Abstract
The enzymatic degradation of l-methionine and the subsequent formation of volatile sulfur compounds (VSCs) are essential for the development of the typical flavor in cheese. In the yeast Yarrowia lipolytica, the degradation of l-methionine was accompanied by the formation of the transamination product 4-methylthio-2-oxobutyric acid. A branched-chain aminotransferase gene (YlBCA1) of Y. lipolytica was amplified, and the l-methionine-degrading activity and the aminotransferase activity were measured in a genetically modified strain and compared to those of the parental strain. Our work shows that l-methionine degradation via transamination is involved in formation of VSCs in Y. lipolytica.
Volatile sulfur compounds (VSCs) are present in many foods (26), and it is estimated that VSCs represent about 10% of the volatile components detected in food and beverages (8). These compounds are commonly found in dairy products, including yogurt (21, 27) and ripened cheeses (13, 19). Their low odor thresholds make important contributions to the odor and aroma of cheeses and may interact with the organoleptic properties of cheeses (7).
It is generally believed that in cheese VSCs are formed exclusively at the late stage of ripening by surface bacteria, the most common of which is Brevibacterium linens (1, 14). However, since they can grow in acidic environment, yeasts develop during the early stage of ripening and may also contribute in a direct way to the formation of VSCs (2). Yarrowia lipolytica, Geotrichum candidum, Kluyveromyces lactis, and Debaryomyces hansenii are the yeasts most frequently isolated from soft cheeses, yet the mechanisms resulting in VSC formation in these yeasts are still controversial.
VSCs arise primarily from the degradation of l-methionine to methanethiol (MTL) in the cheese ecosystem, and they are subsequently converted to other sulfur-bearing compounds, including the MTL oxidation products dimethyl disulfide (DMDS) and dimethyl trisulfide (DMTS) (12). The yeast G. candidum is the most-studied cheese-ripening yeast for production of VSCs (6, 9). This organism has been shown to degrade l-methionine to MTL via a two-step degradation pathway (9) that is probably initiated by an aminotransferase. Aminotransferase, also called transaminase, is a pyridoxal 5′-phosphate (PLP)-dependent enzyme which, in the presence of an amino acceptor (e.g., α-ketoglutarate), catalyzes the formation of the transamination product 4-methylthio-2-oxobutyric acid (KMBA), which is subsequently converted to MTL. Due to the absence of the genome sequence and transformation tools for G. candidum, functional analysis of any gene encoding aminotransferase in this microorganism is not possible. Furthermore, there is no direct evidence for the involvement of any aminotransferase in l-methionine catabolism in yeasts, while such involvement has been reported for branched-chain aminotransferases (BCA) for several bacteria, including Lactobacillus paracasei (32), Lactococcus lactis (17, 28), and Staphylococcus carnosus (24).
Nevertheless, the involvement of aminotransferase in l-methionine catabolism is also suspected in Y. lipolytica (2, 30). The recent availability of the genome sequence of Y. lipolytica (Génolevures: Genomic Exploration of the Hemiascomycete Yeasts [http://cbi.labri.fr/Genolevures/index.php]) enabled us to initiate a functional analysis of a gene encoding a branched-chain aminotransferase.
Due to technological interest for transformation of milk products and due to the generally strong enzymatic potential, our objectives were to investigate food-grade Y. lipolytica strains for production of VSCs and to investigate l-methionine aminotransferase activity that we suspected to be involved in synthesis of VSCs in this yeast. First, we tested the abilities of different Y. lipolytica strains to degrade l-methionine by measuring (i) the formation of the intermediate KMBA and (ii) the production of VSCs. Second, a putative BCA gene was overexpressed in Y. lipolytica, and the consequences for aminotransferase activity and production of VSCs were investigated.
MATERIALS AND METHODS
Microorganisms.
Twelve strains of Y. lipolytica were used in this study. Seven strains (strains 634, 718, 721, 791, 879, 880, and 881) were obtained from Collection de Levures d'Intérêt Biotechnologique (UMR-MGM, INRA, Thiverval-Grignon, France). Three strains (strains 89, 90, and 91), originally isolated from French cheeses, were obtained from the UMR-GMPA laboratory collection. Strains W29 and 136463 were obtained from the laboratory collection of UMR-MGM. Strain W29 (Mat A) is a wild haploid isolate obtained from sewage. Strain 136463 is a laboratory strain (leu2-35 ura3-302 his3-1) (20). All yeast strains were stored in 5% glycerol-nonfat dry milk at −80°C until they were used. Escherichia coli DH5α was used for plasmid preparation.
Culture conditions for strain comparison.
