Abstract
Pseudomonas putida CA-3 is capable of accumulating medium-chain-length polyhydroxyalkanoates (MCL-PHAs) when growing on the toxic pollutant styrene as the sole source of carbon and energy. In this study, we report on the molecular characterization of the metabolic pathways involved in this novel bioconversion. With a mini-Tn5 random mutagenesis approach, acetyl-coenzyme A (CoA) was identified as the end product of styrene metabolism in P. putida CA-3. Amplified flanking-region PCR was used to clone functionally expressed phenylacetyl-CoA catabolon genes upstream from the sty operon in P. putida CA-3, previously reported to generate acetyl-CoA moieties from the styrene catabolic intermediate, phenylacetyl-CoA. However, the essential involvement of a (non-phenylacetyl-CoA) catabolon-encoded 3-hydroxyacyl-CoA dehydrogenase is also reported. The link between de novo fatty acid synthesis and PHA monomer accumulation was investigated, and a functionally expressed 3-hydroxyacyl-acyl carrier protein-CoA transacylase (phaG) gene in P. putida CA-3 was identified. The deduced PhaG amino acid sequence shared >99% identity with a transacylase from P. putida KT2440, involved in 3-hydroxyacyl-CoA MCL-PHA monomer sequestration from de novo fatty acid synthesis under inorganic nutrient-limited conditions. Similarly, with P. putida CA-3, maximal phaG expression was observed only under nitrogen limitation, with concomitant PHA accumulation. Thus, β-oxidation and fatty acid de novo synthesis appear to converge in the generation of MCL-PHA monomers from styrene in P. putida CA-3. Cloning and functional characterization of the pha locus, responsible for PHA polymerization/depolymerization is also reported and the significance and future prospects of this novel bioconversion are discussed.
Polyhydroxyalkanoates (PHAs) are biodegradable polyesters produced by numerous bacterial species, whose varied thermoplastic and elastomeric properties offer the potential for industrial, medical, and bulk consumer applications (34). PHA accumulation typically occurs when a suitable bacterium encounters a relative abundance of utilizable carbon, offset by an inorganic nutrient limitation, (e.g., nitrogen). The physicochemical properties of PHAs depend on their constituent (R)-3-hydroxyacyl-coenzyme A (CoA) monomer compositions; >100 monomers have been identified to date (27, 33). PHA synthase enzymes responsible for (R)-3-hydroxyacyl-CoA monomer polymerization are divided into class I, short-chain-length PHA synthases, polymerizing C4-C5 carbon-length monomers; and class II medium-chain-length (MCL) PHA synthases, whose substrate monomer lengths range between C6 to C12 (23). The subsequent release of (R)-3-hydroxyacyl-CoA monomers from accumulated PHAs is facilitated by a PHA-associated depolymerase enzyme, PhaZ (13, 34).
Pseudomonas putida CA-3 exhibits the unique ability to accumulate MCL-PHAs when grown on the industrial waste pollutant styrene as the sole source of carbon and energy (32). Previously, we reported that styrene degradation in P. putida CA-3 involved an upper pathway converting styrene to phenylacetic acid (PA), and an independently regulated lower pathway initiated via activation of PA to phenylacetyl-CoA (21). A catabolic operon specifically responsible for the metabolism of phenylacetyl-CoA was first identified in P. putida U and Escherichia coli W (22, 5). This pathway, referred to as the PACoA catabolon, reportedly involves oxidation of the aromatic nucleus, followed by ring cleavage and β-oxidation of the alicyclic compound via a multienzyme complex.
In this study, we set out to identify and characterize the genetic apparatus necessary for metabolism of styrene post-phenylacetic acid, the subsequent generation of PHA monomers, and the accumulation of PHAs in P. putida CA-3. This novel conversion of the toxic environmental pollutant styrene to PHA is of biotechnological significance, as it (i) identifies the potential to exploit styrene waste as a low-cost starting material for value-added PHA accumulation and (ii) may also represent an economically attractive incentive for the bioremediation of stored styrene wastes. Ultimately, it will be the generation of recombinant strains capable of PHA overaccumulation from styrene that will dictate the potential application of this technology. In this regard, the identification and molecular characterization of some of the key structural and regulatory components involved, such as those described here, will allow the development of future recombinant strategies.
MATERIALS AND METHODS
Bacterial strains, plasmids, culture media, and growth.
Pseudomonas putida CA-3, a styrene-degrading, bioreactor isolate, has been described previously (18, 20). E. coli CC118λpir hosted the mini-Tn5 derivative pUT-Km1. This suicide plasmid has the R6K origin of replication and encodes resistance to kanamycin and ampicillin (3). Plasmid pRK600 (Cmr) was used as a helper in triparental mating experiments and encodes the tra functions facilitating pUT-Km1 mobilization. PCR2.1-TOPO vector (Invitrogen, California) was used in the cloning of PCR amplification products.
Pseudomonas putida CA-3 was routinely grown on E2-minimal medium (30) containing either 10 mM citrate or 15 mM PA as the sole carbon source. Growth on styrene was facilitated by the addition of 70 μl of liquid styrene to a test tube fixed centrally to the bottom of a 1-liter Erlenmeyer flask. PHA accumulation was achieved by reducing the nitrogen content of the E2 medium in the form of NH4SO4 from 8 mM to 1.5 mM and allowing cultures to grow for 48 h. All E. coli vector hosts were maintained on standard LB agar plates containing the appropriate antibiotic(s) and were inoculated into 10 ml LB overnight broths prior to desired applications.
Nucleic acid manipulations.
Nucleic acid isolation (DNA/RNA), was performed according to Ausubel et al. (2). All oligonucleotide primers used in this study were synthesized by Sigma-Genosys, Ltd. (United Kingdom). Nucleic acid sequence determination involved BigDye Terminator cycle sequencing (Lark Technologies, Inc., England), followed by analysis on 5.75% and 4.75% LongRanger gels for ABI 377.
Amplification of genomic DNA located upstream of the phenylacetyl-CoA ligase paaK gene.
