Abstract
A novel approach combining a flow cytometric in situ viability assay with 16S rRNA gene analysis was used to study the relationship between diversity and activity of the fecal microbiota. Simultaneous staining with propidium iodide (PI) and SYTO BC provided clear discrimination between intact cells (49%), injured or damaged cells (19%), and dead cells (32%). The three subpopulations were sorted and characterized by denaturing gradient gel electrophoresis (DGGE) of 16S rRNA gene amplicons obtained from the total and bifidobacterial communities. This analysis revealed that not only the total community but also the distinct subpopulations are characteristic for each individual. Cloning and sequencing of the dominant bands of the DGGE patterns showed that most of clones retrieved from the live, injured, and dead fractions belonged to Clostridium coccoides, Clostridium leptum, and Bacteroides. We found that some of the butyrate-producing related bacteria, such as Eubacterium rectale and Eubacterium hallii, were obviously viable at the time of sampling. However, amplicons affiliated with Bacteroides and Ruminococcus obeum- and Eubacterium biforme-like bacteria, as well as Butyrivibrio crossotus, were obtained especially from the dead population. Furthermore, some bacterial clones were recovered from all sorted fractions, and this was especially noticeable for the Clostridium leptum cluster. The bifidobacterial phylotypes identified in total samples and sorted fractions were assigned to Bifidobacterium adolescentis, Bifidobacterium longum, Bifidobacterium infantis, Bifidobacterium pseudocatenulatum, and Bifidobacterium bifidum. Phylogenetic analysis of the live, dead, and injured cells revealed a remarkable physiological heterogeneity within these bacterial populations; B. longum and B. infantis were retrieved from all sorted fractions, while B. adolescentis was recovered mostly from the sorted dead fraction.
The human gastrointestinal (GI) tract harbors a complex and dynamic microbial ecosystem in which relationships among bacteria and between these organisms and the host are significant (20). The large number of bacterial species, estimated to be more than 1,000 (53), and the high number of microorganisms that inhabit the GI tract represent enormous biological potential for metabolic conversions (19). These include production of short-chain fatty acids, vitamin synthesis, deconjugation of bile salts, and degradation of mucin (11).
During the last decade, the application of cultivation-independent molecular techniques based on 16S rRNA gene analysis has provided new insights into the microbial ecology of the GI tract. The results of studies have greatly advanced our knowledge by unraveling the complexity (17, 45, 56, 57), structure (15, 27), establishment, and succession (12, 14) of the intestinal microbiota. Yet little is known about the in situ association between the microbial diversity and metabolic activity of a phylogenetically affiliated group. Understanding this relationship is important since not all members of the ecosystem contribute similarly to physiological function, which can be influenced by factors such as nutritional changes, pathogens, stress, and drug intake (19). Therefore, comprehensive in situ analytical approaches that provide simultaneous information about the identity and activity of a microbial cell in its natural environment are essential.
From an ecological point of view, at least three categories of cells can be distinguished within microbial communities (6): (i) viable and active cells that play a functional role and participate in the production of biomass at the time of sampling, (ii) viable and inactive cells (dormant or injured) that might play a role in the future, and (iii) dead cells that may once have been active but no longer play a role in the cycling of chemical elements and hence represent only particulate organic carbon. In the GI tract ecosystem, the members of the latter category may still have some functions, as shown by studies with dead probiotic bacteria (38).
Several innovative approaches are being developed to resolve the linkage between structure, activity, and function in microbial communities. These approaches include methods in which molecular techniques are coupled with substrate labeling, such as stable isotope probing (39, 40), microautoradiography and fluorescent in situ hybridization (28), and labeling with fluorescent functional probes followed by flow cytometry (FCM) and cell sorting (6, 52). FCM has been viewed as a powerful technique for monitoring the metabolic activity of stressed and starved bacteria and identifies microorganisms in their natural habitat (9, 21, 34, 58). One major advantage of FCM is that it allows monitoring of bacterial heterogeneity at the single-cell level and provides a means to sort subpopulations of interest for further molecular analysis (10, 13, 49).
In this paper we report on an application of a viability assessment approach in which FCM was used to monitor the activity of human GI tract microbiota with functional probes. Fecal samples were subjected to fluorescence-activated cell sorting (FACS) followed by denaturing gradient gel electrophoresis (DGGE) analysis of 16S rRNA amplicons to obtain insight into the diversity of the different physiological fractions. Cloning and sequencing of the most abundant DGGE bands from total, viable, dead, and injured cells were performed, and phylogenetic affiliations were assigned to the different metabolic fractions.
MATERIALS AND METHODS
Fecal samples.
Fresh fecal samples were collected from four healthy adults (three females and one male, 25 to 55 years old). These volunteers had not been subjected to any feeding trial, specific diet, or antibiotic treatment for the last year. Samples were processed immediately after collection in an anaerobic chamber with an atmosphere consisting of 10% CO2, 10% H2, and 80% N2. To extract fecal microbial cells, 0.5 g of feces was suspended in 4.5 ml of anaerobic phosphate-buffered saline (PBS) containing 1 mM dithiothreitol and homogenized by vortexing the fecal slurry for 3 min at high speed in the presence of 5 to 10 glass beads with a diameter of 3 μm. After centrifugation at 700 × g for 1 min, 1 ml of supernatant was carefully recovered in an Eppendorf tube and centrifuged at 6,000 × g for 3 min. The supernatant was discarded, and the pellet was washed twice with 1 ml anaerobic PBS. The cell suspensions were sonicated twice for 20 s in a water bath. The cell suspensions were then diluted 10- to 100-fold and kept anaerobically on ice until analysis.
Viability staining.
The following fluorescent probes were obtained from Molecular Probes BV, Leiden, The Netherlands: propidium iodide (PI), SYTO BC, SYTO 9, and SYTO 13. To assess the viability of fecal bacteria based on their membrane integrity, 1 ml of a diluted fecal sample (1 × 108 to 5 × 108 cells) was incubated for 15 min at room temperature in the dark with 14.5 μmol/ml PI in the presence of one of the SYTO dyes. The final concentrations of SYTO 9 and SYTO 13 were 3.34 μM, and 5 μM, respectively; SYTO BC was used according to the manufacturer's instructions. When excited with a 488-nm argon laser, the SYTO dyes fluoresce in the green wavelength range (maximum at 521 nm), and PI fluoresces in the red wavelength range (maximum at 617 nm). All labeled samples were kept on ice for no longer than 1 h before FCM analysis. All the staining steps were carried out under anaerobic conditions.
Flow cytometry and cell sorting.
Flow cytometry was performed with a FACSCalibur (Becton Dickinson, San Jose, Calif.) equipped with an air-cooled 15-mW argon ion laser operating at 488 nm. The green fluorescence of the SYTO dyes (FL1) was collected using a 530- ± 30-nm band-pass filter; the red fluorescence emitted from PI (FL3) was collected using a 650- ± 13-nm band-pass filter. The data were analyzed with the CellQuest software from Becton Dickinson; all parameters were measured using logarithmic amplification. Each sample was acquired between 20 and 30 s at the low rate, and the cell concentration was adjusted to maintain a count of 500 to 600 events/s. Control samples were used for the instrument settings (voltage of the detectors and the compensation) and consisted of unlabeled fecal cells, heat-killed fecal cells stained with PI (FL3), and cells labeled with SYTO BC (FL1). CaliBRITE beads obtained from Becton Dickinson were used to check the instrument sensitivity over time. Fecal cells were discriminated from electronic noise using a double threshold set on both side scatter (SSC) and forward scatter (FSC), with FSC set on E01 and SSC set on 400V. All samples were sonicated twice (20 s each) and thoroughly vortexed prior to FCM analysis. A dual dot plot of FSC versus SSC in combination with a one-parameter histogram representing the SYTO BC fluorescence was used to backgate fecal cells and distinguish them from the background. For total cell enumeration, samples were incubated in the presence of 1 μl SYTO BC for 5 min at room temperature. Unlabeled beads with a diameter of 6.0 μm obtained from a Bacteria Counting Kit (Molecular Probes BV, Leiden, The Netherlands) were added to each sample at a final concentration of 106 beads/ml sample and served as an internal standard to calibrate the sample volume. The ratio of SYTO BC-stained cells to the number of beads was used to calculate the absolute total cell counts. Data were collected in list mode as pulse height signals (four decades in a logarithmic scale each) and were analyzed using the computer program Windows Multiple Document Interface (WinMDI; Joseph Totter, Salk Institute for Biological Studies, La Jolla, Calif.; available at http//:facs.Scripps.edu/software.html).