A preculture of each strain was grown in a 500-ml Erlenmeyer flask containing 100 ml of medium adjusted to pH 5 ± 0.1. The medium was composed of 15 g liter−1 Casamino Acids (Difco, Detroit, MI), 38 ml liter−1 of a 60% sodium lactate stock solution (Prolabo, Fontenay-Sous-Bois, France), 6 g liter−1 yeast extract (Labosi, Oulchy-le-Château, France), 0.1 g liter−1 calcium chloride (Prolabo), 0.5 g liter−1 MgSO4 · 7H2O (Prolabo), 6.8 g liter−1 KH2PO4 (Prolabo), 20 g liter−1 sodium chloride (Prolabo), 20 g liter−1 lactose (Prolabo), and 6 g liter−1 galactose (Sigma-Aldrich, St. Quentin Fallavier, France) and was inoculated with a Y. lipolytica stock suspension (1%, vol/vol). The preculture was agitated (150 rpm) for 48 to 72 h at 25°C. It was used to inoculate (1%, vol/vol) a culture medium (to obtain final concentration of 105 CFU ml−1) having the composition described above supplemented with 1 g liter−1 l-methionine (Sigma-Aldrich) prior to inoculation. Cultures were agitated (150 rpm) at 25°C and harvested after 72 h, which corresponded to the time of optimum production of VSCs. This culture was used for high-performance liquid chromatography (HPLC) analyses of KMBA and l-methionine.
For comparison of strains, the production of VSCs by each Y. lipolytica strain was determined with a model liquid cheese medium (65% cheese curd and 35% water) prepared as previously described (4). The cheese medium was inoculated to obtain a final concentration of 105 CFU g of cheese medium−1.
Culture conditions and media for mutant and parental strain comparison.
The mutant and the parental strains were grown in a potato dextrose broth (PDB) (Difco) recommended for culture of yeasts and molds. Precultures were agitated (150 rpm) 48 to 72 h at 25°C and used to inoculate (1%, vol/vol) PDB culture medium supplemented with 1 g liter−1 l-methionine (Sigma-Aldrich). The cultures were agitated (150 rpm) at 25°C and harvested after 72 h, which corresponded to the time of optimum production of VSCs. These cultures were used for HPLC analyses of KMBA and l-methionine, for analyses of VSCs, and for measurement of enzymatic activities.
Viable cell counts.
Viable cell counts were expressed in CFU per milliliter and were determined by using a standard aerobic plate count procedure with yeast extract glucose chloramphenicol agar (Biokar Diagnostics, Paris, France). Surface inoculation was carried out with a spiral plater (Interscience, St. Nom La Bretèche, France) and 90-mm-diameter petri dishes. The dishes were incubated at 25°C, and colonies were counted after 72 h.
Analysis of volatile sulfur compounds.
Five milliliters of culture broth was analyzed using a headspace analyzer (HP 7695A purge and trap concentrator; Hewlett-Packard, Palo Alto, CA) coupled to a gas chromatograph (HP 6890; Hewlett-Packard) and a mass spectrophotometer detector (HP 6890A quadruple mass spectrometer; Hewlett-Packard), as previously described (25). Volatile compounds were identified by mass spectrometry. The amounts of VSCs possibly formed spontaneously in noninoculated control cultures were subtracted from the total amounts of VSCs produced by inoculated cultures. The amount of each VSC is expressed below as the surface area of the corresponding chromatogram peak, taking into account the dilution used.
HPLC analyses of l-methionine and KMBA.
The culture supernatants were filtered using a polyethersulfone membrane filter (pore size, 0.2 μm; diameter, 25 mm). l-Methionine was quantified by performing HPLC analyses with a Waters column (Symmetry C18 3.5 μm; diameter, 4.6 mm; length, 100 mm; Waters, Saint Quentin-en-Yvelines, France). The operating conditions were as follows: flow rate, 0.6 ml min−1; 20°C; and detection at 210 nm. The mobile phases were H2O and acetonitrile. The gradient was 100% H2O for 2.5 min, 100% to 90% H2O for 0.5 min, 90% to 60% H2O for 7 min, and 60% to 100% H2O for 4 min. l-Methionine (Sigma-Aldrich) was used as the standard. KMBA was quantified by HPLC analyses with a Waters column (IC-Pak; diameter, 7.8 mm; length, 300 mm; Waters). The eluent was 0.1% H3PO4. The flow rate was 1 ml min−1 at 35°C, and detection was at 214 nm. KMBA (Sigma-Aldrich) was used as the standard.
Preparation of cell extracts for enzyme assays.
The fungal biomass was harvested and then ground essentially as previously described (2). The cell extract (CFE) was collected for enzyme assays. The total protein content of the CFE was determined by the method described previously (11) using bovine serum albumin (Sigma-Aldrich) as a standard.
l-Methionine transaminase activity and thiol-producing activity.
Transaminase activity was assayed by measuring the formation of glutamate as a product of the transamination of the substrate l-methionine in the presence of an amino group acceptor, α-ketoglutarate, as described previously (2). The aminotransferase activity was expressed as the amount of glutamate formed in nanomoles per gram of protein per second.
Thiol-producing activity (TPA) was determined as previously described (18, 2) using l-methionine or KMBA as the substrate. Specific activity was expressed in nanomoles of MTL formed from l-methionine or KMBA per second per gram of protein.
DNA techniques.