A modified version of the two-step PCR method reported by Sorensen was used to amplify flanking DNA upstream of the phenylacetyl CoA ligase paaK gene adjoining the styrene degradative operon in P. putida CA-3 (19, 26). A degenerate flanking primer, AFR-Rdm1 (5′-CAGTTCAAGCTTGTCCAGGAATTCNNNNNNNCGCGGT-3′), was combined with a paaK-specific biotinylated primer, S51-Bio (5′-CTGGAACTGGGTGGCAAGAGC-3′) in the first round of PCR. Thermal cycling conditions and reagents were standard; a 52°C annealing temperature was used. Biotinylated fragments were purified from the random amplification pool via overnight binding at 30°C to superparamagnetic Dynabeads M-280 Streptavidin (Dynal), followed by extraction using a magnetic particle concentrator (Dynal). Bound fragments were released by washing with 0.1 M NaOH, followed by neutralization with 0.1 M HCl; 2 μl of this suspension was used as a template for the second round of PCR. The primers in this instance were a nondegenerate flanking primer, AFR-Ndg1 (5′-CAGTTCAAGCTTGTCCAGGAATTC-3′), designed to bind within the 5′ region of amplicons generated with AFR-Rdm1; and AltS-nb1 (5′-ATGTCGTTCTGGGTGTAG-3′), a paaK-specific primer located upstream from the S51-Bio priming site. The annealing temperature of the reaction mixture was 50°C; all other conditions and reagents were standard. PCR products were visualized on 1% agarose gels, and those of >1.5 Kb in size were purified (Qiaex II kit; QIAGEN) and cloned into pCR2.1-TOPO prior to M13 primed sequencing.
Random mini-Tn5 mutagenesis.
A triparental mating approach was used to introduce pUT-Km1 into P. putida CA-3. The mating mixture (1 ml), containing mid-log-phase LB-grown recipient, donor, and helper strains at a ratio of 7:2:1, was centrifuged for 2 min at 16,400 relative centrifugal force, resuspended in 50 μl fresh LB medium, spotted onto an LB agar plate, and incubated at 30°C for 24 h. P. putida CA-3 transconjugants expressing kanamycin resistance were subsequently isolated by plating serial dilutions of the mating mixture onto E2-citrate medium containing kanamycin (50 μg/ml). Approximately 105 isolated colonies were individually transferred to 96-well microtiter plates containing 200 μl of E2-citrate broth in each well. These master plates were subsequently used to identify transposition events, resulting in a loss of the wild-type P. putida CA-3 strain's ability to (i) utilize PA as a sole carbon source and (ii) accumulate PHAs from unrelated carbon sources such as styrene, PA, and citrate. PA-negative phenotype strains were identified by direct transfer to E2-PA agar plates and subsequent screening for the presence or absence of colony growth after 24 h at 30°C. These were subsequently grown overnight in E2-citrate broth, and cells were harvested by centrifugation, washed with E2 medium lacking any carbon source, and used to inoculate E2-styrene broth to assess their ability to utilize styrene. PHA-negative phenotypes were identified by first growing colonies in E2-citrate broth containing 1.5 mM nitrogen (30°C; 24 h), followed by transfer to E2-citrate agar plates lacking any nitrogen, incubation for 48 h at 30°C, and monitoring thereafter for an absence of colony opacity associated with PHA-accumulating microorganisms.
Mapping of transposon insertion sites.
Arbitrarily primed PCR was employed to identify gene disruption sites by using two consecutive rounds of amplification. The primer sequences and appropriate thermal cycling parameters have been published previously (4). The resulting amplicons were visualized on 1% agarose gels prior to purification with a Qiaex II gel extraction kit (QIAGEN, California). Sequencing was performed using a mini-Tn5 internal primer.
Gas chromatography-tandem mass spectrometry (GC-MS) analysis of Tn5 mutant PHA percent cell (dry weight) (% CDW) content.
PHA monomers were extracted from freeze-dried cells by a method previously described (15). Samples were analyzed on a Fisons GC-8000 series gas chromatograph equipped with a 30-m by 0.32-mm HP-1 0.25 μm column (Hewlett Packard) operating in split mode (split ratio, 5:1) with temperature programming (60°C for 2 min, increments of 5°C/min up to 200°C, increments of 40°C/min up to 280°C, and 5 min at 280°C). For peak identification, PHA standards from Pseudomonas oleovorans were used.
Cloning of the PHA operon in P. putida CA-3.
Genomic DNA was isolated from P. putida CA-3 as previously described (2) and PCR screened with oligonucleotide primers designed from existing GenBank PHA gene sequences of various Pseudomonas species to PCR clone phaC1, phaZ, phaC2, and phaG gene homologues from our strain. The primers employed were C1-F (5′-ATGAGTAACAAGAACAACGAT-3′), C1-R (5′-TCAGCGCTCGTGAACGTAGGT-3′), Z-189 (5′-CGGGGTCGGCGGCTCGTCTAC-3′), Z-631 (5′-GCCGGATCTTGTGCAGCCAGTG-3′), C2-F (5′-ATGACAGAAAAACCGGGCAAA-3′), C2-R (5′-TCATCGGGTCAGCACGTAGGT-3′), G-F (5′-ATGAGGCCAGAAATCGCTGT-3′), and G-R (5′-TCAGATGGCCAATGCATGCT-3′). PCR products were cloned into pCR2.1-TOPO (Invitrogen, California), according to the manufacturer's instructions. Sequencing was performed as outlined above using M13 universal primers. The operonic nature of the pha locus was assessed by recombining primers C1-F/C2-R, C1-F/Z-631, and Z-189/C2-R. All PCR products were subsequently cloned and sequenced as described above.
RT-PCR analysis.
