Abstract
A metabolic fingerprint database of enterococci and Escherichia coli from 10 host groups of animals was developed to trace the sources of fecal contamination in surface waters. In all, 526 biochemical phenotypes (BPTs) of enterococci and 530 E. coli BPTs were obtained from 4,057 enterococci and 3,728 E. coli isolates tested. Of these, 231 Enterococcus BPTs and 257 E. coli BPTs were found in multiple host groups. The remaining 295 Enterococcus BPTs and 273 E. coli BPTs were unique to individual host groups. The database was used to trace the sources of fecal contamination in a local creek. The mean diversities (Di) of enterococci (Di = 0.76 ± 0.05) and E. coli (Di = 0.88 ± 0.04) were high (maximum 1) in water samples, indicating diverse sources of fecal contamination. Overall, 71% of BPTs of enterococci and 67% of E. coli BPTs from water samples were identified as human and animal sources. Altogether, 248 Enterococcus BPTs and 282 E. coli BPTs were found in water samples. Among enterococci, 26 (10%) BPTs were identical to those of humans and 152 BPTs (61%) were identical to those of animals (animal BPTs). Among E. coli isolates, 36 (13%) BPTs were identical to those of humans and 151 (54%) BPTs were identical to those of animals. Of the animal BPTs, 101 (66%) Enterococcus BPTs and 93 (62%) E. coli BPTs were also unique to individual animal groups. On the basis of these unique Enterococcus BPTs, chickens contributed 14% of contamination, followed by humans (10%), dogs (7%), and horses (6%). For E. coli, humans contributed 13% of contamination, followed by ducks (9%), cattle (7%), and chickens (6%). The developed metabolic fingerprint database was able to distinguish between human and animal sources as well as among animal species in the studied catchment.
Surface water is frequently contaminated with fecal bacteria. Nonpoint sources such as domestic and wild animal defecation (7, 19), malfunctioning septic trenches (2, 19, 25), storm water drainage, and urban runoff (25, 34) and/or point sources such as industrial effluents and municipal wastes (40) are known to be potential sources of such contamination. It has been reported that various human enteric pathogens such as Salmonella spp., Shigella spp. (13), and hepatitis A (7, 23, 36, 42) have been found in surface waters as a result of human fecal contamination. Defecation from domestic animals may further contribute pathogens such as Escherichia coli O157:H7 and Cryptosporidium spp. (13, 15, 36, 44). Identification of major sources of fecal bacteria, therefore, whether human or animal, is necessary for improved management of surface water quality and the minimization of public health risks associated with such contamination.
Fecal coliforms have been widely used as an indicator of the microbiological quality of surface and ground waters (16, 19, 22, 44). This group of bacteria is commonly found in the gastrointestinal tracts of all warm-blooded animals (21, 35, 52). However, the value of fecal coliforms as an indicator has recently been questioned, because these bacteria can also derive from various sources such as soil, agricultural runoff, composted animals, decaying vegetation, and industrial processes (13, 16, 28). Instead, it has been suggested that E. coli and enterococci are much better indicators of fecal contamination, as these bacteria colonize in the gut of humans and other warm-blooded animals (8, 39). E. coli is widely accepted as a potential fecal indicator bacterium because it is not normally pathogenic, it is easy to detect and culture, and it is found at concentrations much higher than other pathogens in surface waters (50). Fecal streptococci are also considered ideal fecal indicator bacteria because of their ability to survive in the natural environment for lengthy periods (16, 22, 27, 48). However, it has also been noted that the sole presence of these bacteria in surface waters does not provide definitive information regarding their possible source(s) (22, 29, 34, 52).
In recent years, several methods, collectively known as bacterial and microbial source tracking methods, have been developed to distinguish the various sources of animal and/or human fecal contamination (35, 52). These methods include ribotyping (6, 11, 18, 19, 41), pulsed-field gel electrophoresis (45, 46), ribosomal genetic markers (9, 10), repetitive DNA sequences (12, 13), carbon source utilization (17), and antibiotic resistance profiles (22, 40, 53, 54) of fecal indicator bacteria. Chemical methods such as the detection of caffeine (44) and fecal sterols analysis (33) have also been used to detect the source(s) of fecal contamination in surface waters. Most of these methods are based on the hypothesis that phenotypic or genotypic characteristics of specific strains are associated with specific animals (4, 20, 25, 34). On the basis of this hypothesis, a fingerprint database (i.e., phenotypic or genotypic profiles) of strains from known sources has been developed to predict the source(s) of unknown environmental isolates (47, 52). The advantages and disadvantages of these methods have been discussed in various studies (34, 35, 44). For instance, genotypic methods, although highly discriminatory, can be laborious and/or expensive for ecological studies where a large number of isolates needs to be tested (19, 30, 38). The most commonly used phenotypic method, the antibiotic resistance profiles, can be used to test a large number of isolates within a short time and is rather inexpensive. However, it is known that antibiotic resistance genes can be lost from or gained by bacteria under certain conditions (14, 44). In addition, this method does not provide information about fecal indicator bacteria that are not resistant to antibiotics but are derived from different animal species. Chemical methods such as caffeine/pharmaceutical or fecal sterol detection require stringent sampling and can be expensive. In addition, it has also been reported that chemical methods are not sensitive enough to detect recent pollution (44).
A biochemical fingerprinting method known as the PhPlate system (PhPlate AB, Stockholm, Sweden) has been reported and used in many epidemiological and ecological studies (2, 30, 31, 51). It measures the kinetics of bacterial metabolism in microtiter plates. For each bacterial isolate, it yields a biochemical fingerprint made of several quantitative data, which are used with the PhPlate software to calculate the level of similarity between the tested isolates. This system has a high discriminatory ability and reproducibility (30-32) and is shown to be comparable with many genotypic methods in comparative studies (30, 31). The PhPlate system is simple to use and can be applied to studies involving large numbers of isolates and is therefore an excellent tool for studying the diversity (Di) and persistence of fecal indicator bacteria in surface waters (24, 30, 31, 51). In this study, we used the PhPlate system to characterize two fecal indicator bacteria, enterococci and E. coli, from different host groups (i.e., animal species) to develop a metabolic fingerprint database to identify the source(s) of fecal contamination in a local creek.
MATERIALS AND METHODS
Host group sampling.