All DNA manipulations were carried out using standards methods (29). Chromosomal DNA of a Y. lipolytica strain was prepared as previously described (23). The E. coli strain used for plasmid preparation was grown in Luria-Bertani medium containing 10 g liter−1 Bacto tryptone (Difco), 5 g liter−1 Bacto yeast extract (Difco), and 5 g liter−1 NaCl adjusted to pH 7.5 at 37°C with aeration. When needed, ampicillin (50 μg ml−1) was added to the culture medium. All restriction enzymes were used according to the conditions for their use described by their manufacturers (Gibco-BRL, Cergy Pontoise, France, and Biolabs, Saint-Quentin-en-Yvelines, France). The oligonucleotides were synthesized by Eurogentec (Seraing, Belgium). Integrative transformation of Y. lipolytica was performed by the lithium acetate procedure, and clones were selected on YNB (0.17% [wt/wt] yeast nitrogen base, 0.5% [wt/wt] ammonium sulfate, 1% [wt/wt] glucose) (33).
PCR, cloning, and sequencing.
PCR amplification was performed with a Crocodile III thermocycler (Appligene, Illkirch, France) using Ready-to-Go PCR beads (Pharmacia Biotech, Uppsala, Sweden) and the following cycling parameters. DNA denaturation was performed at 95°C for 1 min, and this was followed by annealing at 58°C for 1 min and amplification at 72°C for 2 min. This cycle was performed 30 times before a final amplification at 72°C for 10 min.
A YlBCA1 fragment was amplified by PCR using two oligonucleotides, oligonucleotide 1 (the forward primer) (5′-AAGGAAAAAGCGGCCGCGGATTAGCAGTCCACGGTGG) and oligonucleotide 2 (the reverse primer) (5′-GCGGGCCGCATGGCCAAAATGACCAAACTGTCACCCC). The 1-kb PCR fragment obtained with these oligonucleotides contained the whole BCA1 open reading frame flanked by SfiI and NotI restriction sites. Lambda DNA digested with BstEII was used as the molecular size standard.
The PCR products were extensively restricted by SfiI and NotI. The cleaved products were ligated to an SfiI-NotI digest of pBLCM, a derivative of pBUCM (31) carrying the LEU2 marker instead of the URA3 marker. The transformants were selected for ampicillin resistance and verified by restriction site mapping. Two independent clones were conserved. The plasmids were linearized by AscI to target the terminator of the XPR2 gene (encoding the alkaline extracellular protease [EC3.4.21.14]) of Y. lipolytica in a DNA region that is highly transcribed. The linearized plasmids were used to transform strain 136463. The transformed clones were selected for Leu+ prototrophy. The clones were screened for the presence of correct integration. To do this, we performed PCR with colonies. Mapping with oligonucleotides SJ13 (GGCCTGTCTAGAATCTCTC) and DS102 (TCCCGAAAACGTTCTTCGGGGCG) in the XPR2 locus allowed amplification of a 1.3-kb band characteristic of a right insertion. Strains displaying such bands by colony PCR were retained for further analysis.
Data analyses.
Data were analyzed using the Statgraphics Plus software. The values presented below are means of three replicates. A one-way analysis of variance was performed. Fisher's least-significant difference procedure was applied (α ≤ 0.05) to the individual variables in order to compare means and to assess the significance of differences.
Nucleotide sequence accession number.
The nucleotide sequence of YlBCA1 reported in this paper has been deposited in the DDBJ, EMBL, and GenBank nucleotide sequence databases under accession number XP_502278.
RESULTS
Biosynthesis of KMBA, l-methionine degradation, and production of VSCs by several Y. lipolytica strains.
The abilities of 12 Y. lipolytica strains to degrade l-methionine and to produce KMBA, the l-methionine transamination product, were evaluated. Depending on the strain, 27 to 66% of the initial amount of l-methionine was consumed. Conversely, KMBA was produced by all strains (Table 1), which suggests that a transminase activity is involved in l-methionine catabolism. Furthermore, except for strains 91, W29, and 721, the level of KMBA-specific production remained in the range from 1.2 × 10−8 to 1.6 × 10−8 μmol CFU−1 (Table 1) for all strains. Yields for bioconversion of l-methionine to KMBA ranging from 40 to 43% (strains 91, 634, 880, and 721) to 100% (strain 136463) were obtained. However, the strains which consumed the most l-methionine were not necessarily the strains exhibiting the highest bioconversion yields. For instance, with strain 136463 only 27% of the l-methionine was totally transformed to KMBA, while with strain 634 66% of the l-methionine added initially was converted to KMBA with a 40% bioconversion yield.
TABLE 1.