For the sorting experiment, a FACSVantage flow cytometer (Becton Dickinson) was used, and the sorting criteria were defined by drawing gates in two bivariate dot plots representing FSC versus SSC and red fluorescence (PI) versus green fluorescence (SYTO BC). Filtered, autoclaved PBS (pH 7.2) was used as a sheath fluid; the pressure of the sheath fluid was 45 lb/in2, and the nozzle size was 70 μm. The drop drive frequency was set at 67 kHz/s. Each event was sorted in a sort envelope of 1.2 drops at a rate of 8 × 103 to 1 × 104 events/s. In order to achieve high purity and recovery, the R mode was used to sort 1 × 106 to 5 × 106 cells from each subpopulation into separate sterile polystyrene tubes. Sorted cells were then collected on 0.2-μm polycarbonate filters using a standard 25-mm glass fritted filtration base (Millipore BV, Amsterdam, The Netherlands). The filters were stored at −20°C until analysis.
Fluorescent in situ hybridization.
The Bifidobacterium-specific probe Bif164 (26) and the general bacterial probe Eub338 (2) were purchased from (MWG, Ebersgerg, Germany) and were monolabeled at the 5′ end with Cy5 and Cy3, respectively. Hybridization was carried out as described previously (58).
DNA isolation.
Bacterial DNA from fecal samples and sorted cells was isolated using a QIAamp DNA stool mini kit (QIAGEN, Hilden, Germany), with some modifications. In short, 0.220 g of feces and the sorted filters were placed in different bead-beating tubes filled with 0.3 g of 0.1-mm zirconia/silica beads, and 1.4 ml of ASL solution from the stool mini kit was added. The tubes were then agitated for 3 min at 5,000 rpm in a mini bead beater (Biospec Products, Bartlesville, Okla.). The protocol was then continued from step 3 as described by the manufacturer. DNA samples were stored at −20°C in Tris-EDTA buffer.
PCR amplification.
All primers used in this study are listed in Table 1. DNA isolated from the total fecal community or a sorted fraction was used as a template to amplify the V6 to V8 regions of the 16S rRNA gene with primers 968-GC-f and 1401-r. PCRs were performed using a Taq DNA polymerase kit from Life Technologies (Gaithersburg, Md.). Each reaction mixture (final volume, 50 μl) consisted of 20 mM Tris-HCl (pH 8.4), 3 mM MgCl2, each deoxynucleoside triphosphate at a concentration of 0.2 mM, each primer at a concentration of 0.2 μM, 1.25 U of Taq polymerase, and 1 μl of appropriately diluted template DNA. Samples were amplified with a GenAmp PCR system 9700 (PE Applied Biosystems, Foster City, Calif.) using the following program: predenaturation at 94°C for 5 min; 35 cycles of denaturation 94°C for 30 s, annealing at 56°C for 20 s, and extension at 68°C for 40 s; and a final extension at 68°C for 7 min. The integrity of the nucleic acids was checked visually after electrophoresis on a 1.2% agarose gel containing ethidium bromide.
TABLE 1.
Primers used in the present study
| Primer | Sequence (5′-3′) | Specificity | Reference or source |
|---|---|---|---|
| 1401-r | CGGTGTGTACAAGACCC | Bacterial 16S rRNA | 36 |
| 968-f | AACGCGAAGAACCTTA | Bacterial 16S rRNA | 25 |
| Bact0011-f | AGAGTTTGAT(C/T) (A/C)TGGCTCAG | Bacterial 16S rRNA | 22 |
| Lab0159-f | GGAAACAG(A/G)TGCTAATACCG | Lactobacillus 16S rRNA | 18 |
| Lab0677-r | CACCGCTACACATGGAG | Lactobacillus 16S rRNA | 18 |
| Univ0515-r | ATCGTATTACCGCGGCTGCTGGCA | Universal | 25 |
| Lm26-f | GATTCTGGCTCAGGATGAACG | Bifidobacterium 16S rRNA | 23 |
| Lm3-r | CGGGTGCTICCCACTTTCATG | Bifidobacterium 16S rRNA | 23 |
| Bif164-f | GGGTGGTAATGCCGGATG | Bifidobacterium 16S rRNA | 24 |
| Bif662-r | CCACCGTTACACCGGGAA | Bifidobacterium 16S rRNA | 24 |
| GC-clamp | CGCCGGGGGCGCGCCCCGGGCGGGGCG GGGGCACGGGGGG | 33 | |
| T7 | TAATACGACTCACTATAGG | Sequencing | Promega |
| Sp6 | GATTTAGGTGACACTATAG | Sequencing | Promega |
To investigate the diversity of the Bifidobacterium group, a genus-specific nested PCR was performed using primers lm3 and lm26 (Table 1) to amplify most of the 16S rRNA gene. Following purification of the PCR products with a QIAgen kit (QIAGEN, Hilden, Germany), a second PCR was carried out with a 10-fold-diluted sample as the DNA template with 16S rRNA gene-targeted primers Bif164-f and Bif662-GC-r as described by Satokari et al. (42).
DGGE analysis of PCR results.
Amplicons were separated by DGGE based on the protocol of Muyzer et al. (33) using the Decode system (Bio-Rad Laboratories, Hercules, Calif.) with the following modifications. The polyacrylamide gels consisted of 8% (vol/vol) polyacrylamide (ratio of acrylamide to bisacrylamide, 37.5:1) and 0.5× Tris-acetate-EDTA buffer (pH 8.0). Denaturing acrylamide of 100% was defined as 7 M urea and 40% formamide. The polyacrylamide gels were prepared with denaturing gradients ranging from 38 to 48% and from 45 to 55% to separate the generated amplicons of the total bacterial and Bifidobacterium communities, respectively. The gels were poured from the top using a gradient maker and a pump (Econopump; Bio-Rad Laboratories, Hercules, Calif.) set at a rate of 4.5 ml/min. Prior to polymerization of the denaturing gel (gradient volume, 28 ml), a 7.5-ml stacking gel without denaturing chemicals was added, and the appropriate comb was subsequently inserted. Electrophoresis was performed first for 5 min at 200 V and then for 16 h at 85 V in 0.5× Tris-acetate-EDTA buffer (pH 8.0) at a constant temperature of 60°C. The gels were stained with AgNO3 and dried overnight at 60°C.
Cloning of the PCR-amplified products.