Ten host groups were sampled between July 2003 and August 2004. These groups included horses, cattle, sheep, pigs, ducks, chickens, deer, kangaroos, dogs, and humans (via septic tanks). For each group of farm animals, we initially collected five fecal samples from five individuals within a farm. Up to 32 isolates of both enterococci and E. coli were tested from each sample (i.e., each animal) to determine the diversity of these indicator bacteria. Based on the low diversity (0.41 ± 0.09 for enterococci and 0.53 ± 0.11 for E. coli) (minimum of 0 and maximum of 1) obtained from this assessment, sampling was extended to include multiple (up to 20 farms where possible) farms for each group of farm animals. All septic tanks tested were within the 50- to 100-m distance of the creek (Fig. 1). However, for farm animals, we collected samples from as many farms as we had access to in the studied catchment. In addition, samples were also collected from other catchments within the same geographical area. At each farm, up to three animals were sampled, and from each animal, up to 12 isolates were tested. A total of 234 samples were collected from horses (38 samples), cattle (54 samples), sheep (28 samples), pigs (32 samples), chickens (36 samples), and ducks (46 samples). All samples were collected from fresh feces of individual animals with sterile swabs and inserted into Amies transport medium (Interpath, Melbourne, Australia), transported to the laboratory, and tested within 6 h. Dog samples (47 samples) were collected from two city dog parks on eight occasions. Deer samples (25 samples) were collected from a local deer sanctuary park, and kangaroo samples (20 samples) were collected from the University of the Sunshine Coast, where a large number of kangaroos roam.
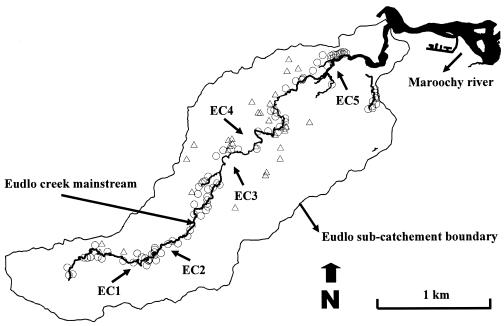
FIG. 1.
Sampling sites (Eudlo Creek site 1 [EC1] to EC5) on Eudlo Creek mainstream. Conventional septic systems (○) within a 50-m distance of the creek and animal farms (▵) are shown. (Reproduced with permission of the Maroochy Shire Council.)
Human samples were collected from the outlet of 39 septic tanks using sterile swabs. Swabs were then inserted into Amies transport medium (Interpath), transported on ice to the laboratory, and tested within 6 h.
Isolation of enterococci and E. coli.
All fecal samples were streaked on m-Enterococcus (Difco) and chromogenic E. coli/coliform (Oxoid, United Kingdom) agar plates and were incubated at 37°C for 24 h (for E. coli) and 48 h (for enterococci). This chromogenic medium allows specific detection of E. coli through substrate cleavage by the enzyme glucuronidase and formation of purple colonies, which are different from other fecal coliforms (rose/pink colonies). All enterococci were tested for esculin hydrolysis on bile esculin agar (Oxoid) to confirm their identification (3) before being tested for biochemical fingerprint with the PhPlate system.
Biochemical fingerprinting with the PhPlate system.
The principle of the biochemical fingerprinting with the PhPlate system has been described previously (2, 37). This method uses quantitative measurements of the kinetics of several biochemical reactions of bacteria in microtiter plates with dehydrated substrates (26, 37). The typing reagents used in this method are specifically chosen for different groups of bacteria to give an optimal discriminatory power and reproducibility (37). For each bacterial isolate, it yielded a biochemical fingerprint made of several quantitative data which are used with the PhPlate software to calculate the level of similarity between the tested isolates. Prepared microtiter plates contained 11 different substrates in each row and allowed the testing of eight isolates per plate. In this study, we used two types of plates specifically developed for typing of E. coli (PhP-RE plates) and enterococci strains (PhP-RF plates). The 11 substrates used for enterococci and E. coli have been described previously (24, 30, 51). The growth medium for PhP-RF contained 0.2% (wt/vol) proteose peptone (Oxoid), 0.05% (wt/vol) yeast extract (Oxoid), 0.5% (wt/vol) NaCl, and 0.011% (wt/vol) bromothymol blue, and for E. coli, it contained 0.1% (wt/vol) proteose peptone and 0.011% (wt/vol) bromothymol blue, according to the manufacturer's instructions.
From each sample, up to 12 single and isolated colonies were randomly selected with sterile toothpicks directly from the chromogenic coliform/E. coli agar plates (for E. coli) and from the bile esculin agar (for enterococci) and suspended into the first well of each row containing only 350 μl of growth medium. Using a multichannel pipette, aliquots of 25 μl of bacterial suspension were transferred into each of the other 11 wells containing 150 μl growth medium. Plates were then incubated at 37°C, and the A620 was measured at 7, 24, and 48 h for E. coli and at 16, 40, and 64 h for enterococci using a microplate reader (Lab-Systems Multiskan, Finland). After the final reading, the mean value for all three readings was calculated for each isolate (biochemical fingerprint). Similarities between the isolates were calculated as correlation coefficients and clustered according to the unweighted-pair group method with arithmetic averages (UPGMA) (49). An identity (ID) level of 0.965 was established based on the reproducibility of the system after testing 20 isolates in duplicate. Isolates with similarity higher than the ID level were regarded as identical and assigned to similar biochemical phenotypes (BPTs). BPTs with identical isolates were called common (C-BPT) and those with one isolate were called single (S-BPT). All S-BPTs and representative isolates of each C-BPT were transferred to McConkey agar (Oxoid) for purity and further tested for indole production and citrate before they were saved on tryptic soy broth (Oxoid) with 15% (vol/vol) glycerol at −80°C.
The phenotypic diversity among the isolates was measured with Simpson's index of diversity (Di) (6). Di in the present study depends on isolate distribution into different BPTs. Diversity is high (maximum of 1) for a population consisting of different BPTs and is low (minimum of 0) if the population consists of few BPTs. The phenotypic similarity between different bacterial populations in two or more samples was calculated as population similarity (Sp) coefficient. The Sp coefficient calculates the proportion of isolates that are identical in two or more compared bacterial populations (29). It is high (maximum of 1) if two populations contain similar BPTs and is low (minimum of 0) if the population contains different BPTs. Clustering of Sp coefficients was also performed according to the UPGMA. All data handling, including optical readings, calculations of correlations and coefficients, diversity indexes, and S values, as well as clustering and printing dendrograms, was performed using the PhPlate software version 4001 (PhPlate system, PhPlate AB, Stockholm).