Production of KMBA, l-methionine consumption, and production of VSCs by 12 strains of Y. lipolytica
| Strain | KMBA production (μmol CFU−1, 108) | l-Methionine consumption (μmol CFU−1, 108) | Yield (%)a | Production of DMDS + DMTS (surface area CFU−1) |
|---|---|---|---|---|
| 136463 | 1.31 ± 0.19 bcb | 1.31 ± 0.06 (27)c h | 100 | 18.80 ± 2.69 a |
| W29 | 1.61 ± 0.15 a | 2.52 ± 0.08 (49) d | 64 | 13.23 ± 0.05 b |
| 881 | 1.17 ± 0.05 c | 2.90 ± 0.09 (60) b | 40 | 1.02 ± 0.01 c |
| 91 | 0.58 ± 0.13 c | 1.40 ± 0.07 (30) h | 41 | 0.81 ± 0.01 c |
| 89 | 1.21 ± 0.01 bc | 1.88 ± 0.03 (38) g | 64 | 0.80 ± 0.00 c |
| 634 | 1.25 ± 0.17 bc | 3.14 ± 0.05 (66) a | 40 | 0.77 ± 0.01 c |
| 880 | 1.28 ± 0.02 bc | 2.97 ± 0.04 (57) b | 43 | 0.65 ± 0.01 d |
| 90 | 1.26 ± 0.03 bc | 2.25 ± 0.09 (47) c | 56 | 0.57 ± 0.00 d |
| 879 | 1.26 ± 0.02 bc | 2.77 ± 0.02 (54) c | 45 | 0.52 ± 0.04 d |
| 718 | 1.17 ± 0.02 c | 2.43 ± 0.03 (49) d | 48 | 0.52 ± 0.00 d |
| 721 | 0.90 ± 0.01 d | 2.30 ± 0.04 (48) e | 39 | 0.34 ± 0.00 d |
| 791 | 1.37 ± 0.01 b | 2.02 ± 0.06 (41) f | 68 | 0.14 ± 0.00 d |
The yield is the number of moles of KMBA formed per mole of l-methionine consumed, expressed as a percentage.
Within a column different single letters indicate that values are significantly different. Two letters indicate that a value is between the values for two groups.
The values in parentheses indicate the amount of l-methionine consumed expressed as a percentage of the amount of l-methionine initially added (6.7 mM).
The production of VSCs was measured for the 12 strains of Y. lipolytica cultivated on a liquid cheese model medium. The major VSCs produced, representing more than 98% of the total VSC production, were DMDS and DMTS (Table 1). For VSC production, strain 136463 proved to be the most efficient of the 12 Y. lipolytica strains. In addition to DMDS and DMTS, five strains of Y. lipolytica (strains W29, 881, 89, 880, and 90) were also able to produce 3-(methylthio)-propanal, also called methional, which represented no more than 2% of the total VSC production. The production of methional by Y. lipolytica is likely a result of the enzymatic decarboxylation of KMBA.
Y. lipolytica 136463 is a laboratory strain which was genetically manipulated and contains selectable genetic markers. Furthermore, due to its efficiency for production of VSCs, we selected this strain to study amplification of a BCA gene in Y. lipolytica.
Identification and genetic characterization of branched-chain aminotransferase genes of Y. lipolytica.
From the results described above, it seemed likely that Y. lipolytica strains produce an aminotransferase possibly involved in l-methionine catabolism. We therefore searched for genes encoding aminotransferases in the full genome of Y. lipolytica (Génolevures: Genomic Exploration of the Hemiascomycete Yeasts [http://cbi.labri.fr/Genolevures/index.php]). Based on other studies, we decided to focus on genes encoding BCA. Several bacterial BCAs have been reported to degrade l-methionine and to play a major role in cheese flavor development (e.g., synthesis of VSCs) as a result of l-methionine degradation (28, 34). However, l-methionine has never been tested as a substrate for any yeast aminotransferase, although several BCAs have been characterized in several yeasts (15, 16, 22); however, their substrate specificities were evaluated only with branched-chain amino acids (22). Two genes encoding BCAs were identified in the Y. lipolytica genome on the basis of sequence homology with the known mitochondrial and cytoplasmic BCA genes (mitochondrial BAT1, cytoplasmic BAT2) of Saccharomyces cerevisiae and Schizosaccharomyces pombe, which were studied and characterized previously (15, 16, 22).
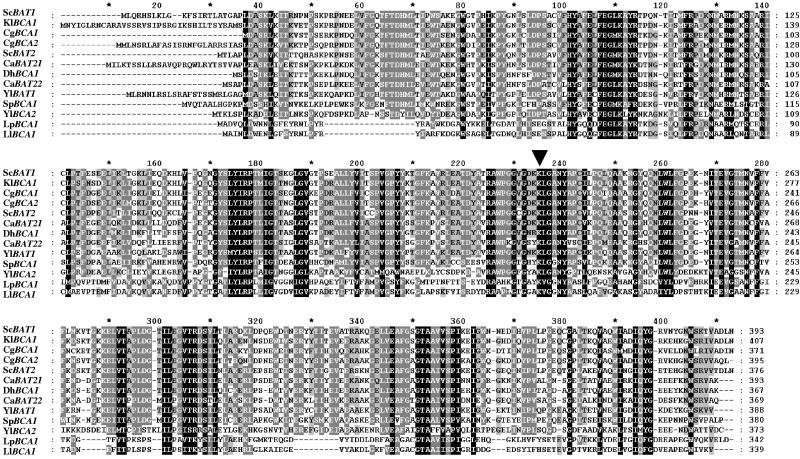
An amino acid sequence alignment of the products of these genes with those of several other organisms is presented in Fig. 1. This alignment was constructed using the ClustalX multiple-sequence alignment program. Residues that are identical or similar in all of these sequences are indicated. Both Y. lipolytica BCA genes exhibited a pyridoxal phosphate attachment site of Cys/Met metabolism enzymes at positions 225 to 240, indicating that the genes really encode a PLP-dependent enzyme. The Lys235 residue (Fig. 1) has been shown to be essential for the formation of the Schiff base intermediate with pyridoxal phosphate (22).
