Abstract
The anaerobic oxidation of methane (AOM) is a key process in the global methane cycle, and the majority of methane formed in marine sediments is oxidized in this way. Here we present results of an in vitro 13CH4 labeling study (δ13CH4, ∼5,400‰) in which microorganisms that perform AOM in a microbial mat from the Black Sea were used. During 316 days of incubation, the 13C uptake into the mat biomass increased steadily, and there were remarkable differences for individual bacterial and archaeal lipid compounds. The greatest shifts were observed for bacterial fatty acids (e.g., hexadec-11-enoic acid [16:1Δ11]; difference between the δ13C at the start and the end of the experiment [Δδ13Cstart-end], ∼160‰). In contrast, bacterial glycerol diethers exhibited only slight changes in δ13C (Δδ13Cstart-end, ∼10‰). Differences were also found for individual archaeal lipids. Relatively high uptake of methane-derived carbon was observed for archaeol (Δδ13Cstart-end, ∼25‰), a monounsaturated archaeol, and biphytanes, whereas for sn-2-hydroxyarchaeol there was considerably less change in the δ13C (Δδ13Cstart-end, ∼2‰). Moreover, an increase in the uptake of 13C for compounds with a higher number of double bonds within a suite of polyunsaturated 2,6,10,15,19-pentamethyleicosenes indicated that in methanotrophic archaea there is a biosynthetic pathway similar to that proposed for methanogenic archaea. The presence of group-specific biomarkers (for ANME-1 and ANME-2 associations) and the observation that there were differences in 13C uptake into specific lipid compounds confirmed that multiple phylogenetically distinct microorganisms participate to various extents in biomass formation linked to AOM. However, the greater 13C uptake into the lipids of the sulfate-reducing bacteria (SRB) than into the lipids of archaea supports the hypothesis that there is autotrophic growth of SRB on small methane-derived carbon compounds supplied by the methane oxidizers.
Since the pioneering work on the anaerobic oxidation of methane (AOM) (2, 33), there has been increasing evidence that the vast majority of methane arising from deeper horizons of marine sediments is recycled by this process into the global pool of inorganic carbon (34). Recently, it was shown that mainly two phylogenetic groups of archaea, termed ANME-1 and ANME-2, are involved (4, 15, 29). Mechanistic insights into AOM as a potential reverse of methanogenesis were obtained from genetic and biochemical studies of natural samples (13, 14, 21). Also, the AOM process, with its 1:1 stoichiometry between methane oxidation and sulfate reduction, could be established in the laboratory (26). However, the mode of coupling between methane oxidation and sulfate reduction is still insufficiently understood. Hoehler et al. (16) proposed that anaerobic methanotrophic archaea oxidize methane with direct coupling to sulfate-reducing bacteria (SRB) based on interspecies hydrogen transfer. Other findings do not indicate that there is this type of syntrophy, and other coupling mechanisms or even the entire reaction sequence in one organism alone has been discussed (26, 42).
Another insufficiently understood aspect of AOM is the assimilation of carbon into microbial cell mass. Neither growth yields (amounts of biomass formed per mole of methane oxidized) nor pathways for substrate carbon assimilation have been investigated. A suitable tool for tracing the incorporation of substrate carbon into cell components such as lipids is the use of 13C-labeled substrates. Recently, this approach was successfully used to link characteristic lipid biomarkers to the biosynthetically active source microorganisms (5, 6, 8, 24) or to investigate the fate of anthropogenic contaminants during microbial biodegradation (1, 35).
Several studies have shown that specific isoprenoid hydrocarbons, fatty acids, isoprenoid and nonisoprenoid dialkyl glycerol diethers (DAGE), and glycerol dialkyl glycerol tetraethers are produced by AOM-performing consortia (3, 15, 25, 31, 32, 40). These lipids are characterized by strong depletion of natural 13C, indicating that there is incorporation of carbon from biogenic methane. Recently, we distinguished ANME-1 associations from ANME-2 associations on the basis of specific lipid patterns (3). However, details concerning lipid biosynthesis for both associations are unknown. The large microbial buildup fostered by methane gas seepage in anoxic waters of the Black Sea is a unique, promising system for the study of biosynthetic processes due to the presence of compact biomass that is essentially free of sediment (25). Molecular analyses based on 16S rRNA and the distribution of group-specific lipid biomarkers (3, 25) revealed the predominance of ANME-1 associations and to a lesser extent the presence of ANME-2 consortia, both accompanied by sulfate-reducing bacteria belonging to the Desulfosarcina-Desulfococcus group.
Here we describe the results of long-term in vitro incubations with 13CH4 of a microbial mat from the Black Sea with high AOM activity. The data obtained allowed us to trace the incorporation of methane carbon into biomass. By using specific lipids methane carbon uptake into the microbially heterogeneous mat could be attributed to distinct microbial groups. Moreover, the data obtained contribute to our understanding of biosynthetic pathways of isoprenoid hydrocarbons and isoprenoid glycerol ethers in naturally relevant populations.
MATERIALS AND METHODS
Mat sample and incubation.
The microbial mat was obtained during a research cruise of the RV Professor Logachev in July 2001 from the anoxic waters of the northwest Black Sea shelf and corresponded to sample LOGA C described previously (3) and the microbial mat studied by Michaelis et al. (25) (44°46.5′N, 31°59.6′E; water depth, 230 m). Mat pieces were taken with the manipulator of the submersible Jago and were placed into a barrel filled with anoxic in situ water. The closed barrel was transported to the surface. On board, samples were immediately transferred into glass bottles with anoxic Black Sea water, sealed with butyl rubber stoppers, and stored at 8°C under a methane or nitrogen atmosphere until further processing.