Database development.
In developing the database, we categorized the BPTs into two distinct types, unique (UQ) and shared (SH) BPTs, on the basis of their occurrence in host groups. The UQ-BPTs are those BPTs that are specific to a single host group, whereas SH-BPTs were found in multiple host groups. To achieve this, all BPTs obtained from each animal were compared with those of other animals within a host group. If identical, a representative of identical BPTs, as well as all nonidentical BPTs, was initially saved in the database and regarded as total BPTs for each host group of animals. Furthermore, total BPTs from each host group were cross-referenced with those of others to calculate the occurrence of BPTs among different host groups. For instance, if a BPT from a host group (e.g., horse) was identical to a BPT from another (e.g., sheep), this BPT was regarded as SH-BPT (i.e., “shared”) between two host groups. If a BPT from a host group was not identical to those of any other group, it was regarded as UQ-BPT (i.e., “unique”).
Surface water sampling.
Water samples were collected from the Eudlo Creek, a subcatchment of the Maroochy River in the Southeast area of Queensland, Australia (Fig. 1). The total area of this largely rural subcatchment is approximately 7,980 ha, of which >85% is not serviced by a centralized sewer system. The creek is approximately 8 km in length and has been reported by the Environmental Protection Agency and Waterwatch (a community-based organization) to be contaminated with fecal bacteria and nitrates. Possible sources of contamination include intensive animal farms and approximately 1,600 conventional septic systems (2).
Samples were collected from up to five sites across the Eudlo subcatchment (Fig. 1) during November 2003 to December 2003 and during August 2004 to September 2004 on seven different occasions. In all, 27 samples were collected and were tested in triplicate. Water samples were collected in 500-ml sterile bottles from 30 cm below the water surface and transported on ice to the laboratory and tested within 6 h. The membrane filtration method was used to process all the water samples (5). Different dilutions of water samples were filtered through 0.45-μm-pore-size membranes (Millipore) and placed on chromogenic E. coli/coliform (Oxoid) and m-Enterococcus agar plates (Difco), and the plates were incubated at 37°C for 24 h (for E. coli) and 48 h (for enterococci). After incubation, from each water sample, up to 40 (where possible) enterococci and E. coli isolates (where possible) were typed with the PhPlate system as described above.
Statistical analysis.
Mann Whitney's nonparametric test was used to determine the significant difference between the mean number of Enterococcus BPTs and E. coli BPTs found in all host groups. In addition, this test was performed on the overall diversity of enterococci and E. coli from all host groups.
RESULTS
A total number of 4,057 enterococci and 3,728 E. coli isolates were typed from 10 host groups. Within each host group, different BPTs were found, some of which were identical. Representatives of the identical BPTs and the nonidentical BPTs were initially included in the database and regarded as total BPTs found in each host group. By applying this approach, a total of 526 BPTs of enterococci and 530 BPTs of E. coli were obtained from all host groups. Table 1 shows the number of isolates tested and the number of total BPTs found in each host group. For enterococci, the ratio of BPTs over the number of total isolates tested from each host group ranged from 7.3% (for sheep) to 18.7% (for horse), yielding a mean value of 13.9 ± 4.0 for all host groups. With E. coli, this ratio ranged from 8.2% (for sheep) to 17% (for ducks), yielding a mean value of 14.4 ± 2.5 (Table 1). The mean number of total Enterococcus and E. coli BPTs found in all host groups did not differ significantly (P = 0.97).
TABLE 1.
Number of samples tested from each host group and the number of total BPTs found
| Host group | No. of samples | No. of isolates tested
|
No. of total BPTs found (% over isolates)
|
||
|---|---|---|---|---|---|
| Enterococci | E. coli | Enterococci | E. coli | ||
| Human | 56 | 1,072 | 621 | 94 (8.8) | 92 (14.8) |
| Animals | |||||
| Horses | 38 | 407 | 407 | 76 (18.7) | 60 (14.7) |
| Dogs | 47 | 404 | 408 | 49 (12.1) | 64 (15.7) |
| Ducks | 46 | 408 | 404 | 58 (14.2) | 69 (17) |
| Cattle | 55 | 411 | 401 | 47 (11.4) | 53 (13.2) |
| Chicken | 36 | 408 | 408 | 74 (18.1) | 59 (14.5) |
| Pigs | 32 | 312 | 400 | 54 (17.3) | 53 (13.3) |
| Sheep | 27 | 287 | 367 | 21 (7.3) | 30 (8.2) |
| Deer | 25 | 204 | 200 | 28 (13.7) | 31 (15.5) |
| Kangaroos | 20 | 144 | 112 | 25 (17.4) | 19 (17.) |
| Total | 382 | 4,057 | 3,728 | 526 (13.9 ± 4)a | 530 (14.4 ± 2.5)a |
Mean and standard deviation.
The mean diversity of both enterococci and E. coli isolates within each host group ranged from 0.41 ± 0.38 (for sheep) to 0.75 ± 0.25 (for horses) and from 0.44 ± 0.27 (for sheep) to 0.85 ± 0.07 (for deer), respectively (Table 2). However, the overall diversities of both indicator bacteria (0.6 ± 0.1 for enterococci versus 0.65 ± 0.1 for E. coli) did not differ significantly (P = 0.36).
TABLE 2.
Mean diversity of fecal indicator bacteria in host groups
| Host group | Mean Dia
|
|
|---|---|---|
| Enterococci | E. coli | |
| Human | 0.50 ± 0.30 | 0.50 ± 0.30 |
| Animals | ||
| Horses | 0.75 ± 0.25a1 | 0.63 ± 0.26b1 |
| Dogs | 0.45 ± 0.32 | 0.57 ± 0.27 |
| Ducks | 0.72 ± 0.23 | 0.77 ± 0.22 |
| Cattle | 0.54 ± 0.34 | 0.53 ± 0.28 |
| Chicken | 0.72 ± 0.26a2 | 0.82 ± 0.18b2 |
| Pigs | 0.68 ± 0.28 | 0.73 ± 0.24 |
| Sheep | 0.41 ± 0.38 | 0.44 ± 0.27 |
| Deer | 0.59 ± 0.32a3 | 0.85 ± 0.07b3 |
| Kangaroos | 0.64 ± 0.20 | 0.72 ± 0.14 |
P < 0.2 for a1 versus b1 and a2 versus b2; P < 0.005 for a3 versus b3.