PCR amplicons generated with the sets of primers for the total community (968-GC-f and 1401-r) and the bifidobacterial community (Bif164-f and Bif662-GC-r) were purified with a Qiaquick PCR purification kit (QIAGEN, Hilden, Germany) used according to the manufacturer's instructions. PCR products were cloned into Escherichia coli JM109 using the Promega pGEM-T vector system (Promega, Madison, Wis.). Colonies of ampicillin-resistant transformants were transferred with a sterile toothpick to 50 μl of Tris-EDTA and were incubated at 95°C for 15 min to lyse the cells. PCRs were performed with cell lysates using pGEM-T-specific primers T7 and Sp6 (Table 1) to check the sizes of the inserts. The plasmids containing inserts that were the right size were used to screen the transformants with the V6-V8 primers (968f-GC-f and 1401r) and the Bifidobacterium-specific primers (Bif164-f and Bif662-GC-r). Clones used for the subsequent sequence analysis were selected according to the migration position of the clone PCR fragment in the DGGE gel compared to the fragments in the original DGGE profile. Insert PCR amplicons of selected transformants were purified with a QIAquick PCR purification kit (QIAGEN, Hilden, Germany) and were subjected to DNA sequence analysis (Westburg, Leusden, The Netherlands).
Sequence similarity was analyzed using the BLAST tool (1) at the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov/BLAST). Alignment and further phylogenetic analysis of the sequences were done using the ARB software package (29), and trees were constructed using the neighbor-joining method (41).
RESULTS
Optimization of fluorescent probes applied to fecal microbiota.
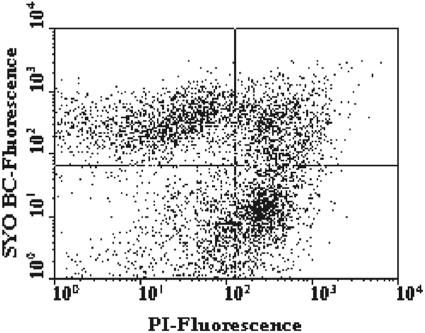
Three SYTO dyes (SYTO BC, SYTO 9, and SYTO 13) were used in combination with PI in order to optimize monitoring of the bacterial viability of the fecal microbiota of four healthy adults (adults A, B, C, and D). The best discrimination between live damaged and dead cells was obtained with the fluorescent probes SYTO BC and PI (data not shown). Figure 1 shows the cytograms of green fluorescence (FL1) versus red fluorescence (FL3) for a fecal sample stained with both SYTO BC and PI. Three cell subpopulations could be identified: SYTO BC-stained viable cells, SYTO BC- and PI-stained injured cells, and PI-stained dead cells. The signal in the lower left quadrant of the dot plot was due to the instrument's electronic noise and the background signal from nonfecal cells present in the sample. The total cell numbers ranged from 1.68 × 1011 to 4.64 × 1010 cells/g (wet weight) of feces for the four adults (Table 2). Cells having an intact membrane (live bacterial cells) accounted for 49.0% ± 8.2% of the total bacterial counts, while dead cells accounted for 31.7% ± 2.2% of the total cell counts for the four adults. The double-stained fraction representing the injured fecal cells showed large variation and accounted for 19.4% ± 8.7% of the total cell counts.
FIG. 1.
FCM analysis of a fecal sample stained with SYTO BC and PI. The two-color dot plot discriminated between SYTO BC-stained viable cells (upper left quadrant), double-stained injured cells (upper right quadrant), PI-stained dead cells (lower right quadrant), and STYO BC- and PI-stained injured cells. Results were obtained with the FACSCalibur.
TABLE 2.
Viability assessment of fecal microbiota using FCM and membrane integrity probes (PI and SYTO BC)
| Adult | Total counta | Eub338 countb | Viability count (%)c
|
||
|---|---|---|---|---|---|
| Viable cells | Dead cells | Injured cells | |||
| A | 1.68 × 1011 | 1.24 × 1011 | 42.9 | 31.3 | 25.8 |
| B | 6.06 × 1010 | 3.94 × 1010 | 51.7 | 28.8 | 19.5 |
| C | 6.88 × 1010 | 5.44 × 1010 | 59.4 | 33.5 | 7.1 |
| D | 4.64 × 1010 | 2.97 × 1010 | 41.8 | 33.2 | 25.0 |
| Mean (SD) | 8.60 × 1010 (5.55 × 1010) | 6.15 × 1010 (4.20 × 1010) | 49.0 ± 8.2 | 31.7 ± 2.2 | 19.4 ± 8.7 |
Number of fecal cells/gram (wet weight) of feces determined by SYTO BC staining.
Number of cells hybridized by the bacterial probe Eub338 labeled with Cy5.
Percentages of viable, dead, and injured cells as monitored by the combination of the fluorescent dyes PI and SYTO BC.
Fluorescence-activated cell sorting.
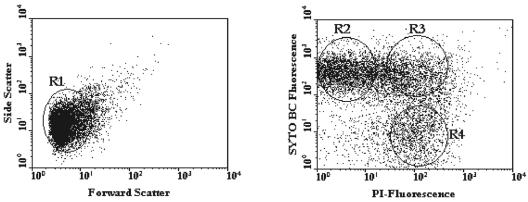
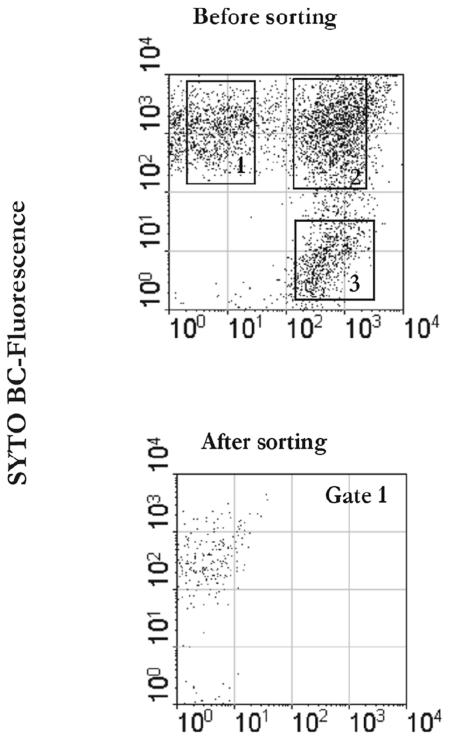
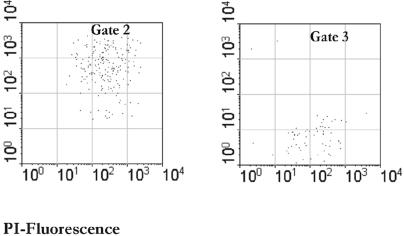
Cells were sorted based on both scatter parameters and fluorescence signals in FL1 and FL3 by gating the subpopulation of interest. This is illustrated by the analysis of the samples from adult A shown in Fig. 2. Approximately 1 × 106 to 5 × 106 cells were recovered from each subpopulation. For adult C, the injured cells represented only 7.1% of the total fecal cells and could not be recovered quantitatively during the sorting experiment. The major fractions in this sample were the live bacteria (59.4%) and dead bacteria (33.5%). The sorted fractions were also checked for purity by reanalyzing a 1-ml aliquot from each sorting tube with both the FACSVantage and the FACSCalibur (Fig. 3). Sorted cells were then concentrated on filters that were immediately rechecked by epifluorescence microscopy for purity based on their fluorescence signal and were subjected to DNA extraction in parallel with the nonsorted, fecal input samples.
FIG. 2.
FACSVantage dual-parameter dot plots of a fecal sample from adult A stained with SYTO BC and PI. A region (R1) was defined around the whole cell population in the dot plot on the left. In the dot plot on the right, regions were set around target populations as follows: SYTO BC-stained cells (R2), double-stained cells (R3), and PI-stained cells. A gate was defined whereby any particle present within R1 and R2 was sorted as a live cell, any particle present within R1 and R3 was sorted as an injured cell, and any particle present within R1 and R4 was sorted as a dead cell. Sorted cells were recovered in separate sterile tubes for a range of 1 × 106 to 5 × 106 cells.
FIG. 3.