Total RNA was isolated from P. putida CA-3 cells grown under differing carbon and nitrogen (C/N) ratios. For each growth condition, 1 μg of RNA was reverse transcribed as previously reported (21), and 2 μl of the resulting cDNA pool was used as a template for PCR amplifications of phaC1, Z, C2, and G with the relevant primer pairs. To assess the transcriptional status of paaX, total RNA was isolated from P. putida CA-3 cells growing on citrate and PA, and reverse transcription-PCR (RT-PCR) was performed with the following paaX internal primers: pX-F1 (5′-CGCCATAACGCCAAACCCCT-3′) and pX-R1 (5′-TGCCCCACTGAACAACCTGAT-3′). In the case of all cDNA generated, PCR with oligonucleotides specific to the citrate synthase housekeeping gene of P. putida CA-3 acted as a positive control for each reverse transcription reaction. Total RNA was used as template in negative-control PCRs to demonstrate the absence of contaminating genomic DNA.
Nucleotide sequence accession numbers.
Nucleotide sequence data generated in this study were deposited in GenBank under accession numbers AY714618, AY714619, and AY726000.
RESULTS
Identification of PACoA catabolon genes in P. putida CA-3.
Amplified flanking-region PCR was used to clone a 2.8-kb region immediately upstream of the phenylacetyl-CoA ligase gene paaK, located 413 bp upstream from the styrene catabolic operon in P. putida CA-3. Sequence analysis identified full-length paaX and paaY gene homologues, which appeared to encode the archetypal PACoA catabolon transcriptional repressor and an associated regulatory enzyme (currently of unknown function), respectively (Table 1). RT-PCR analysis of total RNA from P. putida CA-3 revealed constitutive expression of paaX during growth on E2 minimal salts medium with styrene, phenylacetic acid, or citrate as a sole carbon source (data not shown). A 795-bp paaN gene homologue fragment was also identified, which appeared to encode a putative aldehyde dehydrogenase involved in ring cleavage of 2-hydroxyphenylacetyl-CoA (22). Table 1 shows the results of BLAST P screening of protein databases using the predicted amino acid sequences of the three genes. All three amino acid sequences displayed >99% similarity to reported PACoA catabolon enzymes, located upstream of the styrene degradative operon in Pseudomonas sp. Y2 (1, 29). The paaXYN genes also displayed similar structural organization in both Pseudomonas strains. These findings strongly support the involvement of an archetypal PACoA catabolon in degradation of the styrene catabolic intermediate, phenylacetic acid, in P. putida CA-3.
TABLE 1.
BLAST P GenBank comparison of amino acid sequences of genes identified upstream of paaK in P. putida CA-3
| Gene | Product (no. of amino acids) | Similar polypeptide | % Identification | Organism | Accession no. |
|---|---|---|---|---|---|
| paaX | 307 | PaaX | 99 | Pseudomonas sp. strain Y2 | CAD76909 |
| PaaX2 | 86 | Pseudomonas sp. strain Y2 | CAE45100 | ||
| PaaX | 79 | P. putida U | AAC24342 | ||
| paaX | 41 | E. coli W | CAA66101 | ||
| paaY | 198 | PaaY | 99 | Pseudomonas sp. strain Y2 | CAD76911 |
| PaaY | 76 | P. putida U | AAC24341 | ||
| PaaY2 | 73 | Pseudomonas sp. strain Y2 | CAE45101 | ||
| PaaY | 60 | E. coli W | CAA66102 | ||
| paaN | 265a | PaaN | 99 | Pseudomonas sp. strain Y2 | CAD76913 |
| PaaZ | 52 | Azoarcus evansii- | AAG28964 | ||
| PaaN3 | 19 | Pseudomonas sp. strain Y2 | CAE45118 | ||
| PaaN | 15 | P. putida U | AAC24340 | ||
| PaaZ | 13 | E. coli W | CAA66089 |
Fragment.
Styrene/phenylacetic acid mini-Tn5 mutants.
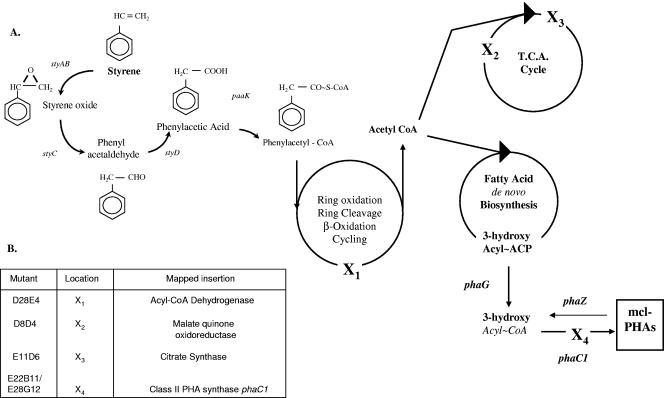
In an attempt to identify functional genes essential to metabolism of the PA intermediate produced during styrene degradation, random mini-Tn5 mutagenesis of P. putida CA-3 was performed. This generated three P. putida CA-3 mutants that were no longer capable of utilizing styrene or phenylacetic acid as a source of carbon and energy but retained the ability to grow on citrate. In two of these mutants (D8D4 and E11D6), mapping of the Tn5 insertion sites revealed the disruption of two tricarboxylic acid (TCA) cycle genes essential for the incorporation of acetyl-CoA into the TCA cycle, namely, the NAD-dependent malate dehydrogenase, responsible for oxaloacetate formation, and the citrate synthase facilitating condensation of acetyl-CoA and oxaloacetate to citrate (Fig. 1A and B). These mutants provide the first evidence that the end product of styrene metabolism in P. putida CA-3 is acetyl-CoA. GC-MS analysis of D8D4 and E11D6 revealed a marked reduction in the PHA accumulation capacity of mutant E11D6 and a complete loss of PHA production in mutant D8D4 (Table 2). These observations are interesting, given the recent report on PHA accumulation from styrene in P. putida CA-3, implicating de novo fatty acid synthesis in the generation of PHA monomers (32). Thus, the potential availability of additional acetyl-CoA moieties to fatty acid synthesis in these TCA cycle mutants offered the potential for increased PHA output from these strains. It is not clear at this point why disruption of the malate dehydrogenase should result in a complete loss of PHA accumulation.