Unique and shared BPTs.
When we compared the total BPTs of all host groups with each other, it was found that certain BPTs were specific to individual host groups. These BPTs were referred to as UQ-BPTs. For enterococci, the range of UQ-BPTs among host groups varied from 7 (in sheep) to 66 (in humans). For E. coli, this figure was 6 (in kangaroos) and 69 (in humans) (Table 3). The mean percentage of total UQ-BPTs among enterococci and E. coli isolates was 56% and 51%, respectively. Certain BPTs were also found in multiple host groups, and they were referred to as SH-BPTs. For instance, of the 76 total Enterococcus BPTs found in horses, 54 were found only in horses (i.e., UQ-BPTs), whereas 22 were found not only in horses but also in other host groups (SH-BPTs).
TABLE 3.
Number of unique and shared BPTs in host groups
| Host source | No. of UQ-BPTsa (% over total BPTs)
|
No. of SH-BPTsb (% over total BPTs)
|
||
|---|---|---|---|---|
| Enterococci | E. coli | Enterococci | E. coli | |
| Human | 66 (70) | 69 (75) | 28 (30) | 23 (25) |
| Animals | ||||
| Horses | 54 (71) | 32 (53) | 22 (29) | 28 (47) |
| Dogs | 24 (49) | 32 (50) | 25 (51) | 32 (50) |
| Ducks | 29 (50) | 32 (46) | 29 (50) | 37 (54) |
| Cattle | 23 (49) | 24 (45) | 24 (51) | 29 (55) |
| Chicken | 41 (55) | 33 (56) | 33 (45) | 26 (44) |
| Pigs | 28 (52) | 25 (47) | 26 (48) | 28 (53) |
| Sheep | 7 (33) | 11 (37) | 14 (67) | 19 (63) |
| Deer | 13 (46) | 9 (29) | 15 (54) | 22 (71) |
| Kangaroos | 10 (40) | 6 (32) | 15 (60) | 13 (68) |
| Total | 295 (56) | 273 (51) | 231 (44) | 257 (49) |
Identical BPTs within each host group are not included.
BPTs found in multiple host groups.
For enterococci, the range of SH-BPTs among host groups varied from 14 (in sheep) to 33 (in chickens), and for E. coli, these figures were 13 (in kangaroos) and 37 (in ducks) (Table 3). Therefore, a total of 295 Enterococcus BPTs and 273 E. coli BPTs occurred only once in the database, while 231 BPTs for enterococci and 257 BPTs for E. coli isolates occurred in multiple host groups. All BPTs (i.e., UQ- or SH-BPTs) from animal groups that were not found in humans were collectively categorized as animal BPTs. The animal BPTs consisted of 432 Enterococcus BPTs and 438 E. coli BPTs, of which 229 (53%) Enterococcus BPTs and 204 (47%) E. coli BPTs were UQ-BPTs (Tables 1 and 3).
Tracking the source of contamination in Eudlo Creek.
A total of 27 water samples were collected from five sites along the Eudlo Creek mainstream (Fig. 1). From each water sample, up to 40 enterococci and E. coli isolates (where possible) were typed and compared with the database. The mean diversities of enterococci (Di = 0.76 ± 0.05) and E. coli (Di = 0.88 ± 0.04) were generally high (maximum of 1) in water samples, indicating diverse sources of these bacteria. A total of 791 enterococci (248 total BPTs) were tested from water samples, of which 26 BPTs (10%) were found only in humans (i.e., UQ-BPTs) and 152 BPTs (61%) belonged to animals (i.e., animal BPTs) tested in this study (Table 4). Of the 550 E. coli isolates (282 total BPTs) tested from the same water samples, 36 BPTs (13%) were of human origin and 151 BPTs (54%) belonged to animals tested (Table 4). The remaining 70 Enterococcus BPTs and 95 E. coli BPTs either belonged to BPTs shared between humans and animals (28 Enterococcus BPTs and 23 E. coli BPTs) or did not match the database and were therefore regarded as unknown BPTs (Table 4).
TABLE 4.
Comparison of BPTs from water samples with the database
| Sampling occasion | Sampling site | No. of isolates tested (no. of total BPTs found)
|
No. of total BPTs identical to database
|
Unknown BPTs
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| Human UQ-BPTs
|
Animal BPTs
|
||||||||
| Enterococci | E. coli | Enterococci | E. coli | Enterococci | E. coli | Enterococci | E. coli | ||
| 1 | EC1 | 32 (10) | 32 (12) | 2 | 1 | 6 | 5 | 2 | 6 |
| EC2 | 38 (15) | 25 (15) | 1 | 4 | 9 | 11 | 5 | 0 | |
| EC3 | 39 (16) | 14 (11) | 1 | 1 | 13 | 8 | 2 | 2 | |
| 2 | EC1 | 29 (9) | 23 (14) | 1 | 3 | 5 | 11 | 3 | 0 |
| EC2 | 39 (13) | 22 (12) | 1 | 2 | 6 | 8 | 6 | 2 | |
| EC3 | 40 (10) | 65 (26) | 2 | 4 | 7 | 17 | 1 | 5 | |
| 3 | EC1 | 38 (12) | 19 (10) | 2 | 2 | 8 | 5 | 2 | 3 |
| EC2 | 36 (14) | 21 (14) | 3 | 3 | 8 | 11 | 3 | 0 | |
| EC3 | 39 (17) | 23 (8) | 3 | 9 | 6 | 5 | 2 | ||
| 4 | EC1 | 22 (9) | 18 (10) | 1 | 1 | 6 | 5 | 2 | 4 |
| EC2 | 23 (10) | 19 (12) | 8 | 5 | 2 | 7 | |||
| EC3 | 23 (8) | 20 (12) | 2 | 3 | 5 | 5 | 5 | ||
| EC4 | 23 (6) | 17 (13) | 3 | 3 | 5 | 3 | 5 | ||
| EC5 | 23 (10) | 10 (16) | 6 | 4 | 4 | 6 | |||
| 5 | EC1 | 23 (7) | 7 (5) | 1 | 4 | 4 | 2 | 1 | |
| EC2 | 23 (7) | 14 (6) | 1 | 4 | 2 | 2 | 4 | ||
| EC3 | 23 (7) | 11 (8) | 1 | 5 | 3 | 1 | 5 | ||
| EC4 | 23 (8) | 6 | 2 | ||||||
| EC5 | 23 (7) | 13 (11) | 2 | 4 | 4 | 3 | 5 | ||
| 6 | EC1 | 23 (9) | 21 (10) | 2 | 7 | 4 | 2 | 4 | |
| EC2 | 23 (6) | 29 (18) | 1 | 2 | 4 | 4 | 1 | 12 | |
| EC3 | 23 (6) | 7 (5) | 5 | 2 | 1 | 3 | |||
| EC4 | 23 (8) | 1 | 5 | 2 | |||||
| EC5 | 23 (6) | 11 (8) | 1 | 3 | 3 | 3 | 4 | ||
| 7 | EC1 | 39 (4) | 33 (10) | 2 | 2 | 2 | 3 | 5 | |
| EC2 | 39 (7) | 33 (7) | 1 | 3 | 6 | 3 | 1 | ||
| EC3 | 39 (7) | 37 (15) | 1 | 1 | 3 | 10 | 3 | 4 | |
| Total | 27 | 791 (248) | 550 (282) | 26 | 36 | 152 | 151 | 70 | 95 |
Comparison of total BPTs found in water samples over the entire sampling period with the database showed that 61% of Enterococcus and 54% of E. coli BPTs were identical to animal BPTs and that some were also unique to individual animal groups. Distribution of UQ-BPTs among animal species ranged between 0% (deer) to 13% (chicken) for enterococci and 0% to 8% (ducks) for E. coli isolates. Ten percent of Enterococcus UQ-BPTs and 13% of E. coli UQ-BPTs found in water samples were identical to those of humans.