Sorting purity of recovered fractions. A fecal sample was stained with PI and SYTO BC. Sorted cells were reanalyzed with the FACSClaibur as shown in the panels at the bottom.
Genetic diversity of dead, live, and injured fecal bacteria assessed by PCR-DGGE.
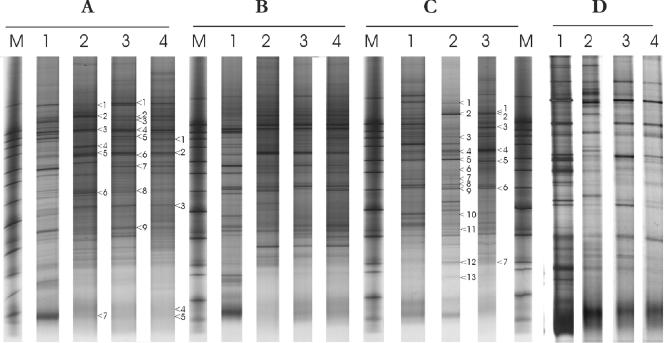
The PCR-DGGE banding patterns of the different amplicons obtained from the total fecal community (before sorting) and the sorted viable, dead, and injured cells revealed different profiles in terms of the number of bands, the position, and the intensity (Fig. 4). Each subpopulation produced a complex fingerprint that in all cases was less complex than that of the total fecal community. Variations in the profiles were observed between the adults, indicating that not only the total fecal community but also the distinct subpopulations are characteristic for each individual.
FIG. 4.
DGGE of PCR amplicons of the V6 to V8 regions of the 16S rRNA gene, obtained from a total fecal sample (lanes 1) and sorted viable (lanes 2), dead (lanes 3), and injured (lanes 4) subpopulations from adults A to D. Lanes M contained the marker. Bands that were sequenced are indicated by numbered arrowheads (see Fig. 7).
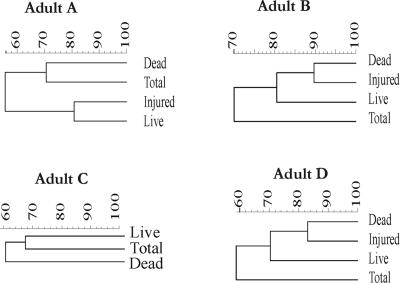
To assess changes in the genetic diversity of bacterial communities within the different sorted cell fractions, a similarity index (SI) was calculated based on the Pearson product-moment correlation coefficient. Subsequently, a cluster analysis was performed using the unweighted-pair group method using arithmetic averages, and dendrograms were generated (Fig. 5). The PCR-DGGE patterns for adults B and D revealed nearly identical profiles for the injured and dead cells (SIs, 89 and 83% for adults B and D, respectively). For the adult C sample, which contained only a small injured fraction that could not be recovered (see above), the SIs of the total bacteria compared with the live and dead communities were 77%, and 52%, respectively. For adult A, the SI for the total bacteria compared to live fecal bacteria was 55%, while the SI for the comparison between the total and dead communities was 71%. For each individual, a few bands were unique in each profile of the sorted viable and dead fractions. In contrast, only a very limited number of bands that were found in either the live, dead, or injured profiles were not detected at the community level.
FIG. 5.
Dendrograms based on the DGGE gel for each individual were generated by using the Pearson correlation index for each pair of lanes within a gel and were used as a measure of similarity between the community fingerprints. The clustering of patterns was calculated using the unweighted-pair group method using average linkages.
Analysis of the bifidobacteria in the sorted populations by specific PCR-DGGE.
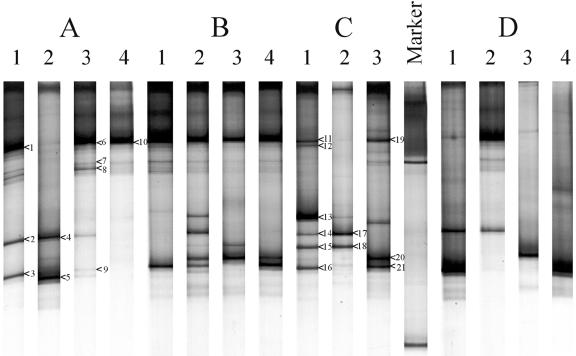
To determine whether the results for the total bacterial population matched the results for a specific bacterial group, we focused on the bifidobacterial populations in the different adults since they represented one of the major groups of the fecal microbiota. Fluorescent in situ hybridization with the Bif164 probe, which is specific for bifidobacteria, revealed that this group accounted for 10.10% ± 0.39%, 1.50% ± 0.09%, 2.84% ± 0.11%, and 3.93% ± 0.15% of the total fecal cells for adults A, B, C, and D, respectively. DGGE analysis of rRNA gene amplicons obtained with Bifidobacterium-specific primers revealed that the bifidobacterial community in the four individuals showed only limited diversity (Fig. 6). Interestingly, bifidobacterial amplicons were detected in the total fecal microbiota of the four adults, as well as in the different sorted fractions isolated from their feces. In general, the variations in the DGGE patterns of the Bifidobacterium-specific amplicons matched those observed with the total bacterial amplicons in the live, dead, and injured fecal microbiota of the four adults (Fig. 5).
FIG. 6.
DGGE profiles of bifidobacterial PCR products of fecal samples from the four healthy adults (A, B, C, and D). Lanes 1, total fecal bacteria before sorting; lanes 2, live fecal bacteria; lanes 3, dead fecal bacteria; lanes 4, injured cells. The dominant fragments indicated by numbered arrowheads were sequenced and compared to known sequences in the GenBank database, as shown in the phylogenetic tree in Fig. 8.
Phylogenetic analysis of dominant bands in the PCR-DGGE patterns.
Of the 40 sequenced clones retrieved from all sorted bacterial fractions of two individuals (Fig. 4), only 19 exhibited 95% or less 16S rRNA sequence identity with their nearest relatives. This indicates that the majority of the sequences were derived from new, undescribed bacterial phylotypes. Most cloned sequences analyzed could be assigned to the major phylogenetic lineages commonly encountered in human fecal clone libraries (Fig. 7) (45, 50). These included Clostridium coccoides (Clostridium cluster XIVa), Clostridium leptum (Clostridium cluster IV) (7), and the Bacteroides group. In general, it was Clostridium cluster XIVa that contained most of sequences (27 clones) derived from both clone libraries. Analysis of the positions of the clones (Fig. 7) revealed that the phylogenetic distribution of sequences from the clone library from adult A was remarkably different from the phylogenetic distribution of sequences from the clone library from adult C. Furthermore, the allocations in the phylogenetic tree of the cloned sequences retrieved from the live, dead, and injured fractions of both libraries (adults A and C) were also different. These observations were supported by the PCR-DGGE analysis, which revealed that the viable and dead communities produced different profiles (Fig. 4).