The experiment was carried out in two glass bottles (bottles A and B; 156 ml each) sealed with butyl rubber stoppers and crimp caps. Pieces of mat material were homogenized in a bowl using a fork under sterile conditions. Three milliliters of a mat suspension was incubated with 53 ml artificial seawater medium (the salinity and sulfate concentrations were adjusted to the in situ conditions [about 22‰ and ∼16 mM], respectively) in bottles A and B. All manipulations were performed under an N2-CO2 (90:10, vol/vol) atmosphere in an anoxic glove chamber (Mecaplex). The remaining headspace (103 ml) was flushed with methane for 4 min, and the overpressure was adjusted to a final value of 0.6 × 105 Pa. Finally, 10 ml of CO2 and 10 ml of 13C-labeled methane (∼7% 13CH4; δ13C, 5,400‰) were added. The bottles were incubated at 12°C and gently shaken horizontally to facilitate mixing of the methane and sulfate. Pure 13CH4 was not used as it was shown previously that extremely high concentrations of a 13C-labeled substrate may retard microbial growth significantly (43).
After 96 days of incubation the mat material in both bottles was pelleted by centrifugation. All of the material from bottle A (wet weight, ∼3 g) was used for analyses of lipids and stable carbon isotope ratios. Bottle B was resupplied with medium and gas as described above. After an additional 97 days of incubation the mat material was pelleted and split into two aliquots (1.5 g [wet weight] each). One aliquot was used for biomarker and δ13C analyses, and the other was resupplied as described above, incubated for an additional 123 days, and then pelleted and analyzed.
For quantification of sulfate, 1.5 ml of a particle-free water sample was mixed with 0.1 ml of 2 M HCl. After the preparation was heated in a boiling water bath, 0.4 ml of a 0.5 M BaCl2 solution was added. Precipitated BaSO4 was quantitatively collected on a nitrocellulose filter (diameter, 25 mm; pore size, 0.2 mm), washed with 10 ml distilled water, dried at 60°C, and quantified by weighing.
Extraction and fractionation of lipids.
For lipid biomarker analyses 1 g (wet weight) of a mat sample was saponified in 6% KOH in methanol (75°C, 3 h) and extracted with n-hexane to obtain the neutral lipids. Carboxylic acid methyl esters were obtained by acidification of the residual phase to pH 1, extraction with CH2Cl2, and subsequent methylation using trimethylchlorosilane reagent in methanol (1:8, vol/vol) for 1 h at 80°C. For carboxylic acids (methyl esters) double bonds were determined by using dimethyl disulfide derivatives (7) and performing coelution experiments with authentic standards. Neutral lipids were separated by thin-layer chromatography, which resulted in a hydrocarbon fraction (Silica Gel 60; 0.25 mm; CH2Cl2; Rf, 0.35 to 1). Double bond positions of the isoprenoid hydrocarbons were not determined. Alcohols were silylated with N,O-bis(trimethylsilyl)-trifluoroacetamide for 1 h at 80°C prior to analyses. To analyze high-molecular-weight ether-bound lipids, aliquots of the neutral lipids were subjected to ether cleavage by HI treatment (2 h at 110°C), and reduction of the resulting iodides using LiAlH4 in dry diethyl ether under an Ar atmosphere was performed by using a procedure modified from the procedure described by Kohnen et al. (20).
Gas chromatography, gas chromatography-mass spectrometry, gas chromatography-combustion isotope ratio mass spectrometry, and elemental analyzer-combustion isotope ratio mass spectrometry.
Compounds were analyzed by combined gas chromatography-mass spectrometry (HP6890 gas chromatograph coupled to a Micromass Quattro II mass spectrometer) and were identified by comparison of the mass spectra and retention times with previously published data and/or data for reference compounds (3, 25). The analytical error for lipid quantification was about 5% using cholestane as an internal standard.
δ13C values of lipids were analyzed (minimum of three replicates) using a ThermoFinnigan Trace GC gas chromatograph coupled to a Finnigan MAT 252 isotope ratio mass spectrometer. Combustion of the components to CO2 was performed with a CuO-Ni-Pt furnace operated at 940°C. The stable carbon isotope compositions are expressed in the delta notation (δ13C) compared with the Vienna-Pee Dee Belemnite standard (the standard deviation was usually less than 0.5‰). Isotopic compositions of alcohols and fatty acids were corrected for addition of trimethylsilane and the addition of the carbon atom during the preparation of methyl esters.
For measurements of carbon isotope ratios (δ13C) of the bulk mat samples the samples were combusted in a Carlo Erba NA-2500 analyzer, from which the evolved CO2 was passed into a continuous flow of helium through a ConFlo II interface to a Finnigan MAT 252 isotope mass spectrometer. The analytical error of this method was ≤0.2‰.
Carbon stable isotope values are expressed in δ notation in parts per thousand compared with V-PDB and were calculated as follows: δ13C = [(Rsample/Rstandard) − 1] × 1,000. The concentration of 13C in any given compound was calculated from δ13C and the total concentration (Ct) using the following equation (with RV-PDB = 0.0112372 ± 0.0000090): 13C concentration = Ct/{1/[(δ13C/1,000 + 1) × RV-PDB] + 1}.
RESULTS
A microbial mat performing anaerobic oxidation of methane was incubated in vitro under strictly anoxic conditions using 13C-enriched methane for 316 days. At three times the mat was sampled and the samples were used for lipid analyses (i.e., to determine compound-specific δ13C values) in order to trace the methane-derived carbon in the biomass and in lipids specific for distinct microorganisms.