FIG. 1.
Alignment of yeast branched-chain amino acid aminotransferases. The alignment was constructed using the Clustal X v1.8 software. The N-terminal extensions observed in several aminotransferases, such as the products ofYlBCA1 and ScBAT1, correspond to mitochondrial presequences. The following sequences were used: Candida albicans CaBAT21 (GenBank accession no. EAL02229) and CaBAT22 (GenBank accession no. EAK95653), Candida glabrata CgBCA1 (GenBank accession no. XP_446368) and CgBCA2 (GenBank accession no. XP_449352), Debaryomyces hansenii DhBCA1 (GenBank accession no. CAG86908), Lactococcus lactis LlBCA1 (GenBank accession no. NP_267444), Lactobacillus plantarum LpBCA1 (GenBank accession no. NP_785849), Kluyveromyces lactis KlBCA1 (GenBank accession no. XP_451451), Saccharomyces cerevisiae ScBAT1 (Swiss-Prot accession no. P38891), ScBAT2 (Swiss-Prot accession no. P47176), Schizosaccharomyces pombe SpBCA1 (GenBank accession no. NP_595180), and Yarrowia lipolytica YlBCA1 (GenBank accession no. XP_502278) and YlBCA2 (GenBank accession no. XP_505642). The arrowhead indicates the lysine residue of the active site of PLP-dependent enzymes.
The mitochondrial localization can be deduced from the presence of an N-terminal extension enriched in serine, threonine, and polar amino acids. Some species, such as Saccharomyces or Yarrowia species, have two BCA genes, one with a mitochondrial targeting signal and one which is cytoplasmic. In other species, there is only one BCA, and it has either cytoplasmic features (as in D. hansenii) or mitochondrial features (as in K. lactis). From these observations, it cannot been concluded that there is preferential compartmentation for BCA. Surprisingly, the branched-chain amino acid aminotransferases from L. lactis and Lactobacillus plantarum aligned fairly well with the other BCAs from yeasts; this could be indicative of horizontal transfer. This observation was especially obvious on the phylogenic tree (data not shown) in which BCAs of lactic acid bacteria were closely related to the other BCAs and in which ScBAT1 and KlBCA1 did not seem to belong to this group.
The analysis of the Y. lipolytica DNA sequence revealed an open reading frame that encodes a 388-amino-acid protein for the mitochondrial gene (YlBCA1) and a 373-amino-acid protein for the cytosolic gene (YlBCA2). Furthermore, mitochondrial BCA has been reported to be a prominent isoenzyme in S. cerevisiae during the growth phase (16). We therefore decided to focus our functional analysis studies on the YlBCA1 gene encoding a mitochondrial BCA in Y. lipolytica.
Effect of amplification of the YlBCA1 gene in Y. lipolytica on conversion of l-methionine to VSCs and catabolic activities.
In the cells, there are several amino acid aminotransferases with overlapping specificities for l-methionine. A gene inactivation strategy would have been cumbersome and most probably would have led to viability defects. An amplification strategy was therefore chosen. If amplification of the activity results in an increase in the VSC yield, it can be concluded that this activity is involved in the pathway leading to the biosynthesis VSCs. This could occur even if there are concurrent activities at this step. Moreover, overexpression of this activity should not have a detrimental effect on the growth of the cells.
The YlBCA1 gene was cloned by PCR and placed under the control of the strong promoter hp4d, and then it was reintroduced by integrative transformation into parental strain 136463 to obtain the BCA1 transformant, which was studied further. In order to assess the role of YlBCA1 in l-methionine catabolism, we compared the aminotransferase activitiy and l-methionine degradation activity of the BCA1 transformant with those of the parental strain cultivated in PDB enriched in l-methionine. A 62% increase in KMBA biosynthesis, which is consistent with an increase in aminotransferase activity, was observed for the BCA1 transformant compared to the parental strain (Table 2). The production of VSCs was also assessed for both strains cultivated in the culture medium described above. DMDS and DMTS accounted for more than 98% of the total VSCs detected by gas chromatography-mass spectrometry. The BCA1 transformant produced 55% more VSCs than the parental strain produced (Table 2). While MTL is expected to be the major reaction product of l-methionine catabolism together with KMBA, DMDS and DMTS are by far the VSCs that are produced most. This is due to the fact that MTL is a highly reactive sulfur compound that quickly reacts with itself, forming the oxidized and more stable compounds DMDS and DMTS (12).
TABLE 2.