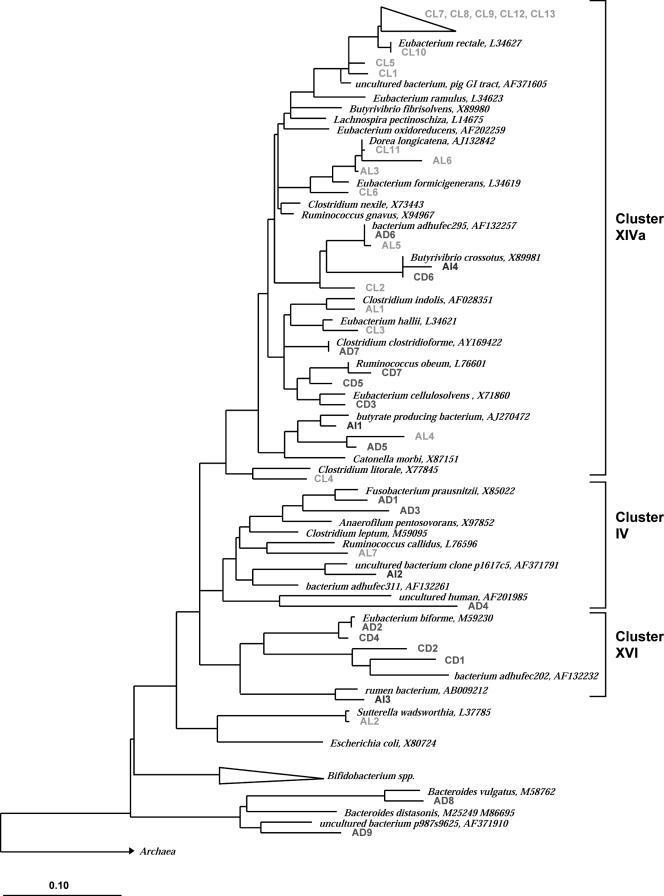
FIG. 7.
Phylogenetic tree of partial 16S rRNA sequences based on E. coli positions 968 to 1376 retrieved from total sorted viable (L), injured (I), and dead (D) fractions from fecal samples from adult A and adult C and full-length reference sequences. The alignment and phylogenetic analysis were performed with the ARB software (29), and the tree was constructed using the neighbor-joining method (41) based on alignment positions conserved in at least 50% of the sequences analyzed. Bar = 10% diversity.
For the clone library of adult C, six of the Clostridium cluster XIVa clones, designated CL7, CL8, CL9, CL10, CL12, and CL13, were affiliated with the Eubacterium rectale subgroup with sequence similarities ranging from 94 to 100%, and they were all recovered from the live fraction. Furthermore, two clones (CL5 and CL1) were retrieved from the live fraction, and they were affiliated with butyrate-producing bacterium L2-21 (accession no. AJ270477) (96% sequence similarity) and with uncultured bacterial clone HuCA2 (accession no. AJ408958) (94% sequence similarity), respectively. Thus, they may belong to novel species. Clones CL11 and CL6, retrieved from the live fraction, were related to Dorea longicatena (98% sequence similarity), a newly reclassified species, and to Eubacterium formicigenerans (95% sequence similarity), respectively (47). In addition, the sequences of clones CL3 and CL2 identified from the live fraction were most similar to the sequences of Eubacterium hallii (96% similarity) and butyrate-producing bacterium SM4/1 (accession no. AY305314) (96% similarity), respectively. However, all three clones (CD1, CD2, and CD4) that fell in the Eubacterium biforme group (Clostridium cluster XVI) were retrieved only from the dead fraction. An additional clone, designated CD6, was also identified in the dead fraction and exhibited a perfect match with Butyrivibrio crossotus. Ruminicoccus obeum-like species represented by clones CD5 and CD7 were also found in the dead fraction of the fecal sample from adult C. Finally, one clone (CD3) retrieved from the dead sorted fraction exhibited only 92% sequence similarity with Clostridium cellulosolvens. Noticeably, the Bacteroides group was not represented in the clone library for adult C, nor was the C. leptum subgroup.
The composition of the clone libraries of adult A differed from the composition of the clone libraries of adult C (Fig. 7), although Clostridium cluster XIVa contained most of the clones, as observed for adult C. However, the distributions of the clones within this cluster differed for the two individuals; for example, no clones from the clone library of adult A were identified as E. rectale. Two clones from the live fraction, AL3 and AL6, were closely related to E. formicigenerans (96% sequence similarity) and to butyrate-producing bacterium SS3/4 (accession no. AY305316) (95% sequence similarity), respectively. Five clones (AD1, AD3, AL7, AI2, and AD4) were located in the C. leptum subgroup (Clostridium cluster IV), in which Fusobacterium prausnitzii was the major fecal species (7). These clones were retrieved from the live, injured, and dead fecal fractions of adult A. Two clones (AL5 and AD6) were more than 98.7% similar to each other, were closely related (99% sequence similarity) to uncultured bacterium adhufec295 isolated by Suau et al. (45), and exhibited only 92 to 94% similarity to the closest cultured relative (Clostridium nexile). These two clones were found in both the dead and live fractions; furthermore, they had the same band position in the PCR-DGGE gel (Fig. 4A, lanes 2 and 3, band 1). Remarkably, the clones that clustered in the Bacteroides phylum were isolated only from the dead fraction; one clone (AD8) was affiliated with the Bacteroides vulgatus subgroup, and another clone (AD9) was affiliated with the Bacteroides distasonis subgroup. Finally, one clone that was closely related to the gram-negative bacterium Sutterella wadsworthensis (99% sequence similarity), which is known to be associated with intestinal infections, was recovered from the live fecal cells (51).
Phylogenetic analysis of the bifidobacterium community.
To identify the major bifidobacterial populations in the viable, dead, and injured populations of fecal microbiota of adults A and C, cloning of approximately 550-bp PCR products generated with primers Bif164-f and Bif662-GC-r was performed. Twenty-three clones that corresponded to the major bands in the PCR-DGGE patterns were sequenced (Fig. 6). Analysis of these sequences revealed that all of the clones retrieved were identified as Bifidobacterium species commonly found in the human GI tract (Fig. 8). Most clones (15 of 23) were located in the Bifidobacterium adolescentis group, five clones showed 96 to 99% sequence similarity to the Bifidobacterium longum-Bifidobacterium infantis group, two clones were identified as Bifidobacterium bifidum (95 to 99% sequence similarity), and one clone had high sequence similarity (99% sequence similarity) with Bifidobacterium pseudocatenulatum.
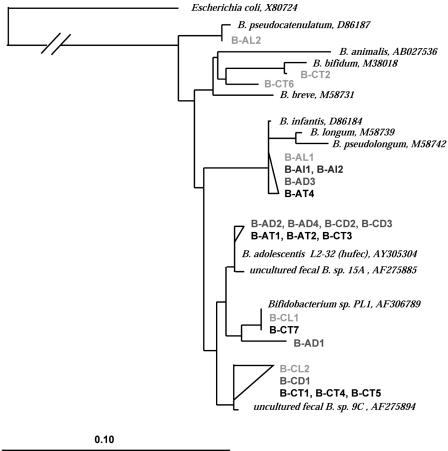
FIG. 8.
Phylogenetic tree of bifidobacterial sequences based on E. coli positions 164 to 694. The alignment and phylogenetic analysis were performed with the ARB software (29), and the tree was constructed using the neighbor-joining method (41) based on alignment positions conserved in at least 50% of the sequences analyzed. Bar = 10% diversity. The GenBank accession numbers of reference sequences are indicated. Letters in designations: A, adult A; C, adult C; T, total fecal cells; L, sorted live fraction; I, sorted injured cells; D, sorted dead cells.
Clones designated B-AT4, B-AL1, B-AD3, B-AI1, and B-AI2 were identified as members of the B. infantis-B. longum group (98 to 99% sequence similarity). These clones were obtained from the fecal sample from adult A and were detected in all cell fractions studied that contained total, live, dead, and injured subpopulations.