In vitro consumption of sulfate.
Consumption of sulfate was measured at the three sampling times. After 96 days the added sulfate (15.42 mM) was nearly completely reduced, and the remaining concentration was 1.29 mM. After addition of 20.28 mM and 16.85 mM sulfate, at the next two times the sulfate was consumed and the residual concentrations were 1.14 mM and 5.43 mM, respectively.
Microbial composition of the mat.
The mat sample used for the experiment was an aliquot of the mat described previously (25) and sample LOGA C described by Blumenberg et al. (3). The mat was reported to consist mainly of ANME-1 archaea, which were accompanied by SRB belonging to the Desulfosarcina-Desulfococcus group. However, ongoing molecular microbiological (18) and biomarker (3) investigations showed that various amounts of ANME-2 archaea, which grew in close association with SRB belonging to the cluster mentioned above, were also present.
Lipid composition of the mat.
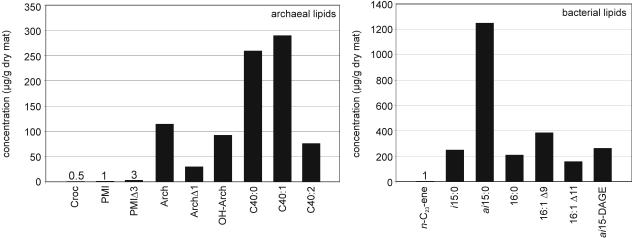
Figure 1 shows the concentrations of selected archaeal and bacterial lipid compounds found at the beginning of the experiment, prior to addition of [13C]methane. Large amounts of bacterial fatty acids with terminally branched 12-methyltetradecanoic acid (ai15:0) were observed, and the concentrations exceeded the concentrations of other fatty acids by more than threefold (ai15:0 concentration, 1,243 μg/g [dry weight]). Also present at a high level (262 μg/g [dry weight]) was the nonisoprenoid glycerol diether ai15DAGE, which presumably is of bacterial origin. The archaeal lipids included isoprenoid hydrocarbons (crocetane, 2,6,10,15,19-pentamethyleicosane [PMI], 2,6,10,15,19-pentamethyleicosenes [PMIΔs]), isoprenoid dialkyl glycerol diethers (saturated and monounsaturated archaeol, sn-2-hydroxyarchaeol), and glycerol dialkyl glycerol tetraethers (analyzed as biphytanes after ether cleavage). The quantitatively dominant archaeal biomarkers were biphytanes, including an acyclic compound (C40:0; 259 μg/g [dry weight]) and two internally cyclized compounds, one with one pentacyclic ring (C40:1; 288 μg/g [dry weight]) and one with two pentacyclic rings (C40:2; 76 μg/g [dry weight]).
FIG. 1.
Concentrations of selected bacterial and archaeal compounds at the beginning of the in vitro experiment. For chemical structures of selected compounds see the Appendix. Abbreviations: Croc, crocetane (2,6,11,15-tetramethylhexadecane); PMI, 2,6,10,15,19-pentamethyleicosane; PMIΔ3, PMIΔ with three double bonds; Arch, archaeol (2,3-di-O-phytanyl-sn-glycerol); OH-Arch, sn-2-hydroxyarchaeol; C40:0, acyclic biphytane; C40:1, internally cyclized biphytane (with one pentacyclic ring); C40:2, internally cyclized biphytane (with two pentacyclic rings); n-C23-ene, n-tricos-10-ene; i15:0, 13-methyltetradecanoic acid; ai15:0, 12-methyltetradecanoic acid; 16:0, hexadecanoic acid; 16:1Δ9, hexadec-9-enoic acid; 16:1Δ11, hexadec-11-enoic acid; ai15-DAGE, 1,2-di-O-12-methyltetradecyl-sn-glycerol.
For all the bacterial and archaeal lipids 13C was strongly depleted, and the δ13C values were between −77.5 and −98‰.
Change of δ13C signatures during the experiment.
The δ13C of the total decarbonated biomass shifted from an initial value of −72.2‰ to −65.5‰ after 96 days and to −62.6‰ after 193 days (there was no data point after 316 days due to sample size limitation). The substantial enrichment of 13C illustrates that there was incorporation of methane carbon into the microbial system.
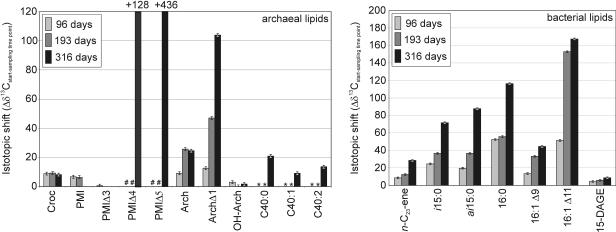
Figure 2 shows the difference between the δ13C at the start of the experiment and the δ13C after 96, 193, or 316 days (Δδ13Cstart-end) for selected compounds. Most of the compounds showed increasing uptake of 13C throughout the experiment. For the major compounds, the greatest δ13C shifts were found for bacterial fatty acids, and the Δδ13Cstart-end for 16:1Δ11 was ∼165‰. Significant enrichment of 13C was also observed for the nonisoprenoid hydrocarbon n-tricos-10-ene (Fig. 2). In contrast, bacterial glycerol diethers (e.g., ai15 DAGE) yielded only low Δδ13C values (about 10‰).
FIG. 2.