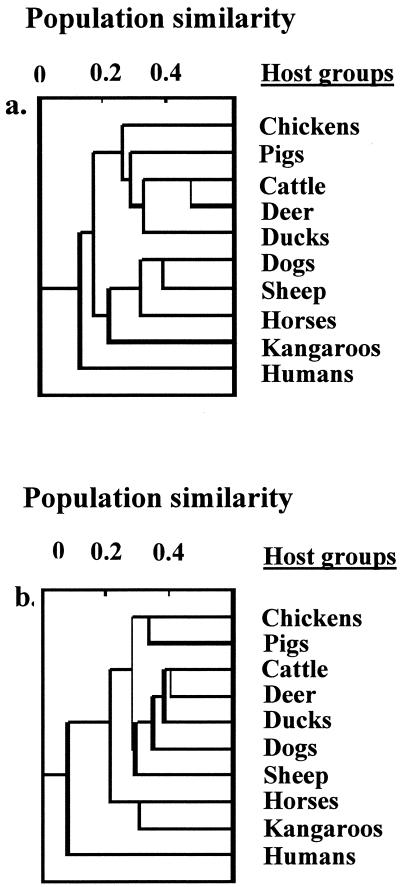
To identify whether there is a fundamental difference between the populations of both fecal indicator bacteria from humans and animals, we also performed a population similarity comparison between all BPTs found in humans and animals. The results indicated that the mean similarity among Enterococcus (0.27 ± 0.1) and E. coli (0.34 ± 0.06) populations between different animal groups was significantly higher (P = 0.003 for enterococci and P = 0.001 for E. coli isolates) than the mean similarity between human and animals (i.e., 0.16 ± 0.03 for enterococci and 0.09 ± 0.02 for E. coli isolates) (Fig. 2).
FIG. 2.
UPGMA dendrogram of population similarity of Enterococcus (a) and E. coli (b) populations from all host groups.
DISCUSSION
Identification of potential sources of fecal contamination in surface waters requires a method that is capable of distinguishing between human and animal sources. Ideally, the method should also be sensitive enough to discriminate different animal species. In recent years, several genotypic and phenotypic methods have been developed to trace the sources of fecal contamination in surface waters by typing fecal indicator bacteria (9-11, 13, 19, 22, 40, 41, 43, 45, 46, 53, 54). In this study, we used a biochemical fingerprinting method to develop a host-specific metabolic fingerprint database of two recommended fecal indicator bacteria, enterococci and E. coli (1, 44), to trace the sources of fecal contamination in surface waters.
It is known that the size and representativeness of the database are two important factors for determining the source(s) of fecal contamination (44, 52). To date, most of the genotypic and phenotypic host origin databases are based on testing up to 500 isolates of either enterococci or E. coli (11-13, 19, 34, 40, 44, 52). In addition, these isolates were usually collected from a small number of samples, which may not sufficiently represent the diverse fecal indicator bacteria found in different host groups. In developing the metabolic fingerprint database, we specifically focused on two important factors. One was the number of isolates to be tested from each animal species, and the other was how well these numbers represent the diversity of indicator bacteria among the animal species. To address this question, we initially tested 160 isolates of both fecal indicator bacteria from five randomly chosen individuals of the same species of animals within a farm (data not shown). This comparison showed that animals of same species within a farm carry many identical BPTs, which could be explained by their frequent contact with each other or dietary similarity, and therefore share a common bacterial population (19, 20, 26). However, we obtained a better diversity when we compared animals of the same species from one farm with those of another within a radius of 20 km of both the inside and outside of the studied catchment.
For this reason, we reduced the number of samples to three individuals of the same species from each farm and increased the number of farms up to 20 where possible within and outside the study area. Using this strategy, we tested an average of 400 isolates of both fecal indicator bacteria from 10 host groups yielding a total number of 4,087 enterococci and 3,728 E. coli isolates from different farms or locations. Based on this experience, however, we suggest that the emphasis should be focused on testing more individual animals (preferably from different farms) rather than testing more bacterial isolates from each individual, so as to obtain diverse phenotypes or genotypes of known sources.