Cloned sequences closely related to B. adolescentis were identified from fecal samples from both subjects. Seven sequences (B-AT1, B-AT2, B-AD2, B-AD4, B-CT3, B-CD2, and B-CD3), although not identical, were more than 97% similar to each other and were 97 to 99% similar to a B. adolescentis human fecal isolate. These sequences were retrieved from the total and dead fecal cells of both adults, indicating that these bacteria were dead at the time of sampling. Other clones closely related to B. adolescentis (B-CT1, B-CT4, B-CT5, B-CL2, and B-CD1) were obtained only from adult C; these clones had high sequence similarity with an uncultured fecal organism, Bifidobacterium sp. strain 9C retrieved from a fecal human clone library (42). These clones were isolated from the live and dead fecal fractions of adult C and were not found in the samples from adult A. Two identical clones, designated B-CT7 and B-CL1, were retrieved from the total community, as well as from the live fecal bacteria of adult C. These sequences were affiliated with uncultured fecal Bifidobacterium sp. strain PL1 with 1% sequence divergence. However, the two latter clones did not migrate at the same position in the DGGE gel (Fig. 6C, bands 14 and 18, respectively). An additional clone (clone B-AD1) recovered from the dead fraction of adult A exhibited 98.82% similarity with clones B-CT7 and B-CL1; however, the corresponding amplicons showed a different migration behavior in the DGGE gel (Fig. 6, band 8A).
Clone B-AL2 corresponding to band 5 in the PCR-DGGE gel (Fig. 6) with 99% sequence similarity to B. pseudocatenulatum was identified in the live fraction of the fecal sample from adult A. Finally, two clones (clones B-CT2 and B-CT6) recovered from the total fecal microbiota of adult C were identified as B. bifidum (99 and 95% sequence similarity, respectively). These clones were recovered from the total fecal bacterial community of adult C and were not detected in any other sorted fractions.
DISCUSSION
Recent microbial GI tract research has focused on techniques such as the use of 16S rRNA gene-based molecular tools to examine community structure and biodiversity. However, it is essential to identify microbes based on their ecophysiological traits (i.e., to identify microbes that are functionally active versus those microbes that are effectively redundant and play little or no role at a particular time or at a given site in the intestinal tract). In this study, we developed a new approach in which flow cytometric in situ viability assessment using functional probes was coupled with molecule-based techniques to obtain insight into the genetic diversity of the total, active, and dead fecal cells.
The live/dead assay (SYTO BC/PI), which addresses membrane integrity, provided resolution of the fecal cell population into viable intact cells, injured cells, and dead cells. Membrane integrity is measured by the capacity of cells to exclude impermeant dyes, of which PI has been the most widely used (21, 43). These dyes do not enter an intact cell because they are insoluble in the hydrophobic membrane phase (44). However, the permenat SYTO dyes are able to enter both viable and dead cells, staining them green (16). In dead cells simultaneous staining with SYTO and PI activates the energy transfer phenomena (the fluorescent emission of SYTO is absorbed by PI and no longer visible, and hence the cells fluoresce red). In cells with a damaged membrane, the energy transfer is not complete due to the low concentration of PI; thus, these cells emit both green and red fluorescence (4). We previously demonstrated the efficiency of an FCM viability assay to monitor the physiological heterogeneity of bile salt-stressed B. adolescentis, a common member of the GI tract, and Bifidobacterium lactis, a widely used probiotic organism (5). Here we showed that for fecal samples, the viable fraction accounted for approximately one-half and the dead cells accounted for almost one-third of the total number of cells in the four volunteers. The remainder of the cells represented the injured cells, but the population size showed some variation among the individuals tested (19.4% ± 8.7%). Recently, in another study, fecal samples from 10 healthy adults were analyzed using PI and were reported to contain fractions of dead bacteria ranging from 17 to 34% (3).
The three different populations identified by the PI/SYTO BC assay were sorted, and the 16S rRNA gene was amplified and subsequently analyzed by PCR-DGGE. The PCR-DGGE banding pattern of each individual revealed remarkable differences between the distinct physiological fractions of the fecal microbiota. The variations between the profiles indicate that not only the total fecal community but also the distinct subpopulations are characteristic for each individual. These observations are in agreement with previous studies in which it was demonstrated that the GI tract microbiota is host specific (26, 46, 48, 57). The SIs for comparisons between the total community and the sorted live, dead, and injured populations were lower than the average SI reported for a stable microbiota of healthy adults over time (56). However, for two adults (adults B and D) the banding patterns of the dead and injured fractions were quite similar (SI > 80%). The presence of common bands in the dead and injured fractions suggests that some populations were physiologically heterogeneous. The death or injury of certain fecal bacteria may be due to a variety of factors, including depletion of the readily metabolized carbohydrates, low availability of free water, a high ammonia concentration as digesta moves from the proximal colon toward the distal colon, and sensitivity to antimicrobial products produced by bacteria or the host (8, 31). Consequently, some bacterial populations may tolerate these challenging conditions in the distal colon better than others. Interestingly, some bands were detected both in the total community and in the fraction of metabolically active cells, but they were not identified in the dead fraction. These observations suggest that these phylotypes were viable at the time and the site of sampling. Inversely, some bands were found only in the dead sorted fraction, indicating that the cells were dead at the time and site of sampling, but they might have been active in the upper part of the colon (31).
The 40 sequenced clones obtained from dominant bands in the DGGE profiles of the sorted viable, dead, and injured populations were assigned to three major phylogenetic lineages, namely, the Bacteroides, C. coccoides, and C. leptum lineages; the two latter lineages correspond to Clostridium rRNA clusters XIVa and IV, respectively (7). This is in agreement with previous studies in which comprehensive fecal clone libraries were obtained from healthy adult fecal samples to obtain insight into the diversity of the GI tract microbial ecosystem (17, 45, 50, 54, 57). The results of these studies showed that a majority (>50%) of the observed diversity was attributable to unknown dominant microorganisms in the human gut. In our study of the 40 sequenced clones retrieved from all sorted bacterial fractions of both individuals, 19 exhibited 95% or less 16S rRNA gene sequence identity with their nearest relatives.
The phylogenetic distribution of sequences from the adult A clone library differed in terms of prevalence and species diversity from the phylogenetic distribution of sequences from the adult C clone library, as well as the allocation in the phylogenetic tree of the cloned sequences retrieved from the live, dead, and injured fractions (Fig. 8). The phylogenetic analysis of the predominant bacteria obtained from the two adult fecal samples showed that some bacterial groups, such as the E. rectale subgroup, E. formicigemerans, D. longicatena, and E. hallii, were metabolically active at the time of sampling. This indicates that these bacteria have an important function at the end of the gastrointestinal tract. Some members of the Eubacteria are known to have a fermentative metabolism with butyrate as a major end product (37). In contrast, phylotypes belonging to the Bacteroides group and the Ruminococcus obeum- and E. biforme-like bacteria were obviously dead at the time and site of sampling, suggesting that these bacteria may have been active in the upper part of the GI tract. Bacteroides spp. are known to be one of the predominant bacterial groups in the gut; they are highly saccharolytic, and they may also play an important role in the metabolism of bile salts and mucin degradation (55). Macfarlane et al. (30) showed that during growth of fecal bacteria under carbon-limiting conditions in an in vitro model mimicking the distal colon, the number of viable Bacteroides declined significantly, especially with a long retention time in the fermentor. Hence, it is possible that these conditions are indeed met in the terminal colon and notably reduce the viability of Bacteroides spp. Furthermore, some fecal populations identified in the clone library of adult A were composed of viable and nonviable bacteria; this was noticeable for the C. leptum cluster. The differences observed between the two adults in terms of genetic diversity of the total, live, and dead fecal bacterial populations might be attributable not only to the host-genotype effect but also to other factors, such as diet and host health.