Production of KMBA, l-methionine consumption, growth, production of VSCs, and l-methionine transaminase activity in the parental strain and in the BCA1 transformant
| Strain | KMBA production (μmol CFU−1, 108) | l-Methionine consumption (μmol CFU−1, 108) | Growth (CFU ml−1, 10−7) | Production of DMDS + DMTS (surface area CFU−1) | l-Methionine transaminase activity (nmol g of protein−1 s−1) | Thiol-producing activity (nmol g of protein−1 s−1)
|
|
|---|---|---|---|---|---|---|---|
| From l-methionine | From KMBA | ||||||
| 136463 | 1.68 ± 0.25 aa | 2.45 ± 0.07 a | 7.21 ± 0.32 b | 12.47 ± 2.13 a | 53.99 ± 4.04 a | 12 ± 3 a | 24 ± 4 a |
| BCA1 transformant | 2.72 ± 0.14 b | 4.42 ± 0.12 b | 5.84 ± 0.16 a | 19.37 ± 2.37 b | 80.06 ± 5.24 b | 29 ± 4 b | 18 ± 2 a |
Within a column different letters indicate that values are significantly different.
However, TPAs were also found in CFE of both the parental strain and the BCA1 transformant. The data showed that the level of TPA was 2.5-fold greater in the transformant strain than in the parental strain when l-methionine was the substrate, while the level of TPA remained unchanged with the substrate KMBA (Table 2).
DISCUSSION
Y. lipolytica is one of the most extensively studied nonconventional yeasts. Its ecological niche includes lipid-rich foods, like cheese, as well as sewage plants. Owing to their efficiency at producing aroma compounds, strains of Y. lipolytica have been used for the preparation of cheese flavor compounds (5, 10) and can also contribute to production of VSCs from l-methionine (30).
In this work, the gene encoding a BCA, YlBCA1, was identified in the yeast Y. lipolytica. Due to its importance in microbiology, aminotransferase activity has been studied in detail in numerous microorganisms, including yeasts and bacteria. In S. cerevisiae, two isoenzyme forms of BCAs have been identified (16, 22). In this microorganism, two genes encode these isoenzymes; one gene encodes the mitochondrial enzyme (ScBAT1), and another gene encodes the cytosolic enzyme (ScBAT2). It was found that ScBAT1 encodes a 393-amino-acid protein with an NH2-terminal extension that directs the protein to the mitochondrial matrix. A highly homologous 376-amino-acid protein, ScBAT2, was found in the cytosol of S. cerevisiae (16, 22). In S. pombe, only one gene encoding a BCA has been identified (15). This gene encodes a 381-amino-acid protein with a calculated molecular mass of 42,521 Da. S. pombe BCA exhibits 47 to 52% identity with both BCAs isolated from S. cerevisiae. In Y. lipolytica, two open reading frames, one that encodes a 388-amino-acid protein (YlBCA1) and one that encodes a 373-amino-acid protein (YlBCA2), were identified. The calculated molecular masses of these proteins were 42,584 Da and 40,914 Da, respectively, which is in good agreement with the molecular masses of other yeast BCAs (41 to 43 kDa) (15, 22). Furthermore, it was anticipated that the BCA1 gene encodes a mitochondrial enzyme as it displayed a mitochondrial targeting presequence, but we have no direct evidence for the localization of the BCA1 gene product. It is expected that amplification of BCA1 would not modify the localization of the protein as the mitochondrial import machinery has a very high transport capacity.
The amino acid sequence derived from the DNA sequence of YlBCA1 had no significant identity with cystathionine-γ-lyase (CYS3) or cystathionine-γ-synthase (MET5) of S. cerevisiae, two other PLP-dependent enzymes possibly involved in l-methionine catabolism. Moreover, there was no significant homology between the YlBCAI gene and the MGL gene, a prokaryotic gene encoding an l-methionine-γ-lyase catalyzing the one-step degradation of l-methionine to MTL, that has recently been identified in B. linens (1). In contrast, the YlBCA1 sequence exhibited 53% and 63% homology with S. pombe SpBCA1 and S. cerevisiae ScBAT1, respectively, and 35% identity with the branched-chain aminotransferase of L. lactis which was shown to be active on l-methionine (34). All this shows that YlBCA1 truly encodes a branched-chain aminotransferase. Indeed, overexpression of the YlBCA1 gene significantly increased l-methionine transamination (e.g., aminotransferase activity and KMBA synthesis), as well as the production of VSCs in Y. lipolytica. Furthermore, the fact that the level of TPA with l-methionine was significantly greater in the BCA1 transformant than in the parental strain, while the level of TPA with KMBA remained unchanged, strongly suggests that l-methionine transamination is a limiting step in conversion of l-methionine to MTL, whereas conversion of KMBA to MTL is not limiting. Transamination is therefore a key step in l-methionine catabolism in Y. lipolytica.
Branched-chain amino acid aminotransferases have been studied in S. cerevisiae with respect to catabolism of branched-chain amino acids (e.g., leucine, isoleucine, and valine) (22). However, l-methionine has never been studied as a possible substrate for BCAs from S. cerevisiae or any other yeast. In the present study, we first studied the l-methionine-degrading activities of Y. lipolytica and demonstrated that a branched-chain aminotransferase could be involved in this process. This is of considerable interest since aminotransferase involvement in production of VSCs by Y. lipolytica was, until now, rather speculative. The fact that yeasts could also develop in the early stage of cheese ripening, in contrast to the cheese-ripening surface bacteria, is also important. Better knowledge of aminotransferase regulation mechanisms could therefore help in promoting biosynthesis of VSCs earlier during the ripening process, as well as biotransformations of other food products. Further studies on regulation of aminotransferase in Y. lipolytica could be of interest. For instance, induction of l-methionine aminotransferase activity by the carbon source which has been reported in the yeast G. candidum (3) could be studied at the level of gene expression in Y. lipolytica.