FIG. 1.
(A) Proposed route of PHA accumulation from styrene in P. putida CA-3 under nitrogen-limiting conditions. Genes responsible for the stepwise conversions are indicated by italics, where known. styAB, two-component styrene mono-oxygenase gene; styC, styrene oxide isomerase gene; styD, phenylacetaldehyde dehydrogenase gene; paaK, phenylacetyl-CoA ligase gene; phaG, 3-hydroxyacyl-ACP-CoA transacylase gene; phaC1, class II MCL-PHA synthase gene; phaZ, PHA depolymerase gene. X's indicate pathway interruptions by mini-Tn5 insertions. (B) Table identifying enzymes disrupted by mini-Tn5 insertion sites and respective locations within proposed pathway. D28E4, acyl-CoA dehydrogenase; D8D4, malate-quinone oxidoreductase; E11D6, citrate synthase; E22B11/E28G12, both mapped to a class II MCL-PHA synthase.
TABLE 2.
GC-MS analysis of P. putida CA-3 mini-Tn5 mutantsa
| P. putida CA-3 | Growth in NH4+ concentration of:
|
||
|---|---|---|---|
| 0 | 0.2 mM | 0.4 mM | |
| Wild type | 2.3 ± 0.9 | 9.35 ± 1.75 | 5.5 ± 0.5 |
| D8D4 | 0 | 0 | 0 |
| E11D6 | 3.45 ± 0.75 | 5.75 ± 0.25 | 1.8 ± 0.8 |
E11D6, citrate synthase-negative mutant; D8D4, malate dehydrogenase mutant. All cells were grown on 10 mM citrate (8 mM NH4+) for 18 h and resuspended in 10 mM PA for 36 h with 0, 0.2, and 0.4 mM NH4+, respectively, before PHA (% CDW) analysis.
In a third mutant displaying the styrene negative phenotype (D28E4), the Tn5 insertion site was located 375 bp downstream from the ATG translational start codon of a 3-hydroxyacyl-CoA dehydrogenase gene typically involved in β-oxidation (Fig. 1). Together, these mutants demonstrated that acetyl-CoA is the end product of styrene metabolism in P. putida CA-3, which involves β-oxidation enzymes. It should be noted that BLAST P comparative protein database analysis of the deduced 125-amino-acid, N-terminal acyl-CoA dehydrogenase sequence did not produce any significant percentage identity (<16%) with previously reported PACoA catabolon-associated acyl-CoA dehydrogenases (Table 3). Thus, it would appear that β-oxidation enzymes other than those encoded by the PACoA catabolon may be involved in styrene/PA catabolism in P. putida CA-3.
TABLE 3.
Amino acid comparison of the P. putida CA-3 mini-Tn5-disrupted 3-hydroxyacyl-CoA dehydrogenase with PACoA catabolon associated 3-hydroxyacyl-CoA dehydrogenases from a variety of Pseudomonas species
| Mini-Tn5-disrupted 3-hydroxyacyl-CoA dehydrogenase | % Identification | PACoA catabolon-3-hydroxyacyl-CoA dehydrogenase (no. of amino acids) | Organism | Accession no. |
|---|---|---|---|---|
| 16.3 | PaaC (507) | Pseudomonas sp. strain Y2 | CAD76920 | |
| 125-amino-acid fragment | 14.7 | PaaC2 (505) | Pseudomonas sp. strain Y2 | CAE45104 |
| 14.3 | PaaC (505) | P. putida U | AAC24331 | |
| 14.3 | PaaH (475) | E. coli W | CAA66097 |
Cloning of a phaG homologue; the link between de novo fatty acid synthesis and 3-hydroxyacyl-CoA PHA monomers.
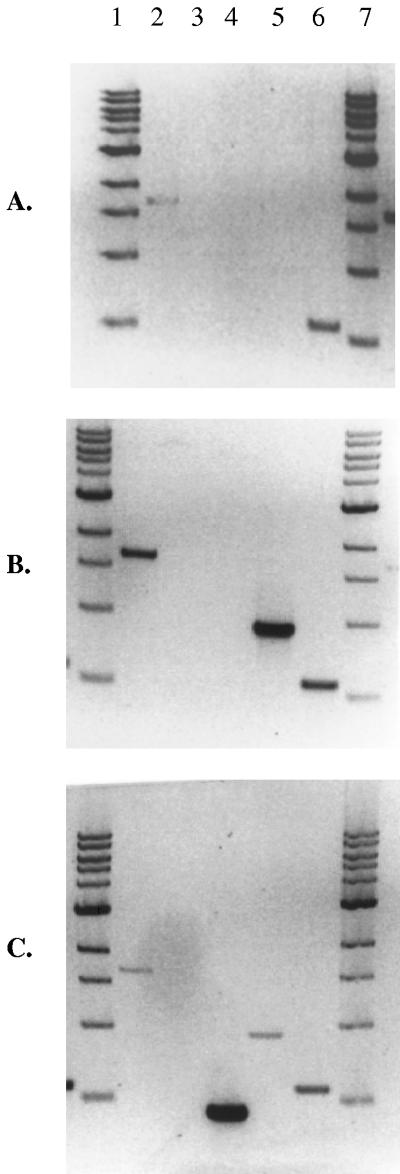
In conjunction with our investigation of styrene metabolism to assess its contribution to PHA accumulation in P. putida CA-3, we also sought to identify the genetic apparatus responsible for PHA monomer sequestration, polymerization, and depolymerization. As mentioned previously, PHA accumulation from styrene in P. putida CA-3 was recently reported to involve de novo fatty acid synthesis, based on biochemical inhibition studies (32). Rehm et al. were the first to report the role of a 3-hydroxyacyl-acyl carrier protein (ACP)-CoA transacylase, PhaG, responsible for PHA monomer sequestration from de novo fatty acid biosynthesis during growth of P. putida KT2440 on the unrelated carbon source, gluconate (24). With this in mind, an 888-bp phaG gene homologue was cloned from the P. putida CA-3 genome, which displayed >98% similarity at the amino acid level to PhaG from P. putida KT2440 (Table 4). In P. putida CA-3, PHA accumulation to 23.2% CDW only occurs under nitrogen-limiting conditions (Table 5, “Wild type”). Analysis of the transcriptional profile of phaG during growth of P. putida CA-3 under limiting and nonlimiting conditions revealed that maximal expression of the gene also occurred only under nitrogen limitation (Fig. 2B, lane 5). This provides a molecular basis for the inhibition of PHA accumulation in P. putida CA-3 by 2-bromo-octanoate, a transacylase inhibitor (32), and suggests that this transcriptionally regulated phaG homologue is likely to play a key role in PHA monomer sequestration during nitrogen-limited growth of P. putida CA-3 on styrene.