The PCR-DGGE and phylogenetic analyses of bifidobacterial communities were in agreement with previous studies which showed that adult fecal bifidobacterial populations are host specific at the strain level (42). Most of the bifidobacterial phylotypes detected in all samples and sorted fractions were identified as B. adolescentis, B. longum, B. infantis, B. pseudocatenulatum, and B. bifidum. Matsuki et al. (32) frequently detected B. longum, B. adolescentis, and B. catenulatum by direct species- and group-specific PCR in fecal samples, while B. infantis and B. breve were detected less frequently in adult human feces (32). Furthermore, analysis of the live, injured, and dead bifidobacterial populations by FCM in combination with PCR-DGGE, cloning, and sequencing revealed a striking physiological heterogeneity within these populations. Indeed, the cloned sequences that belonged to the B. longum-B. infantis group were retrieved from all sorted fractions (viable, injured, and dead), whereas B. adolescentis-related phylotypes were retrieved mostly from the dead fractions. Although bifidobacteria are able to grow in different culture media, certain species, such as B. adolescentis, are difficult to recover from fecal samples by culturing methods, while they could be detected by culture-independent methods (3, 35). This may be explained simply by the fact that a number of B. adolescentis-related species are dead or injured at the time that they are recovered from the feces and hence cannot grow in culture media.
The combination of FCM cell sorting and subsequent 16S rRNA gene analysis that we used here for the first time for the fecal microbiota provides relevant ecological information related to the diversity and activity of the fecal microbiota in situ and highlights the physiological heterogeneity of this complex ecosystem. FCM and cell sorting unquestionably offer novel applications to study the microbial ecophysiology of the GI tract.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R. I., B. Binder, R. Olson, S. W. Chisholm, R. Devereux, and D. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Apajalahti, J. H. A., A. Kettunen, P. H. Nurminen, H. Jatila, and W. E. Holben. 2003. Selective plating underestimates abundance and shows differential recovery of bifidobacterial species from human feces. Appl. Environ. Microbiol. 69:5731-5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Bäckhed, F., R. E. Ley, J. L. Sonnenburg, D. A. Peterson, and J. I. Gordon. 2005. Host-bacterial mutualism in the human intestine. Science 307:1915-1920. [DOI] [PubMed] [Google Scholar]
- 4.Barbesti, S., S. Citterio, M. Labra, M. D. Baroni, M. G. Neri, and S. Sgorbati. 2000. Two- and three-color fluorescence flow cytometric analysis of immunoidentified viable bacteria. Cytometry 40:214-218. [PubMed] [Google Scholar]
- 5.Ben-Amor, K., P. Breeuwer, P. Verbaarschot, F. M. Rombouts, A. D. L. Akkermans, W. M. De Vos, and T. Abee. 2002. Multiparametric flow cytometry and cell sorting for the assessment of viable, injured, and dead Bifidobacterium cells during bile salt stress. Appl. Environ. Microbiol. 68:5209-5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernard, L., C. Courties, C. Duperray, H. Schafer, G. Muyzer, and P. Lebaron. 2001. A new approach to determine the genetic diversity of viable and active bacteria in aquatic ecosystems. Cytometry 43:314-321. [PubMed] [Google Scholar]
- 7.Collins, M. D., P. A. Lawson, A. Willems, J. J. Cordoba, J. Fernandez-Garayzabal, P. Garcia, J. Cai, H. Hippe, and J. A. Farrow. 1994. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int. J. Syst. Bacteriol. 44:812-826. [DOI] [PubMed] [Google Scholar]
- 8.Cummings, J. 1995. The large intestine in nutrition and disease, p. 155. Danone chair monograph, Bruxelles, Belgium.
- 9.Davey, H., and D. Kell. 1996. Flow cytometry and cell sorting of heterogeneous microbial populations: the importance of single-cell analyses. Microbiol. Rev. 60:641-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davey, H., and M. Winson. 2003. Using flow cytometry to quantify microbial heterogeneity. Curr. Issues Mol. Biol. 5:9-15. [PubMed] [Google Scholar]
- 11.Falk, P. G., L. V. Hooper, T. Midtvedt, and J. I. Gordon. 1998. Creating and maintaining the gastrointestinal ecosystem: what we know and need to know from gnotobiology. Microbiol. Mol. Biol. Rev. 62:1157-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Favier, C. F., E. E. Vaughan, W. M. De Vos, and A. D. L. Akkermans. 2002. Molecular monitoring of succession of bacterial communities in human neonates. Appl. Environ. Microbiol. 68:219-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuchs, B. M., M. V. Zubkov, K. Sahm, P. H. Burkill, and R. Amann. 2000. Changes in community composition during dilution cultures of marine bacterioplankton as assessed by flow cytometric and molecular biological techniques. Environ. Microbiol. 2:191-201. [DOI] [PubMed] [Google Scholar]
- 14.Harmsen, H. J., A. C. Wildeboer-Veloo, G. C. Raangs, A. A. Wagendorp, N. Klijn, J. G. Bindels, and G. W. Welling. 2000. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J. Pediatr. Gastroenterol. Nutr. 30:61-67. [DOI] [PubMed] [Google Scholar]
- 15.Harmsen, H. J. M., G. C. Raangs, T. He, J. E. Degener, and G. W. Welling. 2002. Extensive set of 16S rRNA-based probes for detection of bacteria in human feces. Appl. Environ. Microbiol. 68:2982-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haugland, R. 2002. Molecular probes. Handbook of fluorescent probes and research products, 9th ed. [Online.] Molecular Probes, Inc., Eugene, Oreg. http://www.probes.com.
- 17.Hayashi, H., M. Sakamoto, and Y. Benno. 2002. Phylogenetic analysis of the human gut microbiota using 16S rDNA clone libraries and strictly anaerobic culture-based methods. Microbiol. Immunol. 46:535-548. [DOI] [PubMed] [Google Scholar]
- 18.Heilig, H. G. H. J., E. G. Zoetendal, E. E. Vaughan, P. Marteau, A. D. L. Akkermans, and W. M. de Vos. 2002. Molecular diversity of Lactobacillus spp. and other lactic acid bacteria in the human intestine as determined by specific amplification of 16S ribosomal DNA. Appl. Environ. Microbiol. 68:114-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hooper, L. V., T. Midtvedt, and J. I. Gordon. 2002. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu. Rev. Nutr. 22:283-307. [DOI] [PubMed] [Google Scholar]
- 20.Hooper, L. V., M. H. Wong, A. Thelin, L. Hansson, P. G. Falk, and J. I. Gordon. 2001. Molecular analysis of commensal host-microbial relationships in the intestine. Science 291:881-884. [DOI] [PubMed] [Google Scholar]
- 21.Joux, F., and P. Lebaron. 2000. Use of fluorescent probes to assess physiological functions of bacteria at single-cell level. Microb. Infect. 2:1523-1535. [DOI] [PubMed] [Google Scholar]
- 22.Kane, M., L. K. Poulsen, and D. Stahl. 1993. Monitoring the enrichment and isolation of sulfate reducing bacteria by using oligonucleotide hybridization probes designed from environmentally derived 16S rRNA sequences. Appl. Environ. Microbiol. 59:682-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaufmann, P., A. Pfefferkorn, M. Teuber, and L. Meile. 1997. Identification and quantification of Bifidobacterium species isolated from food with genus-specific 16S rRNA-targeted probes by colony hybridization and PCR. Appl. Environ. Microbiol. 63:1268-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kok, R., A. de Waal, F. Schut, G. Welling, G. Weenk, and K. Hellingwerf. 1996. Specific detection and analysis of a probiotic Bifidobacterium strain in infant feces. Appl. Environ. Microbiol. 62:3668-3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Chichester, United Kingdom.
- 26.Langendijk, P., F. Schut, G. Jansen, G. Raangs, G. Kamphuis, M. Wilkinson, and G. Welling. 1995. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl. Environ. Microbiol. 61:3069-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lay, C., L. Rigottier-Gois, K. Holmstrom, M. Rajilic, E. E. Vaughan, M. D. Collins, P. Thiel, P. Namsolleck, M. Blaut, and J. Dore. 2004. Assessment of human faecal microbiota composition using FISH combined with flow cytometry, pan-European comparison, p. 75. In PROEUHELATH: The Food, GI-tract Functionality and Human Health Cluster 3rd Workshop. VTT-Biotechnology, Melia Sitges, Spain.