Δδ13C values of selected compounds at each sampling time in relation to the δ13C value at the beginning, indicating the biosynthetic activity of the source organisms. The error bars indicate the standard deviations for δ13C analyses. A number sign indicates that trace amount was present (not analyzed); an asterisk indicates that a compound was not analyzed. Calculations for PMIΔ4 and PMIΔ5 were based on a theoretical δ13C starting value of −90‰ (as observed for PMI). For abbreviations see the legend to Fig. 1.
Uptake of methane carbon was also observed for individual archaeal lipids. For major archaeal lipids we observed uptake of 13C into archaeol (Δδ13Cstart-end, 24.5‰), tetraether-released biphytanes (Δδ13Cstart-end, ∼9 to 20‰), and a monounsaturated archaeol (Δδ13Cstart-end, 103.4‰). sn-2-Hydroxyarchaeol and several isoprenoid hydrocarbons (crocetane, PMI, PMIΔ3) exhibited only slight changes in the δ13C (Fig. 2). No clear 13C uptake was found for crocetane over time (Fig. 2). Exceptionally high δ13C values were detected for polyunsaturated 2,6,10,15,19-pentamethyleicosenes with four double bonds (PMIΔ4) and five double bonds (PMIΔ5) (δ13C for PMIΔ5, 346‰). However, PMIΔ4 and PMIΔ5 were found at significant concentrations only at the last sampling time (PMIΔ4, 6.1 μg/g [dry weight]; PMIΔ5, 0.7 μg/g [dry weight]). In samples from the other times these compounds were present only in trace amounts, which did not allow analyses of their δ13C values (the data shown in Fig. 2 and 3 are based on a theoretical δ13C starting value of −90‰).
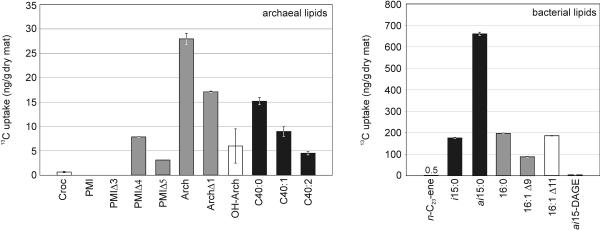
FIG. 3.
Net 13C uptake for selected compounds after completion of the experiment, calculated for 1 g (dry weight) of mat. The error bars indicate the maximum and minimum uptake of 13C using the analytical error for δ13C analyses. Calculations for PMIΔ4 and PMIΔ5 were based on a theoretical δ13C starting value of −90‰ (as observed for PMI) and concentrations measured at the end point. For abbreviations see the legend to Fig. 1. Different kinds of bars indicate the most likely source organisms of the lipid components (based on a previous study [3]), as follows: solid bars, ANME-1 associations; open bars, ANME-2 associations; gray bars, mixed and/or unknown sources.
DISCUSSION
Uptake of methane-derived carbon into microbial biomass.
The bulk decarbonated biomass was clearly enriched in 13C during the experiment, illustrating that carbon was taken up from the added tracer methane. Methane concentrations were not analyzed, but they could be deduced from the sulfate reduction data, assuming a 1:1 stoichiometry between methane oxidation and sulfate reduction (27). Based on this analysis and the isotopic shift of total biomass, 1.9% of the methane C consumed was incorporated into microbial biomass (without considering fractionation effects and assuming no change in the total biomass) during the first period of the experiment (0 to 96 days bottle A), and 1.1% was incorporated during the second period (0 to 193 days, bottle B).
Uptake of methane carbon into individual lipids.
In general, the extent of the δ13C shift of an individual component corresponds directly to the portion of it that is synthesized during exposure of the source organism to the 13C-labeled substrate. High δ13C shifts are to be expected for compounds with short life cycles, while relatively stable compounds should exhibit only minor shifts. Thus, the δ13C shifts reflect the biosynthetic activity of the organisms, while the net 13C uptake values indicate the total carbon uptake by the source organisms.
Figure 3 shows that the uptake of methane carbon into the major bacterial compounds (fatty acids) was much greater than the incorporation into major archaeal lipids (e.g., archaeol, sn-2-hydroxyarchaeol, glycerol dialkyl glycerol tetraether-released biphytanes) (also see Fig. 2). The low energy yield of the anaerobic oxidation of methane (a free energy change of −10 to −40 kJ mol−1 was estimated for various in situ conditions [42]) requires high methane turnover rates for a sufficient energy yield (25) and thus led to high 13CO2 production during our experiment. It is assumed that both SRB and archaea use CO2 as a substrate for biosynthesis in the acetyl-coenzyme A pathway (14, 42). The higher uptake rates of methane-derived carbon into the sulfate reducers than into the methanotrophic archaea imply that the biosynthetic (anabolic) activity of the former organisms is enhanced. An unequal energy share within the coupled AOM process was proposed based on theoretical studies, with the sulfate-reducing partner demanding about 60% of the total energy yield (17). Consequently, more energy accessible to the SRB also increases the energy available for biosynthetic purposes (23), which provides a suitable explanation for the observed higher net 13C uptake of methane-derived carbon by SRB. An enhanced demand for the biosynthesis of lipids by the SRB compared to the archaea might also be explained by the higher chemical and enzymatic resistance of archaeal ether lipids than of bacterial ester-bound lipids (41).