TABLE 4.
BLAST P comparison of the deduced amino acid sequences of pha genes cloned from P. putida CA-3a
| Gene | Product (no. of amino acids) | Similar polypeptide | % Identification | Organism | Accession no. |
|---|---|---|---|---|---|
| phaC1 | 559 | PhaC1 | 98 | P. putida KT2440 | AAM63407 |
| PhaA | 95 | P. oleovorans | M58445 | ||
| PhaC1 | 94 | P. putida U | AF150670 | ||
| PhaC1 | 77 | Pseudomonas sp. strain 61-3 | AB014758 | ||
| phaZ | 256 | PhaZ | 97 | P. putida KT2440 | AAM63408 |
| PhaB | 96 | P. oleovorans | M58445 | ||
| PhaZ | 96 | P. putida U | AF150670 | ||
| PhaZ | 89 | Pseudomonas sp. strain 61-3 | AB014758 | ||
| phaC2 | 560 | PhaC2 | 97 | P. putida KT2440 | AAM63409 |
| PhaC | 92 | P. oleovorans | M58445 | ||
| PhaC2 | 87 | P. putida U | AF150670 | ||
| PhaC2 | 73 | Pseudomonas sp. strain 61-3 | AB014758 | ||
| phaG | 296 | PhaG | 98 | P. putida KT2440 | AF052507 |
| PhaG | 95 | P. oleovorans | AF169252 | ||
| PhaG | 72 | Pseudomonas sp. strain 61-3 | AB047080 |
TABLE 5.
GC-MS analysis of P. putida CA-3 mini-Tn5 mutants
| P. putida CA-3 | Growth substrate (mM) | PHA (% CDW) |
|---|---|---|
| Wild type | PA (10) | 23.2 |
| E28G12a | PA (10) | 0 |
| E22B11a | PA (10) | 0 |
E28G12 and E22B11 are both PhaC1 synthase-negative mutants. Cells were grown on PA for 48 h on E2 medium-8 mM NH4+ before assessment for PHA accumulation.
FIG. 2.
Influence of varied C/N ratios on transcriptional activity of the pha operon genes. Medium conditions are as follows: +C/+N (nonlimited) (A), +C/−N (nitrogen limited) (B), −C/+N (carbon limited) (C). In the case of all growth conditions, lanes 1 and 7 show 10-kb molecular weight markers; lanes 2 to 5 represent the results of phaC1/C2/Z (lanes 2 to 4) and phaG (lane 5) RT-PCRs; lane 6 shows the citrate synthase positive-control RT-PCR.
Cloning and characterization of the pha operon.
With degenerate PCR primer pairs, the PHA gene cluster was cloned from the wild-type CA-3 strain; sequencing revealed the commonly reported operon structure of two class II MCL-PHA synthases (phaC1 and phaC2) flanking the PHA depolymerase-encoding phaZ gene (Table 4) (23). To further characterize the functionality of these PHA synthetic genes, P. putida CA-3 was subjected to mini-Tn5 random mutagenesis, and colonies were screened for a loss of opacity during growth on N-limiting E2-citrate medium. This approach has been used previously to examine the role of de novo fatty acid biosynthesis in PHA and rhamnolipid synthesis by pseudomonads (25). Two PHA-negative P. putida CA-3 mutants, E22B11 and E28G1, were isolated by this method (Fig. 3), and mapping of the transposon insertion sites revealed disruption of the phaC1 gene in both (Fig. 1A and B). GC-MS analysis of these cells grown under PHA-accumulating conditions (N limitation) revealed a complete loss of detectable PHA accumulation (Table 5), confirming the solitary role of PhaC1 in MCL-PHA monomer polymerization in our strain.
FIG. 3.
P. putida CA-3 mini-Tn5 mutants grown on E2-N-limited media for 72 h. The framed colony demonstrates the loss of opacity used as a phenotypic screen for PHA nonaccumulation.
Transcriptional analysis of pha genes.
A transcriptional profile for each of the PHA operon genes was generated by RT-PCR analysis of total RNA from P. putida CA-3 cultures grown under nonlimiting, nitrogen-limiting, and carbon-limiting growth conditions with styrene as the sole carbon source. Figure 2A to C, lanes 2, revealed constitutive expression of phaC1; however, PHA accumulation was only observed under nitrogen limitation (Fig. 2B, lane 2; Table 5), in conjunction with maximal expression of the transacylase homologue PhaG (Fig. 2B, lane 5). Transcription of the phaZ depolymerase was detectable only under carbon limitation (Fig. 2C, lane 4) and coincided with the disappearance of detectable PHAs, previously accumulated under nitrogen limitation (21). Transcription of phaC2 was not detected under any of the growth conditions tested (Fig. 2A to C, lanes 3).
DISCUSSION
Metabolism of phenylacetic acid.
