Abstract
The cells and tissues of many aphids contain bacteria known as “secondary symbionts,” which under specific environmental circumstances may be beneficial to the host insect. Such symbiotic bacteria are traditionally described as intractable to cultivation in vitro. Here we show that two types of aphid secondary symbionts, known informally as T type and U type, can be cultured and maintained in three insect cell lines. The identities of the cultured bacteria were confirmed by PCR with sequencing of 16S rRNA gene fragments and fluorescence in situ hybridization. In cell lines infected with bacteria derived from aphids harboring both T type and U type, the U type persisted, while the T type was lost. We suggest that the two bacteria persist in aphids because competition between them is limited by differences in tropism for insect tissues or cell types. The culture of these bacteria in insect cell lines provides a new and unique research opportunity, offering a source of unibacterial material for genomic studies and a model system to investigate the interactions between animal cells and bacteria. We propose the provisional taxon names “Candidatus Consessoris aphidicola” for T type and “Candidatus Adiaceo aphidicola” for U type.
Symbiotic bacteria in insects are generally regarded as nonpathogenic and include both mutualists (i.e., bacteria advantageous to the insect) and commensals (bacteria that have no apparent significance to the insect). Many symbiotic bacteria are intracellular with tropisms for specific insect cells, and some persist over generations by transovarial vertical transmission (involving the transfer of bacteria to the unfertilized eggs in the maternal ovaries) (2). Due to the intimacy of the relationship, such symbiotic bacteria are not generally amenable to axenic cultivation, which constrains research progress.
Axenic culture may be an unrealistic goal for some insect-symbiotic bacteria which probably require a complex cellular environment that cannot be easily replicated in vitro. Cultivation in animal cells of a defined genotype and differentiated state can offer a valuable alternative to axenic cultivation and allow indefinite maintenance of a single microbial taxon under broadly uniform conditions. Several intracellular bacteria previously considered to be unculturable (e.g., Sodalis, Arsonophonus, and Wolbachia) have now been maintained successfully in insect cell lines (18, 25, 36).
The specific purpose of this study was to culture bacteria that are known generically as the “secondary symbionts” of one group of insects, the aphids. These bacteria are the focus of considerable research interest as they may modulate ecologically important traits of aphids, including plant utilization characteristics (5, 12, 17, 33, 41), natural enemy resistance (12, 27, 29), and thermal tolerance (5). They are identified by their 16S rRNA gene sequences and are described by an informal nomenclature; some examples are R-type (or pea aphid secondary symbiont), T type (or pea aphid Bemisia symbiont), and U type (or pea aphid U-type symbiont) in the γ-proteobacteria and S-type (or pea aphid Rickettsia) in the α-proteobacteria (4, 8, 27). The secondary symbionts are phylogenetically and functionally distinct from the primary symbionts (the γ-proteobacterium Buchnera in most aphids). While Buchnera is obligately vertically transmitted and is required by the insect for sustained growth and reproduction (10), the prevalence of secondary symbionts in aphids is variable and these organisms are transmitted both vertically and horizontally (4, 9, 27).
In this study we focused on two secondary symbionts, T type and U type. Our specific goal was to obtain persistent cultures of these bacteria from three aphid species, principally in an established mosquito cell line. To detect the bacteria in insect cell culture, we used both PCR-dependent diagnostic assays and PCR-independent in situ hybridization analysis.
MATERIALS AND METHODS
Insect material.
The following three sources of secondary symbionts were used as inocula: (i) black bean aphids, Aphis fabae Scop. line CRT01/43, containing the secondary symbiont T type; (ii) pea aphids, Acyrthosiphon pisum Harris line JF 99/14, containing U type; and (iii) rose aphids, Macrosiphum rosae L. line ACD03/01, containing both T- and U type. All aphids were wingless summer parthenogenetic female morphs. Aphids were surface sterilized in 70% ethanol, rinsed in sterile water, air dried on sterile blotting paper, and then subjected to UV radiation (CL-1000 UV cross-linker; UVP, Cambridge, United Kingdom) for 10 min. Individual insects were homogenized with a hand-held glass homogenizer with 100 μl Mitsuhashi and Maramorosch insect medium (MMI) (22) supplemented with 20% fetal bovine serum (FBS) and centrifuged at 100 × g for 2 min. Ten microliters of the supernatant was applied to a 90% confluent monolayer of Aedes albopictus C6/36 cells (European Collection of Cell Cultures reference no. 89051705) grown in a 24-well plate with 1 ml of MMI plus 20% FBS. The preparation was centrifuged at 1,000 × g at room temperature for 10 min to bring the bacteria into contact with the cells and incubated at 26.5°C for 16 h. Cells were washed twice in phosphate-buffered saline (PBS), pH 8.5, incubated in MMI containing 20% FBS and 50 μg ml−1 gentamicin for 1 h at 26.5°C, rinsed twice in PBS, and then incubated in fresh MMI containing 20% FBS for 10 days at 26.5°C. At 10-day intervals, medium taken from the cell layer described above was passaged onto a new 90% confluent C6/36 cell monolayer. All investigations of aphid-derived bacteria in association with the insect cells were conducted with 10-day-old cultures.
The insect cell cultures were also tested routinely for microorganisms cultivable on 5% sheep blood agar plates incubated at 26.5°C, and they were inspected daily for 10 days at a magnification of ×600 for microbial growth using an inverted M100 microscope (Swift-Microtec, Oxford, United Kingdom). Surface-sterilized aphids (prepared as described above) were homogenized and plated directly onto 5% sheep blood agar plates that were incubated at 26.5°C and inspected daily for 10 days.