Nevertheless, our results do not exclude the possibility of decoupling of SRB and anaerobic methanotrophic archaea during AOM, with methane oxidation as well as sulfate reduction taking place in the archaeal cells (42). Recently, genomic features of both sulfate-reducing and methanogenic archaea have been detected in ANME-1 archaea (14). However, the higher level of 13C uptake by SRB excludes the possibility that heterotrophic consumption of archaeal biomass by the bacterial partners is a prominent process.
All fatty acids investigated became enriched in 13C, although to different extents (Fig. 2 and 3). The greatest shift in δ13C within major components was found for the monounsaturated 16:1Δ11 fatty acid (Δδ13Cstart-end, 167‰). However, the net 13C uptake of this compound (184.4 ng 13C/g [dry weight]) was lower than that for the ai15:0 fatty acid (661 ng 13C/g [dry weight]). Both compounds are attributed to different sulfate-reducing bacteria belonging to the Desulfosarcina-Desulfococcus group. 16:1Δ11 fatty acid is suggested to originate from AOM-specific SRB (10) prevailing in sediment with ANME-2 dominance (4), and the ai15:0 fatty acid is a prominent lipid of ANME-1-associated SRB (3, 25). The net 13C uptake of compounds attributed to ANME-1-associated SRB exceeded that of compounds attributed to ANME-2-associated bacteria by about threefold (Fig. 3). However, greater δ13C shifts were observed for lipids specific for ANME-2-associated bacteria (Fig. 2). The distribution pattern of bacterial lipids (Fig. 1) revealed a clear predominance of ANME-1-linked SRB over ANME-2-linked SRB within the mat investigated (3). A rough estimate of SRB from the relative concentration of lipid biomarkers resulted in a mass ratio of ANME-1-associated SRB to ANME-2-associated SRB of 6:1. Based on this estimate, while about threefold more methane carbon is incorporated by ANME-1-associated SRB than by ANME-2 associated SRB, the latter organisms appear to be biosynthetically about twice as active as the former organisms.
Nonisoprenoid ether lipids have been described for several AOM sites (25, 30), and it has been suggested that they are bacterial biomarkers (for instance, for members of the deeply branching phylum Thermodesulfobacterium) (22). A recent investigation of AOM-performing mats from the Black Sea suggested that distinct nonisoprenoid ether lipids originate from the same source bacteria (SRB) as the structurally corresponding fatty acids (e.g., ai15-DAGE and ai15:0 fatty acid) (3). However, the nonisoprenoid ai15-DAGE had a Δδ13Cstart-end of only about 10‰, and the uptake of 13C was 3.7 ng/g (dry weight), values much lower than those found for the corresponding ai15:0 fatty acid (Fig. 2 and 3). This indicates that the biosynthesis rates of DAGEs are lower than those of structurally related fatty acids within the source organism.
The source organism of the nonisoprenoid hydrocarbon n-tricos-10-ene is still unknown, although it has been suggested that this compound originates from bacteria found only at AOM sites (39). The observed 13C uptake for this compound (Δδ13Cstart-end, 28.6‰) revealed biosynthetic activity of the organism in the AOM-performing mat. For the major archaeal lipids, archaeol, with a Δδ13Cstart-end of 24.6‰, showed the strongest uptake of 13C. However, archaeol is a substantial lipid compound in all archaea investigated so far (19) and thus provides no specific information about the biosynthetic activity of certain phylogenetic groups. The information for a suite of 13C-depleted C40 isoprenoids which are proposed to originate from archaea belonging to the ANME-1 cluster is more precise (3). These tetraether-released biphytanes indicate that there is biosynthetic activity of the source archaea with significant uptake of methane carbon (Δδ13Cstart-end, 9.1 to 20.9‰). For sn-2-hydroxyarchaeol and the isoprenoid hydrocarbon crocetane, biomarkers most probably originating from ANME-2 archaea (3), there was almost no change in δ13C (Fig. 2). Apparently, no substantial biosynthetic activity of ANME-2 archaea took place. This might have been due to the relatively low methane partial pressure used during the experiment (0.16 MPa). The methane-dependent sulfate reduction in ANME-2-dominated samples could be increased five times by increasing the methane pressure from 0.1 MPa to 1.1 MPa. The same treatment of ANME-1-dominated associations resulted in only a twofold increase (26, 27). Hence, different reactions to methane partial pressure might be responsible for the marginal biosynthetic activity of ANME-2 archaea compared to ANME-1 archaea during our in vitro experiment with the relatively low methane partial pressure. However, considerable 13C incorporation occurred within lipids of sulfate-reducing bacteria suggested to be linked to ANME-2 archaea (e.g., 16:1Δ11) (3, 10, 38). This indicates that these SRB may not rely solely on their ANME-2 archaeal partners.
Biosynthetic pathways of selected archaeal lipids.
It is well known that AOM-performing archaea contain a suite of one saturated and several unsaturated 2,6,10,15,19-pentamethyleicosenes (11, 12). At the beginning of our experiment, only PMIΔs with up to three double bonds were detected. For these compounds uptake of 13C was not observed. Significant concentrations of PMIΔs with four and five double bonds were found only after 316 days, and both exhibited substantial 13C enrichment (Δδ13C of PMIΔ4, 128‰; δ13C of PMIΔ5, 436‰) (Fig. 2). This suggests that polyunsaturated PMIΔs represent early steps in PMI biosynthesis by methanotrophic archaea and are subsequently reduced. This biosynthetic pathway was also suggested for methanogenic archaea (37).