The comparison of total BPTs in each host group with others showed that many identical BPTs were shared in multiple host groups. Bacteria are ubiquitous in the environment and can be found transitionally in many animal species simultaneously. Similar shared fingerprints (ribotypes) have also been reported among different host groups in other studies (19, 20, 34). However, in our study, the percentage of shared BPTs among host groups was quite high. This is due to the fact that we not only tested a large number of isolates from each host group but also tested a wide range of host groups and therefore found more shared BPTs among host groups. In contrast, a number of BPTs were also specific to their individual host group and were therefore regarded as UQ-BPTs. A recent molecular-based study (25) defined unique genotypes on the basis of specificity to individual host group rather than by comparing these genotypes to those found in other host groups. However, in our study, we defined UQ-BPTs as those BPTs that occurred only once in each host group after being compared with all other total BPTs found in other host groups. The number of UQ-BPTs in our study varied among different host groups. Some host groups (i.e., sheep, deer, and kangaroos) contained a smaller number of UQ-BPTs than others. This may be explained by the fact that a smaller number of samples tested from these host groups are from limited locations, and therefore, our sampling effort could not capture the diversity found among these host groups. Nevertheless, we found that these UQ-BPTs can be used as specific fingerprints to pinpoint the sources of fecal contamination in surface waters. In contrast, some SH-BPTs were found in two or more animal species including humans. For instance, we found 28 BPTs of enterococci and 23 BPTs of E. coli, which occurred in both human and animal host groups. These BPTs could not be used to distinguish the various sources of fecal contamination and were excluded from our database. However, we also found that certain SH-BPTs, though found among different animal species, were not found in humans and could therefore be categorized as animal BPTs (including UQ- and SH-BPTs among nine host groups of animals).
To evaluate the ability of our database to identify sources of fecal contamination, we collected a total of 27 water samples from five different sites along the Eudlo Creek mainstream on several occasions and compared the total BPTs of both fecal indicator bacteria isolated from these sites with our database. Ten percent of Enterococcus BPTs and 13% of E. coli BPTs were identified as human UQ-BPTs. It should be noted that in our study, human samples were obtained from septic tanks rather than fresh human fecal samples, and therefore, some unique and/or shared strains may have not survived in the septic tanks and may therefore not have been detected. Of the animal BPTs, 101 (66%) Enterococcus BPTs and 93 (62%) E. coli BPTs were unique to individual host groups. On the basis of UQ-BPTs for enterococci, chickens contributed 13% of contamination, followed by humans (13%). For E. coli, humans contributed 13%, followed by ducks (9%). Both the Enterococcus and E. coli databases were in close agreement in terms of identifying the sources of contamination (i.e., 10% of enterococci and 13% of E. coli isolates for humans and 6% of enterococci and 7% of E. coli isolates for cattle were identified from the same water samples), although it was not quite consistent for certain host groups (i.e., for chickens, 14% of enterococci and 6% of E. coli isolates). However, interestingly, total BPTs from deer were not identical to those found in the water samples which can be explained by the fact that deer are normally kept in a sanctuary, which restricted their access to nearby creeks, and in addition, our study area did not contain any wild deer. On the basis of this, we conclude that both fecal indicator bacteria can be used alone or in combination with each other, which provides much better insight regarding the contributing sources. Using this PhPlate system and the same indicator bacteria, we have recently shown that a combination of both fecal bacteria provides a better understanding of the sources of human contamination in surface waters through failed septic systems (2).
Certain BPTs of both fecal indicator bacteria found in water samples did not match our database. This may be due to the fact that either our database was not large enough to capture the diversity of these indicator bacteria or these unknown BPTs might have originated from other nonpoint sources or a combination of both. It has been suggested that a library size of up to 40,000 isolates may be needed to capture the genetic diversity present among E. coli isolates (25). It is also recommended that databases should ideally be developed from the animal species residing in the study area, as they are more likely to contribute fecal contamination to surface waters in the study area (25). In our study, although we tried to collect samples from as many farms as possible within and outside the studied area, we did not have access to all farms in the studied catchment. Another important factor that has to be considered is that the number and the types of animals within a study area may vary over time due to agricultural practices and/or animal migration (25), and therefore, it may not be possible to include samples from all animals that reside in a study area. This will restrict the ability of a database to trace the sources of contamination from all animals within a watershed. In addition, it is known that geographical variability exists among indicator bacteria (19), which limits the efficiency of a database to identify unknown environmental isolates when these bacteria are collected from another geographical area. Therefore, it has to be noted that the universal use of such a database which is developed for a limited geographical area should be interpreted with care.
In conclusion, we developed a host-specific metabolic fingerprint database of two fecal indicator bacteria and used it successfully to trace the source of fecal contamination in the studied creek. The database was capable of identifying the sources of more than 65% of fecal bacteria in the studied creek. We also found that while this system could differentiate between human and animal sources of fecal contamination, it was also capable of further differentiating between animal species and can therefore be used as a potential tool to trace the sources of fecal contamination in a confined geographical area.
Acknowledgments
We thank Anthony Weston, Sandra Hipwood, and Daniel Morgan for their assistance in collecting samples. We also thank Maroochy Shire Council for their cooperation.
This study was financially supported by an internal grant, SRG 03/5, from the University of the Sunshine Coast.
REFERENCES
- 1.Abbot, S., B. Caughley, and G. Scott. 1993. Evaluation of Enterolert for the enumeration of enterococci in the marine environment. N. Z. J. Mar. Freshwater Res. 32:505-513. [Google Scholar]
- 2.Ahmed, W., R. Neller, and M. Katouli. 2004. Evidence of septic systems failure determined by a bacterial biochemical fingerprinting method. J. Appl. Microbiol. 98:910-920. [Online.] doi: 10.1111/j.1365-2672.2004.02522.x. [DOI] [PubMed] [Google Scholar]
- 3.American Public Health Association. 1995. Standard method for the examination of water and wastewater, 19th ed. American Public Health Association, Inc., Washington, D.C.
- 4.Amor, K., D. E. Heinrichs, E. Frirdich, K. Ziebell, R. P. Johnson, and C. Whitfield. 2000. Distribution of core oligosaccharide types in lipopolysaccharides from Escherichia coli. Infect. Immun. 68:1116-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anonymous. 1997. Water quality—detection and enumeration of coliform organisms, thermotolerant coliform organism and presumptive Escherichia coli. Part 1: membrane filtration method. International Organization for Standardization document 9308/1. ISO, Geneva, Switzerland.
- 6.Atlas, R. 1984. Use of microbial diversity measurements to assess environmental stress, p. 540-545. In M. J. Klug and C. A. Reddy (ed.), Current perspectives in microbial ecology. American Society for Microbiology, Washington, D.C.
