carbon catabolite repression control. J. Bacteriol. 176:2354-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duetz, W. A., S. Marqués, B. Wind, J. L. Ramos, and J. G. van Andel. 1996. Catabolite repression of the toluene degradation pathway in Pseudomonas putida harboring pWWO under various conditions of nutrient limitation in chemostat culture. Appl. Environ. Microbiol. 62:601-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallegos, M.-T., S. Marqués, and J. L. Ramos. 1996. Expression of the TOL plasmid xylS gene in Pseudomonas putida occurs from a σ70-dependent promoter or from σ70- and σ54-dependent tandem promoters according to the compound used for growth. J. Bacteriol. 178:2356-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garmendia, J., and V. de Lorenzo. 2000. The role of the interdomain B linker in the activation of the XylR protein of Pseudomonas putida. Mol. Microbiol. 38:401-410. [DOI] [PubMed] [Google Scholar]
- 20.González-Pérez, M. M., J. L. Ramos, and S. Marqués. 2004. Cellular XylS levels are a function of transcription of xylS from two independent promoters and the differential efficiency of translation of the two mRNAs. J. Bacteriol. 186:1898-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 22.Herrmann, S., Q. Ma, M. S. Johnson, A. V. Repik, and B. L. Taylor. 2004. PAS domain of the Aer redox sensor requires C-terminal residues for native-fold formation and flavin adenine dinucleotide binding. J. Bacteriol. 186:6782-6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hester, K. L., J. Lehman, F. Najar, L. Song, B. A. Roe, C. H. MacGregor, P. W. Hager, P. V. Phibbs, Jr., and J. R. Sokatch. 2000. Crc is involved in catabolite repression control of the bkd operons of Pseudomonas putida and Pseudomonas aeruginosa. J. Bacteriol. 182:1144-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hugouvieux-Cotte-Pattat, N., T. Köhler, M. Rekik, and S. Harayama. 1990. Growth phase dependent expression of the Pseudomonas putida TOL plasmid pWWO catabolic genes. J. Bacteriol. 172:6651-6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jishage, M., K. Kvint, V. Shingler, and T. Nystrom. 2002. Regulation of sigma factor competition by the alarmone ppGpp. Genes Dev. 16:1260-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaniga, K., I. Delor, and G. R. Cornelis. 1991. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene 109:137-141. [DOI] [PubMed] [Google Scholar]
- 27.Kessler, B., V. de Lorenzo, and K. N. Timmis. 1992. A general system to integrate lacZ fusions into the chromosomes of gram-negative eubacteria: regulation of the Pm promoter of the TOL plasmid studied with all controlling elements in monocopy. Mol. Gen. Genet. 233:293-301. [DOI] [PubMed] [Google Scholar]
- 28.Laurie, A. D., L. M. Bernardo, C. C. Sze, E. Skarfstad, A. Szalewska-Palasz, T. Nystrom, and V. Shingler. 2003. The role of the alarmone (p)ppGpp in sigma N competition for core RNA polymerase. J. Biol. Chem. 278:1494-1503. [DOI] [PubMed] [Google Scholar]
- 29.Marqués, S., M. T. Gallegos, M. Manzanera, A. Holtel, K. N. Timmis, and J. L. Ramos. 1998. Activation and repression of transcription at the double tandem divergent promoters for the xylR and xylS genes of the TOL plasmid of Pseudomonas putida. J. Bacteriol. 180:2889-2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marqués, S., A. Holtel, K. N. Timmis, and J. L. Ramos. 1994. Transcriptional induction kinetics from the promoters of the catabolic pathways of TOL plasmid pWW0 of Pseudomonas putida for metabolism of aromatics. J. Bacteriol. 176:2517-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marqués, S., and J. L. Ramos. 1993. Transcriptional control of the Pseudomonas putida TOL plasmid catabolic pathways. Mol. Microbiol. 9:923-929. [DOI] [PubMed] [Google Scholar]
- 32.Miller, J. H. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 33.Minagawa, J., H. Nakamura, I. Yamato, T. Mogi, and Y. Anraku. 1990. Transcriptional regulation of the cytochrome b562-o complex in Escherichia coli. Gene expression and molecular characterization of the promoter. J. Biol. Chem. 265:11198-11203. [PubMed] [Google Scholar]