The ability of P. putida CA-3 to metabolise styrene to PA with subsequent activation to PACoA has been reported previously (18, 21). Currently, we are interested in investigating metabolism beyond this point to understand the influence of this pathway on PHA accumulation from styrene in this strain. The discovery in E. coli W and P. putida U of a novel catabolic route solely for PACoA metabolism (5, 22) is of particular interest for styrene metabolism in P. putida CA-3, as PACoA ligase activities have been reported previously in our strain during growth on styrene or its metabolic intermediate, phenylacetic acid (21). The route, referred to as the PACoA catabolon, reportedly involves oxidation of the PACoA aromatic nucleus, ring cleavage, and β-oxidation cycling of the alicyclic compound. The catabolon genes appear widespread among bacterial genera, although their structural organization varies considerably (16). Investigation of the genomic region directly upstream of the PACoA ligase paaK gene in P. putida CA-3 identified three PACoA catabolon gene homologues, paaXYN (Table 1). The genes encode a PACoA catabolon, negative transcriptional regulator, an associated regulatory protein of unknown function, and an aldehyde dehydrogenase, reportedly involved in ring cleavage, respectively. With E. coli W, it has previously been shown that constitutively expressed PaaX inhibits transcription of the PACoA catabolon in the absence of its inducer, PACoA (5, 6). In this study, RT-PCR analysis revealed that the paaX homologue identified in P. putida CA-3 is constitutively expressed when grown on E2 minimal salts medium with citrate, styrene, or phenylacetic acid as the sole carbon source (results not shown). Furthermore, PACoA ligase activity and paaK gene transcription are only detectable in CA-3 when cultures are grown on substrates generating a PACoA intermediate, i.e., styrene and phenylacetic acid (21). Thus, PaaX may have a similar regulatory role in transcription of a PACoA catabolic operon in P. putida CA-3. As Table 1 indicates, the PACoA catabolon genes identified in P. putida CA-3 are almost identical (>99%) with those reported in the styrene degrading Pseudomonas sp. Y2. The similarity between the CA-3 and Y2 PACoA catabolon genes also extends to their unique genetic organization upstream of the styrene degradative operon. On the basis of these similarities, the recent cloning of a second, functional phenylacetic acid catabolic gene cluster in Pseudomonas sp. Y2 indicated the potential existence of additional PACoA degradation routes in P. putida CA-3 (1). Therefore, in an attempt to functionally define the metabolic fate of PA in P. putida CA-3, mini-Tn5 mutagenesis was used to generate mutants no longer capable of styrene or phenylacetic acid utilization.
Styrene- or phenylacetic acid-negative mini Tn5 mutants.
Two mutants, D8D4 and E11D6, carried insertions in two tricarboxylic acid cycle enzymes involved in acetyl-CoA metabolism (Fig. 1A and B), providing the first definitive proof that styrene is degraded to acetyl-CoA in P. putida CA-3. This finding, together with the identification of functionally expressed PACoA catabolon genes, strongly suggested that a PACoA β-oxidation mechanism may be involved in the generation of acetyl-CoA moieties in P. putida CA-3. Indeed, a third styrene/phenylacetic acid-negative mutant, D28E4, carried a Tn5 insertion in a (S)-3-hydroxyacl-CoA dehydrogenase, typical of β-oxidation and also found in the multienzyme complex of the PACoA catabolon (22). However, the BLAST P results shown in Table 3 demonstrate that this Tn5-disrupted 3-hydroxyacyl-CoA dehydrogenase does not appear to be encoded by the PACoA catabolon, as the deduced, N-terminal amino acid sequence does not share significant (<16%) similarity with the N termini of reported PACoA catabolon-associated 3-hydroxyacyl-CoA dehydrogenases. To our knowledge, this is the first study to demonstrate the involvement of non-PACoA catabolon β-oxidation enzyme activity in aerobic styrene-phenylacetic acid degradation in a Pseudomonas species harboring functionally expressed PACoA catabolon genes. It also demonstrates that noncatabolon β-oxidation is involved in PHA accumulation from these unrelated carbon sources in P. putida CA-3.
With respect to PHA accumulation from unrelated carbon sources, the role of de novo fatty acid biosynthesis in generating (R)-3-hydroxyacyl-CoA monomers for PHA accumulation has been widely reported (34). The metabolism of styrene to acetyl-CoA is supportive of such a route, in conjunction with the inability of P. putida CA-3 cells to accumulate PHAs from styrene in the presence of the fatty acid synthesis inhibitor cerulenin (31). Furthermore, the monomer composition of the PHA accumulated from styrene in P. putida CA-3 clearly indicates the involvement of an anabolic process such as fatty acid synthesis, with 3-hydroxyhexanoic acid, 3-hydroxyoctanoic acid, and 3-hydroxydecanoic acid present at a ratio of 3:27:70 (32). Rehm et al. were the first to report on the activity of a 3-hydroxyacyl ACP-CoA transacylase encoded by phaG, responsible for the transfer of 3-hydroxydecanoate moieties from the fatty acid biosynthesis acyl carrier protein to coenzyme A, which were subsequently packaged by a class II PhaC1 synthase into MCL-PHAs (24). Several groups have subsequently reported an essential role for the enzyme in the accumulation of MCL-PHAs from unrelated carbon sources in wild-type and recombinant Pseudomonas species (7, 9, 10, 11, 17). In this study, a phaG homologue was cloned in P. putida CA-3, which was almost identical to its P. putida KT2440 equivalent (Table 4). Transcriptional analysis revealed that this putative transacylase was maximally expressed under nitrogen-limiting conditions (Fig. 2), with concomitant PHA accumulation from styrene in P. putida CA-3. These findings, together with the earlier report of 2-bromo-octanoate inhibition of PHA synthesis from styrene in CA-3 (32), strongly suggest that this enzyme facilitates MCL-PHA substrate sequestering from fatty acid de novo synthesis when styrene-grown P. putida CA-3 cells are subjected to nitrogen-limiting conditions.
Identification and transcriptional regulation of the polyhydroxyalkanoate pha operon.