Furthermore, although the l-methionine transamination pathway is highly active in Y. lipolytica, the subsequent degradation of KMBA to MTL or to other intermediate sulfur compounds still remains to be elucidated, although it is probably not a limiting step for conversion of l-methionine to MTL. One possibility is the demethiolation of KMBA to MTL and α-ketobutyrate. A KMBA-demethiolating activity has already been measured in Y. lipolytica (2), and the enzyme(s) responsible for this activity could be searched for in this microorganism. Another possible KMBA degradation pathway is the decarboxylation to methional. Since this compound was found in several of the yeasts tested, we suspect that this pathway is active in Y. lipolytica, and KMBA-decarboxylating activities could be investigated.
Acknowledgments
Daniela Cernat Bondar is grateful to the European Community for a 12-month Ph.D. scholarship (YETI program grant QLK3-CT-2000-60055).
Jérôme Delettre is gratefully acknowledged for excellent technical assistance.
REFERENCES
- 1.Amarita, F., M. Yvon, M. Nardi, E. Chambellon, J. Delettre, and P. Bonnarme. 2004. Identification and functional analysis of the gene encoding methionine-γ-lyase in Brevibacterium linens. Appl. Environ. Microbiol. 70:7348-7354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arfi, K., H. E. Spinnler, R. Tâche, and P. Bonnarme. 2002. Production of volatile compounds by cheese-ripening yeasts: requirement for a methanethiol donor for S-methyl thioacetate synthesis by Kluyveromyces lactis, Appl. Microbiol. Biotechnol. 58:503-510. [DOI] [PubMed] [Google Scholar]
- 3.Arfi, K., R. Tâche, H. E. Spinnler, and P. Bonnarme. 2003. Dual influence of the carbon source and l-methionine on the synthesis of sulphur compounds in the cheese-ripening yeast Geotrichum candidum. Appl. Microbiol. Biotechnol. 61:359-365. [DOI] [PubMed] [Google Scholar]
- 4.Arfi, K., M. N. Leclercq-Perlat, A. Baucher, R. Tâche, J. Delettre, and P. Bonnarme. 2004. Contribution of several cheese-ripening microbial associations to aroma compound production. Lait 84:435-447. [Google Scholar]
- 5.Banoo, Z. L. February. 1967. Process for the preparation of cheese flavour compositions, Great Britain patent 1057170.
- 6.Berger, C., J. A. Khan, P. Molimard, N. Martin, and H. E. Spinnler. 1999. Production of sulfur flavors by ten strains of Geotrichum candidum. Appl. Environ. Microbiol. 65:5510-5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger, C., N. Martin, S. Collin, L. Gijs, J. A. Khan, G. Piraprez, H. E. Spinnler, and E. N. Vulfson. 1999. Combinatorial approach to flavor analysis. II. Olfactory investigation of a library of S-methylthioesters and sensory evaluation of selected components. J. Agric. Food. Chem. 47:3247-3279. [DOI] [PubMed] [Google Scholar]
- 8.Boelans, M. H., and L. J. van Gemert. 1993. Sensory properties of optical isomers. Perfum. Flavor. 18:2-16. [Google Scholar]
- 9.Bonnarme, P., K. Arfi, C. Dury, S. Helinck, M. Yvon, and H. E. Spinnler. 2001. Sulfur compound production by Geotrichum candidum from l-methionine: importance of the transmination step. FEMS Microbiol. Lett. 205:247-252. [DOI] [PubMed] [Google Scholar]
- 10.Boudreaux, D. P. June. 1987. Cheese-flavored substance and method of producing same. U.S. patent 4675193.