- 34.Morales, G., J. F. Linares, A. Beloso, J. P. Albar, J. L. Martínez, and F. Rojo. 2004. The Pseudomonas putida Crc global regulator controls the expression of genes from several chromosomal catabolic pathways for aromatic compounds. J. Bacteriol. 186:1337-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pérez-Martín, J., and V. de Lorenzo. 1995. The amino-terminal domain of the prokaryotic enhancer-binding protein XylR is a specific intramolecular repressor. Proc. Natl. Acad. Sci. USA 92:9392-9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pérez-Martín, J., and V. de Lorenzo. 1995. The σ54-dependent promoter Ps of the TOL plasmid of Pseudomonas putida requires HU for transcriptional activation in vivo by XylR. J. Bacteriol. 177:3758-3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pérez-Martín, J., K. N. Timmis, and V. de Lorenzo. 1994. Co-regulation by bent DNA. Functional substitutions of the integration host factor site at sigma 54-dependent promoter Pu of the upper-TOL operon by intrinsically curved sequences. J. Biol. Chem. 269:22657-22662. [PubMed] [Google Scholar]
- 38.Petruschka, L., G. Burchhardt, C. Muller, C. Weihe, and H. Herrmann. 2001. The cyo operon of Pseudomonas putida is involved in carbon catabolite repression of phenol degradation. Mol. Genet. Genomics 266:199-206. [DOI] [PubMed] [Google Scholar]
- 39.Phillips, A. T., and L. M. Mulfinger. 1981. Cyclic adenosine 3′,5′-monophosphate levels in Pseudomonas putida and Pseudomonas aeruginosa during induction and carbon catabolite repression of histidase synthesis. J. Bacteriol. 145:1286-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramos, J. L., S. Marqués, and K. N. Timmis. 1997. Transcriptional control of the Pseudomonas TOL plasmid catabolic operons is achieved through an interplay of host factors and plasmid- encoded regulators. Annu. Rev. Microbiol. 51:341-373. [DOI] [PubMed] [Google Scholar]
- 41.Rojo, F., and A. Dinamarca. 2004. Catabolite repression and physiological control, p. 365-387. In J. L. Ramos (ed.), Pseudomonas. Kluwer Academic/Plenum Publishers, London, United Kingdom.
- 42.Ruíz, R., M. I. Aranda-Olmedo, P. Domínguez-Cuevas, M. I. Ramos-González, and S. Marqués. 2004. Transcriptional regulation of the toluene catabolic pathways, p. 509-537. In J. L. Ramos (ed.), Pseudomonas. Kluwer Academic/Plenum Publishers, London, United Kingdom.
- 43.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 44.Siegel, L. S., P. B. Hylemon, and P. V. Phibbs, Jr. 1977. Cyclic adenosine 3′,5′-monophosphate levels and activities of adenylate cyclase and cyclic adenosine 3′,5′-monophosphate phosphodiesterase in Pseudomonas and Bacteroides. J. Bacteriol. 129:87-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suh, S. J., L. J. Runyen-Janecky, T. C. Maleniak, P. Hager, C. H. MacGregor, N. A. Zielinski-Mozny, P. V. Phibbs, Jr., and S. E. West. 2002. Effect of vfr mutation on global gene expression and catabolite repression control of Pseudomonas aeruginosa. Microbiology 148:1561-1569. [DOI] [PubMed] [Google Scholar]
- 46.Sze, C. C., L. M. D. Bernardo, and V. Shingler. 2002. Integration of global regulation of two aromatic-responsive σ54-dependent systems: a common phenotype by different mechanisms. J. Bacteriol. 184:760-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sze, C. C., and V. Shingler. 1999. The alarmone (p)ppGpp mediates physiological-responsive control at the sigma 54-dependent Po promoter. Mol. Microbiol. 31:1217-1228. [DOI] [PubMed] [Google Scholar]
- 48.Velázquez, F., I. di Bartolo, and V. de Lorenzo. 2004. Genetic evidence that catabolites of the Entner-Doudoroff pathway signal C source repression of the σ54 Pu promoter of Pseudomonas putida. J. Bacteriol. 186:8267-8275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.West, S. E., A. K. Sample, and L. J. Runyen-Janecky. 1994. The vfr gene product, required for Pseudomonas aeruginosa exotoxin A and protease production, belongs to the cyclic AMP receptor protein family. J. Bacteriol. 176:7532-7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Worsey, M. J., and P. A. Williams. 1975. Metabolism of toluene and xylenes by Pseudomonas putida (arvilla) mt-2: evidence for a new function of the TOL plasmid. J. Bacteriol. 124:7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yuste, L., and F. Rojo. 2001. Role of the crc gene in catabolic repression of the Pseudomonas putida GPo1 alkane degradation pathway. J. Bacteriol. 183:6197-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]