The source of the unsaturated archaeol, which had a Δδ13Cstart-end of 103.4‰, is still being debated. Monounsaturated archaeols were shown to originate from hydroxyarchaeol under acidic conditions as laboratory conversion products (9). However, recent studies described this compound as a constitutive membrane lipid of the hyperthermophilic methanogen Methanopyrus kandleri (28). Under the conditions used in this study (mild alkaline hydrolysis), conversion of hydroxyarchaeol to unsaturated archaeol has not been reported and was successfully applied to analysis of hydroxyarchaeol-containing samples (25, 36). Moreover, the large difference in Δδ13C values between the monounsaturated archaeol (Δδ13Cstart-end, 103.4‰) and sn-2-hydroxyarchaeol (Δδ13Cstart-end, ∼2‰) strongly argues against formation of unsaturated archaeol from sn-2-hydroxyarchaeol as an analytical artifact (Fig. 2). Therefore, we believe that the monounsaturated archaeol observed is a natural lipid constituent of specific methanotrophic Euryarchaeota.
With regard to biphytanes, Fig. 3 shows that there was a decrease in 13C uptake from the acyclic compound via the monocyclic compound to the bicyclic compound. Either this indicates that there was a shift within the biphytane distribution of the archaeal cells, with the cyclic biphytanes being discriminated, or it is explained by differences in the individual biosynthesis rates.
Conclusion.
In vitro incubation of an AOM-performing microbial mat from the Black Sea with 13C-enriched methane resulted in considerable incorporation of the 13C label into biomass.
In general, the uptake of methane carbon was greater for sulfate-reducing bacteria than for methanotrophic archaea, suggesting that the bacteria grew on low-molecular-weight methane-derived organic and/or inorganic carbon compounds (e.g., CO2).
Within the bacteria, SRB possibly associated with ANME-1 archaea exhibited higher net 13C incorporation, while SRB believed to be associated with ANME-2 archaea appeared to be biosynthetically more active. Within the archaea, lipid biomarkers specific for ANME-1 archaea became more enriched in 13C than lipid biomarkers specific for ANME-2 archaea, which might have been a result of an adaptation of ANME-1 archaea to low methane partial pressure.
Enhanced incorporation of 13C into unsaturated PMIΔ derivatives with an increasing number of double bonds implies that in the biosynthetic pathway of PMI and PMIΔs in methanotrophic archaea polyunsaturated compounds are precursors of compounds containing fewer or no double bonds. Moreover, the labeling experiment indicated that unsaturated archaeol is a natural constituent of the lipid inventory of specific methanotrophic Euryarchaeota.
The data showed that in vitro incubation experiments using 13C-labeled substrates are powerful tools for investigating the substrate carbon flow into lipid biosynthesis, even in complex microbial communities. However, further work is needed to understand the specific mode of microbial association within anaerobic methanotrophic consortia.
Appendix.
Chemical structures of selected compounds described in the legend to Fig. 1 are shown in Fig. A1.
FIG.A1.
Chemical structures of compounds described in the legend to Fig. 1.
Acknowledgments
We thank the crews of the RV Professor Logachev and the research submersible Jago for excellent collaboration during the field work and Sabine Beckmann for analytical work. We especially acknowledge three anonymous reviewers for comments and suggestions that considerably improved the manuscript. Friedrich Widdel (MPI Bremen), Martin Krüger (BGR Hannover), and Andrea Wieland are gratefully thanked for discussions and helpful comments on the manuscript.
This study received financial support through the GHOSTDABS (03G0559A) and MUMM (03G0554A) programs of the Bundesministerium für Bildung und Forschung (BMBF), the Universität Hamburg, and the Max-Planck-Gesellschaft (Germany).
Footnotes
Publication GEOTECH-119 of the GEOTECHNOLOGIEN program of the BMBF and the DFG and publication 14 of the GHOSTDABS research program.
REFERENCES
- 1.Annweiler, E., G. Antranikian, W. Francke, C. Garms, S. Hebenbrock, H. H. Richnow, and W. Michaelis. 2000. Naphthalene degradation and incorporation of naphthalene-derived carbon into biomass by the thermophile Bacillus thermoleovorans. Appl. Environ. Microbiol. 66:518-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnes, R. O., and E. D. Goldberg. 1976. Methane production and consumption in anaerobic marine sediments. Geology 4:297-300. [Google Scholar]
- 3.Blumenberg, M., R. Seifert, J. Reitner, T. Pape, and W. Michaelis. 2004. Membrane lipid patterns typify distinct anaerobic methanotrophic consortia. Proc. Natl. Acad. Sci. USA 101:11111-11116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boetius, A., K. Ravenschlag, C. J. Schubert, D. Rickert, F. Widdel, A. Gieseke, R. Amann, B. B. Jørgensen, U. Witte, and O. Pfannkuche. 2000. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407:623-626. [DOI] [PubMed] [Google Scholar]