- 7.Barnes, D., and D. M. Gordon. 2004. Coliform dynamics and the implications for source tracking. Environ. Microbiol. 6:501-509. [DOI] [PubMed] [Google Scholar]
- 8.Baudisöva, D. 1997. Evaluation of Escherichia coli as the main indicator of fecal pollution. Water Sci. Tech. 35:333-356. [Google Scholar]
- 9.Bernhard, A. E., and K. G. Field. 2000. A PCR assay to discriminate human and animal feces on the basis of the host difference in Bacteroides-Prevotella genes encoding 16S rRNA. Appl. Environ. Microbiol. 66:4571-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernhard, A. E., and K. G. Field. 2000. Identification of nonpoint sources of fecal pollution in coastal waters by using host-specific 16S ribosomal DNA genetic markers from fecal anaerobes. Appl. Environ. Microbiol. 66:1587-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carson, S. A., B. L. Shear, M. R. Ellersieck, and A. Afsaw. 2001. Identification of fecal Escherichia coli from humans and animals by ribotyping. Appl. Environ. Microbiol. 67:1503-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carson, S. A., B. L. Shear, M. R. Ellersieck, and J. D. Schnell. 2003. Comparison of ribotyping and repetitive extragenic palindromic-PCR for identification of fecal Escherichia coli from humans and animals. Appl. Environ. Microbiol. 69:1836-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dombek, P. E., L. K. Johnson, M. B. Brown, and M. J. Sadowsky. 2000. Use of repetitive DNA sequences and the PCR to differentiate Escherichia coli isolates from human and animal sources. Appl. Environ. Microbiol. 66:2572-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freter, R. 1984. Factors affecting conjugal plasmid transfer in natural bacterial communities, p. 105-114. In M. J. Klug and C. A. Reddy (ed.), Current perspectives in microbial ecology. American Society for Microbiology, Washington, D.C.
- 15.Fujioka, R. S. 2002. Microbial indicators of water quality, p. 234-243. In C. J. Hurst, R. L. Crawford, G. R. Knudsen, K. J. McInerney, and L. D. Stetzenbach (ed.), Manual of environmental microbiology, 2nd ed. ASM Press, Washington, D.C.
- 16.Hagedorn, C., S. L. Robinson, J. R. Filtz, S. M. Grubbs, T. A. Angier, and R. B. Reneau, Jr. 1999. Determining sources of fecal pollution in a rural Virginia watershed with antibiotic resistance patterns in fecal streptococci. Appl. Environ. Microbiol. 65:5522-5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hagedorn, C., J. B. Crozier, K. A. Mentz, A. M. Booth, A. K. Graves, N. J. Nelson, and R. B. Reneau, Jr. 2002. Carbon source utilization profiles as a method to identify sources of faecal pollution in water. J. Appl. Microbiol. 94:792-799. [DOI] [PubMed] [Google Scholar]
- 18.Hartel, P. G., W. I. Segars, N. J. Stern, J. Steiner, and A. Buchan. 1999. Ribotyping to determine host origin of E. coli isolates in different water samples, p. 377-382. In D. S. Olsen and J. P. Potyondy (ed.), Wild land hydrology. Technical publication series TPS-99-3. American Public Health Association, Washington, D.C.
- 19.Hartel, P. G., J. D. summer, J. L. Hill, J. C. Collins, J. A. Entry, and W. I. Segers. 2002. Geographic variability of Escherichia coli isolates from animals in Idaho and Georgia. J. Environ. Qual. 31:1273-1278. [DOI] [PubMed] [Google Scholar]
- 20.Hartel, P. G., J. D. Summer, and W. I. Segars. 2003. Deer diet affects ribotype diversity of Escherichia coli for bacterial source tracking. Water Res. 37:3263-3268. [DOI] [PubMed] [Google Scholar]
- 21.Harwood, V. J., B. D. Parrish, and V. Wagner. 1999. Isolation of fecal coliform bacteria from the diamondback terrapin (Malaclemys terrapain centrata). Appl. Environ. Microbiol. 65:865-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harwood, V. J., J. Whitlock, and V. Withington. 2000. Classification of antibiotic resistance patterns of indicator bacteria by discriminant analysis: use in predicting the source of fecal contamination in tropical waters. Appl. Environ. Microbiol. 66:3698-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hurst, C. J. 1997. Overview of water microbiology as it relates to public health, p. 133-135. In C. J. Hurst, G. R. Knudsen, M. J. McInerney, L. D. Stetzenbach, and M. V. Walter (ed.), Manual of environmental microbiology. American Society for Microbiology, Washington, D.C.
- 24.Iverson, A., I. Kühn, A. Franklin, and R. Möllby. 2002. High prevalence of vancomycin-resistant enterococci in Swedish sewage. Appl. Environ. Microbiol. 68:2838-2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson, L. K., M. B. Brown., E. A. Carruthers., J. A. Ferguson., P. E. Dombek, and M. J. Sadowsky. 2004. Sample size, library composition, and genotypic diversity among natural populations of Escherichia coli from different animals influence accuracy of determining sources of fecal pollution. Appl. Environ. Microbiol. 70:4478-4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katouli, M., A. Lund., P. Wallgren., I. Kühn., O. Söderlind, and R. Möllby. 1997. Metabolic fingerprinting and fermentative capacity of the intestinal flora of pigs during pre- and post-weaning periods. J. Appl. Microbiol. 83:147-154. [DOI] [PubMed] [Google Scholar]
- 27.Kibbey, H. J., C. Hagedorn, and E. L. McCoy. 1978. Use of fecal streptococci as indicators of pollution in soil. Appl. Environ. Microbiol. 35:711-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kreader, C. A. 1995. Design and evaluation of Bacteroides DNA probes for the specific detection of human fecal pollution. Appl. Environ. Microbiol. 61:1171-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kühn, I., G., Allestam, T. A. Strenström, and R. Möllby. 1991. Biochemical fingerprinting of water coliform bacteria, a new method for measuring the phenotypic diversity and for comparing different bacterial populations. Appl. Environ. Microbial. 57:3171-3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kühn, I., M. Katouli, P. Wallgren, O. Söderlind, and R. Möllby. 1995. Biochemical fingerprinting as a tool to study the diversity and stability of intestinal microfloras. Microecol. Ther. 23:140-148. [Google Scholar]
- 31.Kühn, I., G. Allestam, M. Engdhal, and T. A. Stenström. 1997. Biochemical fingerprinting of coliform bacterial populations—comparisons between polluted river water and factory effluents. Water Sci. Tech. 35:343-350. [Google Scholar]
- 32.Kühn, I., J. Albert, M. Ansaruzzaman, S. A. Alabi, N. A. Bhuiyanand, G. Huys, M. S. Islam, P. Jansen, K. Kersters, P. K. B. Neogi, and R. Möllby. 1997. Characterization of Aeromonas spp. isolated from humans with diarrhea, from healthy controls, and from surface water in Bangladesh. J. Clin. Microbiol. 35:369-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leeming, R., A. Ball., N. Ashbolt, and P. D. Nicholas. 1996. Using faecal sterols from humans and animals to distinguish faecal pollution in receiving waters. Water Res. 30:2893-2900. [Google Scholar]
- 34.McLellan, S. L. 2004. Genetic diversity of Escherichia coli isolated from urban rivers and beach water. Appl. Environ. Microbiol. 70:4658-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meays, C. L., K. Broersma, R. Nordin, and A. Majumder. 2004. Source tracking fecal bacteria in water: a critical review of current methods. J. Environ. Manage. 73:71-79. [DOI] [PubMed] [Google Scholar]
- 36.Moe, C. 2002. Waterborne transmission of infectious agents, p. 184-204. In C. J. Hurst, R. L. Crawford, G. R. Knudsen, K. J. McInerney, and L. D. Stetzenbach (ed.), Manual of environmental microbiology, 2nd ed. ASM Press, Washington, D.C.