Random mini-Tn5 mutagenesis was used to generate PHA-negative mutants (Fig. 3), and two of these (E22B11 and E28G12) were found to involve disruption of a class II MCL-PHA synthase encoded by phaC1 (Fig. 1A and B). GC-MS analysis revealed a complete loss of PHA accumulation in these mutants, identifying the essential role of PhaC1 in polyester production in P. putida CA-3. The pha operon was subsequently cloned and found to contain the typical organization of class II pha genes, a PhaC1 synthase, a phaZ-encoded depolymerase, and a second synthase, PhaC2 (Table 4) (24). Previous characterizations of Pseudomonas pha gene loci have reported the initiation of transcription of the operon genes from promoter elements upstream of phaC1 (12, 28). It has also been demonstrated with P. putida U that PHA accumulation is facilitated by both synthases (8). These observations raised concerns that polar effects of the phaC1::Tn5 disruption might inhibit transcription of the intact phaC2, thus masking its ability to accumulate PHAs in P. putida CA-3 grown on styrene. However, RT-PCR analyses revealed that in the wild-type strain, phaC2 gene transcripts could not be detected under any of the growth conditions examined (Fig. 2A to C, lanes 3). Thus, PhaC1, which appears to be constitutively expressed, is the sole synthase involved in PHA packaging under nitrogen-limiting conditions in P. putida CA-3, (Table 5; Fig. 2A to C, lane 2). Only transcription of the phaZ depolymerase gene was observed under carbon-limiting conditions, concurrent with the absence of detectable PHAs (Fig. 2C, lane 4). The cotranscription of phaC1 was also observed under this condition and similar findings have been reported in P. oleovorans and P. aeruginosa with the detection of dual transcripts by Northern blotting corresponding to phaC1 mRNA alone and cotranscripts of phaC1 and phaZ (12, 28). This has been attributed to altered transcription initiation from σ54 and σ70 consensus sequences localized upstream of phaC1 in these strains; our observations suggest a similar control mechanism may operate in P. putida CA-3 (28).
Conclusions.
The ability of P. putida CA-3 to synthesize biodegradable polyesters with potential medical and industrial applicability from the toxic industrial pollutant styrene represents a significant, undiscovered potential within toxic waste bioremediation. The recent identification of the phenylacetyl-CoA pathway as one of the four main pathways for the catabolism of core aromatic intermediates, based on sequence analysis of the metabolically versatile P. putida KT2440 genome, is also very significant in this regard (14). It highlights the potential for a wide variety of aromatic compounds producing a phenylacetic acid metabolic intermediate to be converted to MCL-PHAs. However, our discovery of the involvement of a non-PACoA catabolon β-oxidation enzyme indicates that the complete mechanism of PACoA metabolism has yet to be fully understood. We expect that the identification of essential structural and regulatory genes involved in accumulation of PHAs from styrene will allow us to further exploit this novel bioconversion through the generation of P. putida CA-3 PHA-overproducing strains, via targeted recombination strategies.
Acknowledgments
This project was funded under the PRTLI Programme for Irish Third Level Institutions, administered by the Higher Education Authority.
REFERENCES
- 1.Alonso, S., D. Bartolomé-Martín, M. del Álamo, E. Díaz, J. L. García, and J. Perera. 2003. Genetic characterisation of the styrene lower catabolic pathway of Pseudomonas sp. Strain Y2. Gene 319:71-83. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. Greene Publishing Associates-Wiley Interscience, New York, N.Y.
- 3.De Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Espinosa-Urgel, M., A. Salido, and J. L. Ramos. 2000. Genetic analysis of functions involved in adhesion of Pseudomonas putida to seeds. J. Bacteriol. 182:2363-2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferrández, A., B. Miñambres, B. García, E. R. Olivera, J. M. Luengo, J. L. García, and E. Díaz. 1998. Catabolism of phenylacetic acid in Escherichia coli. J. Biol. Chem. 273:25974-25986. [DOI] [PubMed] [Google Scholar]
- 6.Ferrández, A., J. L. García, and E. Díaz. 2000. Transcriptional regulation of the divergent paa catabolic operons for phenylacetic acid degradation in Eschericha coli. J. Biol. Chem. 275:12214-12222. [DOI] [PubMed] [Google Scholar]
- 7.Fielder, S., A. Steinbüchel, and B. H. A. Rehm. 2000. PhaG-mediated synthesis of poly-(3-hydroxyalkanoates) consisting of medium-chain-length constituents from nonrelated carbon sources in recombinant Pseudomonas fragi. Appl. Environ. Microbiol. 66:2117-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.García, B., E. R. Olivera, B. Miñambres, M. Fernández-Valverde, L. M. Cañedo, M. A. Prieto, J. L. García, M. Martínez, and J. M. Luengo. 1999. Novel biodegradable aromatic plastics from a bacterial source. J. Biol. Chem. 274:29228-29241. [DOI] [PubMed] [Google Scholar]
- 9.Hoffmann, N., A. A. Amara, B. Beermann, Q. Qi, H. Hinz, and B. H. A. Rehm. 2002. Biochemical characterisation of the Pseudomonas putida 3-hydroxyacyl ACP:CoA transacylase, which diverts intermediates of fatty acid de novo biosynthesis. J. Biol. Chem. 277:42926-42936. [DOI] [PubMed] [Google Scholar]
- 10.Hoffmann, N., A. Steinbüchel, and B. H. A. Rehm. 2000. Homologous functional expression of cryptic phaG from Pseudomonas oleovorans establishes the transacylase-mediated polyhydroxyalkanoate biosynthetic pathway. Appl. Microbiol. Biotechnol. 54:665-670. [DOI] [PubMed] [Google Scholar]