- 11.Bradford, M. M. 1976. A rapid and sensitive method for quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:54-63. [DOI] [PubMed] [Google Scholar]
- 12.Chin, H. W., and R. C. Lindsay. 1994. Ascorbate and transition-metal mediation of methanethiol oxidation to dimethyl disulfide and dimethyl trisulfide. Food Chem. 49:387-392. [Google Scholar]
- 13.Cuer, A., G. Dauphin, A. Kergomard, J. P. Dumont, and J. Adda. 1979. Flavour properties of some sulfur compounds isolated from cheeses. Lebensm.-Wiss. Technol. 12:258-261. [Google Scholar]
- 14.Dias, B., and B. Weimer. 1998. Purification and characterisation of l-methionine γ-lyase from Brevibacterium linens BL2. Appl. Environ. Microbiol. 64:3327-3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eden, A., and N. Benvenisty. 1998. Characterization of a branched-chain amino-acid aminotransferase from Schizosaccharomyces pombe. Yeast 14:189-194. [DOI] [PubMed] [Google Scholar]
- 16.Eden, A., G. Simchen, and N. Benvenisty. 1996. Two yeast homologs of ECA39, a target for c-Myc regulation, code for cytosolic and mitochondrial branched-chain amino acid aminotransferases. J. Biol. Chem. 271:20242-20245. [DOI] [PubMed] [Google Scholar]
- 17.Engels, W. J. M., A. C. Alting, M. M. T. G. Arntz, H. Gruppen, A. G. J. Voragen, G. Smit, and S. Wisser. 2000. Partial purification and characterization of two aminotransferases of Lactococcus lactis subsp. cremoris B78 involved in the catabolism of methionine and branched-chain amino acids. Int. Dairy J. 10:443-452. [Google Scholar]
- 18.Ferchichi, M., D. Hemme, M. Nardi, and N. Pamboukdjian. 1985. Production of methanethiol from methionine by Brevibacterium linens CNRZ 918. J. Gen. Microbiol. 131:715-723. [DOI] [PubMed] [Google Scholar]
- 19.Grill, H., S. Patton, and J. F. Cone. 1966. Aroma significance of sulfur compounds in surface-ripening cheese. J. Dairy Sci. 49:409-412. [Google Scholar]
- 20.He, F., J. M. Beckerich, and C. Gaillardin. 1992. A mutant of 7SL RNA in Yarrowia lipolytica affecting the synthesis of a secreted protein. J. Biol. Chem. 276:1932-1937. [PubMed] [Google Scholar]
- 21.Imhof, R., and J. O. Bosset. 1994. Quantitative GC-SM analysis of volatile flavour compounds in pasteurized milk and fermented milk products applying a standard addition method. Lebensm.-Wiss. Technol. 27:241-248. [Google Scholar]
- 22.Kispal, G., H. Steiner, D. A. Courts, B. Rolinski, and R. Lill. 1996. Mitochondrial and cytosolic branched-chain amino acid transaminases from yeast, homologs of the myc oncogene-regulated Eca39 protein. J. Biol. Chem. 271:24458-24464. [DOI] [PubMed] [Google Scholar]
- 23.Le Dall, M. T., J. M. Nicaud, and C. Gaillardin. 1994. Multiple-copy integration in the yeast Yarrowia lipolytica. Curr. Genet. 227:680-685. [DOI] [PubMed] [Google Scholar]
- 24.Madsen, S. M., H. C. Beck, P. Ravn, A. Vrang, A. M. Hansen, and H. Israelsen. 2002. Cloning and inactivation of a branched-chain amino acid aminotransferase gene from Staphylococcus carnosus and characterization of the enzyme. Appl. Environ. Microbiol. 68:4007-4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin, N., C. Berger, C. LeDu, and H. E. Spinnler. 2001. Aroma compound production in cheese curd by coculturing with selected yeast and bacteria. J. Dairy Sci. 84:2125-2135. [DOI] [PubMed] [Google Scholar]
- 26.Mussinan, C., and M. E. Keelan. 1994. Sulfur compounds in foods: an overview, p. 1-6. In C. J. Mussinan and M. E. Keelan (ed.), Sulfur compounds in foods. American Chemical Society, Washington, D.C.
- 27.Ott, A., L. B. Fay, and A. Chaintreau. 1997. Determination and origin of the aroma impact compounds of yogurt flavor. J. Agric. Food Chem. 45:850-858. [Google Scholar]
- 28.Rijnen, L., M. Yvon, R. van Kranenburg, P. Courtin, A. Verheul, E. Chambellon, and G. Smit. 2003. Lactococcal aminotransferases AraT and BcaT are key enzymes for the formation of aroma compounds from amino acids in cheese. Int. Dairy J. 13:805-812. [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 30.Spinnler, H. E., C. Berger, C. Lapadatescu, and P. Bonnarme. 2001. Production of sulfur compounds by several yeasts of technological interest for cheese ripening, Int. Dairy J. 11:242-252. [Google Scholar]
- 31.Swennen, D., M. F. Paul, L. Vernis, J. M. Beckerich, A. Fournier, and C. Gaillardin. 2002. Secretion of active anti-Ras single-chain Fv antibody by the yeasts Yarrowia lipolytica and Kluyveromyces lactis. Microbiology 148:41-50. [DOI] [PubMed] [Google Scholar]
- 32.Thage, B. V., F. P. Rattray, M. W. Laustsen, Y. Ardo, V. Barkholt, and U. Houlberg. 2004. Purification and characterization of a branched-chain amino acid aminotransferase from Lactobacillus paracasei subsp. paracasei CHCC 2115. J. Appl. Microbiol. 96:593-602. [DOI] [PubMed] [Google Scholar]
- 33.Wang, H. J., M. T. Le Dall, Y. Wach, C. Laroche, J. M. Velin, C. Gaillardin, and J. M. Nicaud. 1999. Evaluation of acyl coenzyme A oxidase (Aox) isoenzyme function in the n-alkane-assimilating yeast Yarrowia lipolytica. J. Bacteriol. 181:5140-5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yvon, M., E. Chambellon, A. Bolotin, and F. Roudot-Algaron. 2000. Characterization and role of the branched-chain aminotransferase (BcaT) isolated from Lactococcus lactis subsp. cremoris NCDO 763. Appl. Environ. Microbiol. 66:571-577. [DOI] [PMC free article] [PubMed] [Google Scholar]