- 37.Möllby, R., I. Kühn, and M. Katouli. 1993. Computerized biochemical fingerprinting—a new tool for typing of bacteria. Rev. Med. Microbiol. 4:231-241. [Google Scholar]
- 38.Olive, D. M., and P. Bean. 1999. Principles and applications of methods for DNA-based typing of microbial organisms. J. Clin. Microbiol. 37:1661-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ørskov, F. 1984. Genus I. Escherichia Castellani and Chalmers 1919, 941AL, p. 420-423. In N. R. Kreig and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 1. Williams and Wilkins, Baltimore, Md.
- 40.Parveen, S., R. L. Murphree, L. Edmiston, C. W. Kaspar, K. M. Portier, and M. L. Tamplin. 1997. Association of multiple-antibiotic-resistance profiles with point and nonpoint sources of Escherichia coli in Apalachicola Bay. Appl. Environ. Microbiol. 63:2607-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parveen, S., K. M. Portier, K. Robinson, L. Edmiston, and M. Tamplin. 1999. Discrimination analysis of ribotype profiles of Escherichia coli for differentiating human and nonhuman sources of fecal pollution. Appl. Environ. Microbiol. 65:3142-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Puech, M. C., J. M. McAnulty, M. Lesjak, N. Shaw, L. Heron, and J. M. Watson. 2001. A statewide outbreak of cryptosporidiosis in New South Wales associated with swimming at public pools. Epidemiol. Infect. 126:389-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Samadpour, M., and N. Checowitz. 1995. Little Soos Creek microbial source tracking: a survey. University of Washington Department of Environmental Health, Seattle, Wash.
- 44.Scott, T. M., J. B. Rose, T. M. Jenkins, S. R. Farrah, and J. Lukasik. 2002. Microbial source tracking: current methodology and future directions. Appl. Environ. Microbiol. 68:5796-5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simmons, G. M., Jr., S. A. Herbain, and C. M. James. 1995. Managing non-point fecal coliform sources to tidal inlets. Water Res. Upd. 1995:64-74. [Google Scholar]
- 46.Simmons, G. M., Jr., and S. A. Herbain. 1998. Potential sources of Escherichia coli (E. coli) to children's pool in La Jolla, California. Final report for the City of San Diego and the County of San Diego Dep. of Environmental Health, San Diego, CA. County of San Diego Department of Environmental Health, San Diego, Calif.
- 47.Simpson, J. M., J. W. Santo Domingo, and D. J. Reasoner. 2003. Microbial source tracking: state of the science. Environ. Sci. Technol. 36:5280-5288. [DOI] [PubMed] [Google Scholar]
- 48.Sinton, L. W., A. M. Donnison, and C. M. Hastie. 1993. Faecal streptococci as faecal pollution indicators: a review. Part II. Sanitary significance, survival and use. N. Z. J. Mar. Freshwater Res. 27:117-137. [Google Scholar]
- 49.Sneath, P. H. A., and R. R. Sokal. 1973. Numerical taxonomy: the principles and practice of numerical classification. W. H. Freeman, San Francisco, Calif.
- 50.Toranzos, G. A., and G. A. McFeters. 1997. Detection of indicator microorganisms in environmental fresh waters and drinking waters, p. 184-194. In C. J. Hurst, G. R. Knudsen, M. J. McInerney, L. D. Stetzenbach, and M. V. Walter (ed.), Manual of environmental microbiology. American Society for Microbiology, Washington, D.C.
- 51.Vilanova, X., A. Manero, M. C. Cuéller, and A. R. Blanch. 2002. The effect of a sewage treatment plant effluent on the faecal coliforms and enterococci populations of the reception river waters. J. Appl. Microbiol. 92:210-214. [DOI] [PubMed] [Google Scholar]
- 52.Whitlock, J. R., D. T. Jones, and V. J. Harwood. 2002. Identification of the sources of fecal coliforms in an urban watershed using antibiotic resistance analysis. Water Res. 36:4273-4282. [DOI] [PubMed] [Google Scholar]
- 53.Wiggins, B. A. 1996. Discriminant analysis of antibiotic resistance patterns in fecal streptococci, a method to differentiate human and animal sources of fecal pollution in natural waters. Appl. Environ. Microbiol. 62:3997-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wiggins, B. A., R. W. Andrews, R. A. Conway, C. L. Corr, E. J. Dobratz, D. P. Dougherty, J. R. Eppard, S. R. Knupp, M. C. Limjoco, J. M. Mettenburg, J. M. Rinehardt, J. Sonsino, R. L. Torrijos, and M. E. Zimmerman. 1999. Use of antibiotic resistance analysis to identify nonpoint sources of fecal pollution. Appl. Environ. Microbiol. 65:3483-3486. [DOI] [PMC free article] [PubMed] [Google Scholar]