- 5.Boschker, H. T. S., S. C. Nold, P. Wellsbury, D. Bos, W. de Graaf, R. Pel, R. J. Parkes, and T. E. Cappenberg. 1998. Direct linking of microbial populations to specific biogeochemical processes by 13C-labelling of biomarkers. Nature 392:801-805. [Google Scholar]
- 6.Bull, I. D., N. R. Parekh, G. H. Hall, P. Ineson, and R. P. Evershed. 2000. Detection and classification of atmospheric methane oxidizing bacteria in soil. Nature 405:175-178. [DOI] [PubMed] [Google Scholar]
- 7.Buser, H. R., H. Arn, P. Guerin, and S. Rauscher. 1983. Determination of double bond position in mono-unsaturated acetates by mass spectrometry of dimethyl disulfides adducts. Anal. Chem. 55:818-822. [Google Scholar]
- 8.Crossman, Z. M., N. McNamara, N. Parekh, P. Ineson, and R. P. Evershed. 2001. A new method for identifying the origins of simple and complex hopanoids in sedimentary materials using stable isotope labelling with 13C-CH4 and compound specific stable isotope analyses. Org. Geochem. 32:359-364. [Google Scholar]
- 9.Ekiel, I., and G. D. Sprott. 1992. Identification of degradation artifacts formed upon treatment of hydroxydiether lipids from methanogens with methanolic HCl. Can. J. Microbiol. 38:764-768. [Google Scholar]
- 10.Elvert, M., A. Boetius, K. Knittel, and B. B. Jørgensen. 2003. Characterization of specific membrane fatty acids as chemotaxonomic markers for sulfate-reducing bacteria involved in anaerobic oxidation of methane. Geomicrobiol. J. 20:403-419. [Google Scholar]
- 11.Elvert, M., E. Suess, J. Greinert, and M. J. Whiticar. 2000. Archaea mediating anaerobic methane oxidation in deep-sea sediments at cold seeps of the eastern Aleutian subduction zone. Org. Geochem. 31:1175-1187. [Google Scholar]
- 12.Elvert, M., E. Suess, and M. J. Whiticar. 1999. Anaerobic methane oxidation associated with marine gas hydrates. Superlight C-isotopes from saturated and unsaturated C20 and C25 irregular isoprenoids. Naturwissenschaften 86:295-300. [Google Scholar]
- 13.Hallam, S. J., P. R. Girguis, C. M. Preston, P. M. Richardson, and E. F. DeLong. 2003. Identification of methyl coenzyme M reductase A (mcrA) genes associated with methane-oxidizing archaea. Appl. Environ. Microbiol. 69:5483-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hallam, S. J., N. Putnam, C. M. Preston, J. C. Detter, D. Rokhsar, P. M. Richardson, and E. F. DeLong. 2004. Reverse methanogenesis: testing the hypothesis with environmental genomics. Science 305:1457-1462. [DOI] [PubMed] [Google Scholar]
- 15.Hinrichs, K.-U., R. E. Summons, V. J. Orphan, S. P. Sylva, and J. M. Hayes. 2000. Molecular and isotopic analysis of anaerobic methane-oxidizing communities in marine sediments. Org. Geochem. 31:1685-1701. [Google Scholar]
- 16.Hoehler, T. M., M. J. Alperin, D. B. Albert, and C. S. Martens. 1994. Field and laboratory studies of methane oxidation in an anoxic marine sediment: evidence for a methanogen-sulfate reducer consortium. Global Biogeochem. Cycles 8:451-463. [Google Scholar]
- 17.Hoehler, T. M., M. J. Alperin, D. B. Albert, and C. S. Martens. 2001. Apparent minimum free energy requirements for methanogenic archaea and sulfate-reducing bacteria in an anoxic marine sediment. FEMS Microbiol. Ecol. 38:33-41. [Google Scholar]
- 18.Knittel, K., T. Lösekann, A. Boetius, R. Kort, and R. Amann. 2004. Diversity and distribution of methanotrophic archaea (ANME) at cold seeps. Appl. Environ. Microbiol. 71:467-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koga, Y., H. Morii, M. Akagawa-Matsushita, and M. Ohga. 1998. Correlation of polar lipid composition with 16S rRNA phylogeny in methanogens. Further analysis of lipid component parts. Biosci. Biotechnol. Biochem. 62:230-236. [DOI] [PubMed] [Google Scholar]
- 20.Kohnen, M. E. L., S. Schouten, J. S. Sinninghe Damsté, J. W. De Leeuw, D. A. Merritt, and J. M. Hayes. 1992. Recognition of paleobiochemicals by a combined molecular sulfur and isotope geochemical approach. Science 256:358-362. [DOI] [PubMed] [Google Scholar]
- 21.Krüger, M., A. Meyerdierks, F. O. Glöckner, R. Amann, F. Widdel, M. Kube, R. Reinhardt, J. Kahnt, R. Böcher, R. K. Thauer, and S. Shima. 2003. A conspicuous nickel protein in microbial mats that oxidize methane anaerobically. Nature 426:878-881. [DOI] [PubMed] [Google Scholar]
- 22.Langworthy, T. A., G. Holzer, J. G. Zeikus, and T. G. Tornabene. 1983. Iso- and anteiso-branched glycerol diethers of the thermophilic anaerobe Thermodesulfotobacterium commune. Syst. Appl. Microbiol. 4:1-17. [DOI] [PubMed] [Google Scholar]
- 23.Lengeler, J. W., G. Drews, and H. G. Schlegel. 1999. Biology of the prokaryotes. Thieme, Stuttgart, Germany.