- 28.Lee, N., P. H. Nielsen, K. H. Andreasen, S. Juretschko, J. L. Nielsen, K.-H. Schleifer, and M. Wagner. 1999. Combination of fluorescent in situ hybridization and microautoradiography—a new tool for structure-function analyses in microbial ecology. Appl. Environ. Microbiol. 65:1289-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Y. Adhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic. Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Macfarlane, S., M. E. Quigley, M. J. Hopkins, D. F. Newton, and G. T. Macfarlane. 1998. Polysaccharide degradation by human intestinal bacteria during growth under multi-substrate limiting conditions in a three-stage continuous culture system. FEMS Microbiol. Ecol. 26:231-243. [Google Scholar]
- 31.Marteau, P., P. Pochart, J. Dore, C. Bera-Maillet, A. Bernalier, and G. Corthier. 2001. Comparative study of bacterial groups within the human cecal and fecal microbiota. Appl. Environ. Microbiol. 67:4939-4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsuki, T., K. Watanabe, J. Fujimoto, Y. Kado, T. Takada, K. Matsumoto, and R. Tanaka. 2004. Quantitative PCR with 16S rRNA gene-targeted species-specific primers for analysis of human intestinal bifidobacteria. Appl. Environ. Microbiol. 70:167-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muyzer, G., E. de Waal, and A. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nebe-von-Caron, G., P. J. Stephens, C. J. Hewitt, J. R. Powell, and R. A. Badley. 2000. Analysis of bacterial function by multi-colour fluorescence flow cytometry and single cell sorting. J. Microbiol. Methods 42:97-114. [DOI] [PubMed] [Google Scholar]
- 35.Nielsen, D. S., P. L. Moller, V. Rosenfeldt, A. Paerregaard, K. F. Michaelsen, and M. Jakobsen. 2003. Case study of the distribution of mucosa-associated Bifidobacterium species, Lactobacillus species, and other lactic acid bacteria in the human colon. Appl. Environ. Microbiol. 69:7545-7548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nubel, U., B. Engelen, A. Felske, J. Snaidr, A. Wieshuber, R. Amann, W. Ludwig, and H. Backhaus. 1996. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J. Bacteriol. 178:5636-5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pryde, S. E., S. H. Duncan, G. L. Hold, C. S. Stewart, and H. J. Flint. 2002. The microbiology of butyrate formation in the human colon. FEMS Microbiol. Lett. 217:133-139. [DOI] [PubMed] [Google Scholar]
- 38.Rachmilewitz, D., K. Katakura, F. Karmeli, T. Hayashi, C. Reinus, B. Rudensky, S. Akira, K. Takeda, J. Lee, K. Takabayashi, and E. Raz. 2004. Toll-like receptor 9 signaling mediates the anti-inflammatory effects of probiotics in murine experimental colitis. Gastroenterology 126:520-528. [DOI] [PubMed] [Google Scholar]
- 39.Radajewski, S., P. Ineson, N. R. Parekh, and J. C. Murrell. 2000. Stable-isotope probing as a tool in microbial ecology. Nature 403:646-649. [DOI] [PubMed] [Google Scholar]
- 40.Radajewski, S., I. R. McDonald, and J. C. Murrell. 2003. Stable-isotope probing of nucleic acids: a window to the function of uncultured microorganisms. Curr. Opin. Biotechnol. 14:296-302. [DOI] [PubMed] [Google Scholar]
- 41.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 42.Satokari, R. M., E. E. Vaughan, A. D. L. Akkermans, M. Saarela, and W. M. de Vos. 2001. Bifidobacterial diversity in human feces detected by genus-specific PCR and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 67:504-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shapiro, H. M. 2000. Microbial analysis at the single-cell level: tasks and techniques. J. Microbiol. Methods 42:3-16. [DOI] [PubMed] [Google Scholar]
- 44.Shapiro, H. M. 1995. Practical flow cytometry, 3d ed. Wiley-Liss Inc., New York, N.Y.
- 45.Suau, A., R. Bonnet, M. Sutren, J.-J. Godon, G. R. Gibson, M. D. Collins, and J. Dore. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tannock, G. W., K. Munro, H. J. M. Harmsen, G. W. Welling, J. Smart, and P. K. Gopal. 2000. Analysis of the fecal microflora of human subjects consuming a probiotic product containing Lactobacillus rhamnosus DR20. Appl. Environ. Microbiol. 66:2578-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taras, D., R. Simmering, M. D. Collins, P. A. Lawson, and M. Blaut. 2002. Reclassification of Eubacterium formicigenerans Holdeman and Moore 1974 as Dorea formicigenerans gen. nov., comb. nov., and description of Dorea longicatena sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 52:423-428. [DOI] [PubMed] [Google Scholar]
- 48.Vaughan, E. E., H. G. H. J. Heilig, E. G. Zoetendal, R. Satokari, J. K. Collins, A. D. L. Akkermans, and W. M. de Vos. 1999. Molecular approaches to study probiotic bacteria. Trends Food Sci. Technol. 10:400-404. [Google Scholar]
- 49.Wallner, G., B. Fuchs, S. Spring, W. Beisker, and R. Amann. 1997. Flow sorting of microorganisms for molecular analysis. Appl. Environ. Microbiol. 63:4223-4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, X., S. P. Heazlewood, D. O. Krause, and T. H. Florin. 2003. Molecular characterization of the microbial species that colonize human ileal and colonic mucosa by using 16S rDNA sequence analysis. J. Appl. Microbiol. 95:508-520. [DOI] [PubMed] [Google Scholar]
- 51.Wexler, H. M., D. Reeves, P. H. Summanen, E. Molitoris, M. McTeague, J. Duncan, K. H. Wilson, and S. M. Finegold. 1996. Sutterella wadsworthensis gen. nov., sp. nov., bile-resistant microaerophilic Campylobacter gracilis-like clinical isolates. Int. J. Syst. Bacteriol. 46:252-258. [DOI] [PubMed] [Google Scholar]
- 52.Whiteley, A. S., R. I. Griffiths, and M. J. Bailey. 2003. Analysis of the microbial functional diversity within water-stressed soil communities by flow cytometric analysis and CTC+ cell sorting. J. Microbiol. Methods 54:257-267. [DOI] [PubMed] [Google Scholar]
- 53.Reference deleted.
- 54.Wilson, K. H., and R. B. Blitchington. 1996. Human colonic biota studied by ribosomal DNA sequence analysis. Appl. Environ. Microbiol. 62:2273-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu, J., H. C. Chiang, M. K. Bjursell, and J. I. Gordon. 2004. Message from a human gut symbiont: sensitivity is a prerequisite for sharing. Trends Microbiol. 12:21-28. [DOI] [PubMed] [Google Scholar]
- 56.Zoetendal, E. G., A. D. L. Akkermans, W. M. Akkermans-van Vliet, J. A. G. M. de Visser, and W. M. de Vos. 2001. The host genotype affects the bacterial community in the human gastronintestinal tract. Microb. Ecol. Health Dis. 13:129-134. [Google Scholar]
- 57.Zoetendal, E. G., A. D. L. Akkermans, and W. M. De Vos. 1998. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl. Environ. Microbiol. 64:3854-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zoetendal, E. G., K. Ben-Amor, H. J. M. Harmsen, F. Schut, A. D. L. Akkermans, and W. M. de Vos. 2002. Quantification of uncultured Ruminococcus obeum-like bacteria in human fecal samples by fluorescent in situ hybridization and flow cytometry using 16S rRNA-targeted probes. Appl. Environ. Microbiol. 68:4225-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]