- 11.Hoffmann, N., A. Steinbüchel, and B. H. A. Rehm. 2000. The Pseudomonas aeruginosa phaG gene product is involved in the synthesis of polyhydroxyalkanoic acid consisting of medium-chain-length constituents from non-related carbon sources. FEMS Microbiol. Lett. 184:253-259. [DOI] [PubMed] [Google Scholar]
- 12.Huisman, G. W., E. Wonink, R. Meima, B. Kazemier, P. Terpstra, and B. Witholt. 1991. Metabolism of poly(3-hydroxyalkanoates) (PHAs) by Pseudomonas oleovorans. J. Biol. Chem. 266:2191-2198. [PubMed] [Google Scholar]
- 13.Jendrossek, D., and R. Handrick. 2002. Microbial degradation of polyhydroxyalkanoates. Annu. Rev. Microbiol. 56:403-432. [DOI] [PubMed] [Google Scholar]
- 14.Jiménez, J. I., B. Miñambres, J. L. García, and E. Diaz. 2002. Genomic analysis of the aromatic catabolic pathways from Pseudomonas putida KT2440. Environ. Microbiol. 4:824-841. [DOI] [PubMed] [Google Scholar]
- 15.Lageveen, R. G., G. W. Huisman, H. Preusting, P. Ketelaar, G. Eggink., and B. Witholt. 1988. Formation of polyesters by Pseudomonas oleovorans: effect of substrates on formation and composition of poly-(R)-3-hydroxyalkanoates and poly-(R)-3-hydroxyalkenoates. Appl. Environ. Mirobiol. 54:2924-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luengo, J. M., J. L. García, and E. R. Olivera. 2001. The phenylacetl-CoA catabolon: a complex catabolic unit with broad biotechnological applications. Microbiology 39:1434-1442. [DOI] [PubMed] [Google Scholar]
- 17.Matsumoto, K., H. Matsusaki, S. Taguchi, M. Seki, and Y. Doi. 2001. Cloning and characterization of the Pseudomonas sp. 61-3 phaG gene involved in polyhydroxyalkanoate biosynthesis. Biomacromolecules 2:142-147. [DOI] [PubMed] [Google Scholar]
- 18.O'Connor, K. E., C. M. Buckley, S. Hartmans, and A. D. W. Dobson. 1995. Possible regulatory role for non-aromatic carbon sources in styrene degradation by Pseudomonas putida CA-3. Appl. Environ. Microbiol. 61:544-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Leary, N. D. 2001. Physiology and genetics of styrene degradation in Pseudomonas putida CA-3. Ph.D. thesis. National University of Ireland, Cork, Ireland.
- 20.O'Leary, N. D., K. E. O'Connor, and A. D. W. Dobson. 2002. Biochemistry, genetics and physiology of microbial styrene degradation. FEMS Microbiol. Rev. 26:403-417. [DOI] [PubMed] [Google Scholar]
- 21.O'Leary, N. D., K. E. O'Connor, W. Duetz, and A. D. W. Dobson. 2001. Transcriptional regulation of styrene degradation in Pseudomonas putida CA-3. Microbiology 147:973-979. [DOI] [PubMed] [Google Scholar]
- 22.Olivera, E. R., B. Miñambres, B. García, C. Muniz, M. A. Moreño, A. Ferrández, E. Díaz, J. L. García, and J. M. Luengo. 1998. Molecular characterisation of the phenylacetic acid catabolic pathway in Pseudomonas putida U: the phenylacetyl-CoA catabolon. Proc. Natl. Acad. Sci. USA 95:6419-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rehm, B. H. A., and A. Steinbüchel. 1999. Biochemical and genetic analysis of PHA synthases and other proteins required for PHA synthesis. Int. J. Biol. Macromol. 25:3-19. [DOI] [PubMed] [Google Scholar]
- 24.Rehm, B. H. A., N. Kruger, and A. Steinbüchel. 1998. A new metabolic link between fatty acid de novo synthesis and polyhydroxyalkanoic acid synthesis. J. Biol. Chem. 37:24044-24051. [DOI] [PubMed] [Google Scholar]
- 25.Rehm, B. H. A., T. A. Mitsky, and A. Steinbüchel. 2001. Role of fatty acid de novo biosynthesis in polyhydroxyalkanoic acid (PHA) and rhamnolipid synthesis by pseudomonads: establishment of the transacylase (PhaG)-mediated pathway for PHA biosynthesis in Escherichia coli. Appl. Environ. Microbiol. 67:3102-3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sorensen, A. B., M. Duch, P. Jorgensen, and F. S. Pederson. 1993. Amplification and sequence analysis of DNA flanking integrated proviruses by a simple two-step polymerase chain reaction method. J. Virol. 67:7118-7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steinbüchel, A., and H. E. Valentin. 1995. Diversity of bacterial polyhydroxyalkanoic acids. FEMS Microbiol. Lett. 128:219-228. [Google Scholar]
- 28.Timm, A., and A. Steinbüchel. 1992. Cloning and molecular characterisation of the poly(3-hydroxyalkanoic acid) gene locus in Pseudomonas aeruginosa PA01. Eur. J. Biochem. 209:15-30. [DOI] [PubMed] [Google Scholar]
- 29.Velasco, A., S. Alonso, J. L. García, J. Peréra, and E. Díaz. 1998. Genetic and functional analysis of the styrene catabolic cluster of Pseudomonas sp. strain Y2. J. Bacteriol. 180:1063-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vogel, H. J., and D. M. Bonner. 1956. Acetylornithinase of E. coli: partial purification and some properties. J. Biol. Chem. 218:97-106. [PubMed] [Google Scholar]
- 31.Ward, P. 2004. Ph.D. thesis. National University of Ireland, Dublin, Ireland.
- 32.Ward, P. G., G. de Roo, and K. E. O'Connor. 2005. Accumulation of polyhydroxyalkanoate from styrene and phenylacetic acid by Pseudomonas putida CA-3. Appl. Environ. Microbiol. 71:2046-2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Witholt, B., and B. Kessler. 1999. Perspectives of medium chain length poly(hydroxyalkanoates), a versatile set of bacterial bioplastics. Curr. Opin. Biotechnol. 10:279-285. [DOI] [PubMed] [Google Scholar]
- 34.Zinn, M., B. Witholt, and T. Egli. 2001. Occurrence, synthesis and medical application of bacterial polyhydroxyalkanoate. Adv. Drug Deliv. Rev. 53:5-21. [DOI] [PubMed] [Google Scholar]