- 24.Londry, K. L., L. L. Jahnke, and D. J. Des Marais. 2004. Stable carbon isotope ratios of lipid biomarkers of sulfate-reducing bacteria. Appl. Environ. Microbiol. 70:745-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michaelis, W., R. Seifert, K. Nauhaus, T. Treude, V. Thiel, M. Blumenberg, K. Knittel, A. Gieseke, K. Peterknecht, T. Pape, A. Boetius, R. Amann, B. B. Jørgensen, F. Widdel, J. Peckmann, N. V. Pimenov, and M. B. Gulin. 2002. Microbial reefs in the Black Sea fueled by anaerobic oxidation of methane. Science 297:1013-1015. [DOI] [PubMed] [Google Scholar]
- 26.Nauhaus, K., A. Boetius, M. Krüger, and F. Widdel. 2002. In vitro demonstration of anaerobic oxidation of methane coupled to sulphate reduction in sediment from a marine gas hydrate area. Environ. Microbiol. 4:296-305. [DOI] [PubMed] [Google Scholar]
- 27.Nauhaus, K., T. Treude, A. Boetius, and M. Krüger. 2005. Environmental regulation of the anaerobic oxidation of methane: comparison of ANME-I and ANME-II-communities. Environ. Microbiol. 7:98-106. [DOI] [PubMed] [Google Scholar]
- 28.Nishihara, M., H. Morii, K. Matsuno, M. Ohga, K. O. Stetter, and Y. Koga. 2002. Structural analysis by reductive cleavage with LiAlH4 of an allyl ether choline-phospholipid, archaetidylcholine, from the hyperthermophilic methanoarchaeaon Methanopyrus kandleri. Archaea 1:123-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orphan, V. J., C. H. House, K.-U. Hinrichs, K. D. McKeegan, and E. F. DeLong. 2002. Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments. Proc. Natl. Acad. Sci. USA 99:7663-7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pancost, R. D., I. Bouloubassi, G. Aloisi, J. S. Sinninghe Damsté, and the Medinaut Shipboard Party. 2001. Three series of non-isoprenoidal dialkyl glycerol diethers in cold-seep carbonate crusts. Org. Geochem. 32:695-707. [Google Scholar]
- 31.Pancost, R. D., E. C. Hopmans, J. S. Sinninghe Damsté, and the Medinaut Shipboard Party. 2001. Archaeal lipids in Mediterranean cold seeps: molecular proxies for anaerobic methane oxidation. Geochim. Cosmochim. Acta 65:1611-1627. [Google Scholar]
- 32.Pancost, R. D., J. S. Sinninghe Damsté, S. De Lint, M. J. E. C. Van der Maarel, J. C. Gottschal, and the Medinaut Shipboard Party. 2000. Biomarker evidence for widespread anaerobic methane oxidation in Mediterranean sediments by a consortium of methanogenic archaea and bacteria. Appl. Environ. Microbiol. 66:1126-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reeburgh, W. S. 1976. Methane consumption in Cariaco Trench waters and sediments. Earth Planet. Sci. Lett. 28:337-344. [Google Scholar]
- 34.Reeburgh, W. S., S. C. Whalen, and M. J. Alperin. 1993. The role of methylotrophy in the global methane budget, p. 1-14. In J. C. Murrell and P. D. Kelly (ed.), Microbial growth on C1 compounds. Proceedings of the 7th International Symposium. American Society for Microbiology, Washington, D.C.
- 35.Richnow, H. H., E. Annweiler, M. Koning, J.-C. Lüth, R. Stegmann, C. Garms, W. Francke, and W. Michaelis. 2000. Tracing the transformation of labelled [1-13C]-phenanthrene in a soil bioreactor. Environ. Pollut. 108:91-101. [DOI] [PubMed] [Google Scholar]
- 36.Schouten, S., S. G. Wakeham, E. C. Hopmans, and J. S. Sinninghe Damsté. 2003. Biogeochemical evidence that thermophilic archaea mediate the anaerobic oxidation of methane. Appl. Environ. Microbiol. 69:1680-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sinninghe Damsté, J. S., S. Schouten, N. H. van Vliet, R. Huber, and J. A. J. Geenevasen. 1997. A polyunsaturated irregular acyclic C25 isoprenoid in a methanogenic archaeon. Tetrahedron Lett. 38:6881-6884. [Google Scholar]
- 38.Sturt, H. F., R. E. Summons, K. Smith, M. Elvert, and K.-U. Hinrichs. 2004. Intact polar membrane lipids in prokaryote and sediments deciphered by ESI-HPLC-MS—new biomarkers for biogeochemistry and microbial ecology. Rapid Commun. Mass Spectrom. 18:617-628. [DOI] [PubMed] [Google Scholar]
- 39.Thiel, V., J. Peckmann, O. Schmale, J. Reitner, and W. Michaelis. 2001. A new straight-chain hydrocarbon biomarker associated with anaerobic methane cycling. Org. Geochem. 32:1019-1023. [Google Scholar]
- 40.Thiel, V., J. Peckmann, R. Seifert, P. Wehrung, J. Reitner, and W. Michaelis. 1999. Highly isotopically depleted isoprenoids: molecular markers for ancient methane venting. Geochim. Cosmochim. Acta 63:3959-3966. [Google Scholar]
- 41.van de Vossenberg, J. L. C. M., A. J. M. Driessen, and W. N. Konings. 2000. The special composition of the cell membrane allows Archaea to live in extreme environments, p. 303. In K. B. Storey and J. M. Storey (ed.), Cell and environmental responses to stress, vol. 1. Environmental stressors and gene responses. Elsevier Science Ltd., Amsterdam, The Netherlands. [Google Scholar]
- 42.Widdel, F., A. Boetius, and R. Rabus. 3 May 2004, posting date. Anaerobic biodegradation of hydrocarbons including methane. In M. Dworkin, S. Falkow, E. Rosenberg, K. H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes, electronic edition. Springer, New York, N.Y. [Online.] http://141.150.157.117:8080/prokPUB/chaprender/jsp/showchap.jsp?chapnum=489.
- 43.Zengler, K., H. H. Richnow, R. Rosselló-Mora, W. Michaelis, and F. Widdel. 1999. Methane formation from long-chain alkanes by anaerobic microorganisms. Nature 401:266-269. [DOI] [PubMed] [Google Scholar]