Compatibility of symbionts with dipteran and lepidopteran cell lines.
Ten milliliters of culture medium from C6/36 cell cultures infected with secondary symbionts from A. fabae (T type) or A. pisum (U type) was passed though a 5-μm filter to remove the insect cells. The filtrate, which contained extracellular bacterial cells, was added to a 90% confluent cell monolayer of the Drosophila melanogaster S2 cell line (Drosophila Genetic Resource Centre stock number 6) or the Spodoptera frugiperda SF9 cell line (European Collection of Cell Cultures reference number 89070101). Each symbiont-cell line combination was replicated three times and maintained as described above for the C6/36 cell cultures.
PCR assays and sequencing.
For PCR-based detection and identification of secondary symbionts, total DNA was extracted from single aphids and 10-day-old insect cell cultures using a DNeasy tissue kit (QIAGEN, United Kingdom) by following the manufacturer's protocol for cultured animal cells, and this DNA was used as a template for amplification of 16S rRNA gene fragments. The specific forward primers were PABSF (5′-AGC GCA GTT TAC TGA GTT CA-3′) for T type (8) and U99F (5′-ATC GGG GAG TAG CTT GCT AC-3′) for U type (28), and these primers were combined with the general reverse primer for γ-proteobacteria, 16SB1 (5′-TAC GGY TAC CTT GTT ACG T-3′) (13); the diagnostic products were 1,660 nucleotides (nt) long for T type and 1,500 nt long for U type. Shorter products (630 nt) were generated using the γ-proteobacterium-specific primers 16SC1 Forward (5′-GAA TTC TAG GTG TAG CGG TGA) and 16SD1 Reverse (5′-GCG ATT CCG ACT TCG TGG A-3′) (this study). The PCR mixtures contained PCR buffer, 2 mM MgCl2 (Invitrogen, United Kingdom), each deoxynucleoside triphosphate (Promega, United Kingdom) at a concentration of 0.24 mM, each primer (MWG Biotech, GmbH) at a concentration of 20 μM, and 1 U Platinum Taq polymerase (Invitrogen, United Kingdom). The reaction conditions were 30 cycles of 94°C for 1 min, 54°C for 1 min, and 72°C for 2 min; however, the denaturing step was extended to 5 min for the first cycle, and the extension time was increased to 8 min for the last cycle. PCR products were purified with a QIAquick PCR purification kit (QIAGEN, United Kingdom), ligated into a pGEM-T vector (Promega, United Kingdom), and transformed into Escherichia coli strain DH5α (Promega, United Kingdom) according to the manufacturer's instructions, except that the competent bacteria were pretreated with 0.1% (vol/vol) β-mercaptoethanol before transformation. Plasmid DNA was purified using a QIAprep miniprep kit (QIAGEN) according to manufacturer's instructions, and consensus sequences were derived from the sequences of three clones, determined in both directions.
Microorganisms were identified by cultivation on 5% sheep blood agar plates. DNA was extracted from single colonies using the bacterial protocol of the DNeasy tissue kit (QIAGEN, United Kingdom) and was used as a template for amplification of 16S rRNA gene fragments with the general primer pair for γ-proteobacteria, 16SA1 (5′-AGA GTT TGA TCM TGG CTC AG-3′) (13) and 16SB1 (see above), by using the amplification conditions described above. A fragment of each PCR product was sequenced (Lark Technologies, United Kingdom) using the general γ-proteobacterial internal forward primer p1 (5′-CCT ACG GGA GGC AGC AG-3′) (24).
Phylogenetic analysis.
Clustal_X version 1.8 (32) was used for multiple-sequence alignment with manual adjustment to exclude gaps and ambiguous bases from the analysis. Phylogenetic trees were generated with PAUP* 4.0b2 (31). The following two methods of tree drawing were used: (i) neighbor joining using the Kimura two-parameter model (19) with correction for multiple substitutions and (ii) an optimal maximum-likelihood model for the data found using Modeltest v3.06 (26). The optimum model for the 1,480-nt 16S rRNA gene fragment data was found to be a HKY85+G+I model, and the optimum model for the 635-nt 16S rRNA gene fragment data set was found to be a HKY+G model (16). These models contained estimates (generated from the data) for transition/transversion ratios, nucleotide frequencies, the proportion of invariable sites, and among-site rate variation. The “distribution of rates at variable sites” used discrete approximations of gamma and shape parameters set at 0.527 and 0.113 for the 1,480-nt and 635-nt data sets, respectively. Bootstrap analyses (11) were performed with 1,000 replications for both data sets to assess the reliability of the resulting phylogenies.
In situ hybridization.
All hybridization procedures were conducted under RNase-free conditions. Insect cells from a routine culture were suspended in MMI plus 20% FBS. A sample (500 μl) of a cell suspension was applied to Superfrost-Plus slides (BDH, United Kingdom) and allowed to settle for 30 min at 25°C. Adhering cells were fixed for 10 min in 4% (wt/vol) formaldehyde in PBS (Invitrogen), and this was followed by cell permeabilization in 70% ethanol for 10 min and three washes in PBS. Postfixation in situ hybridization was performed using 5′-end-labeled oligonucleotide probes (high-performance liquid chromatography purified; MWG Biotech, GmbH) that specifically hybridize to 16S rRNA of γ-proteobacteria (EUB338; fluorescein isothiocyanate [FITC] labeled; 5′-GCT GCC TCC CGT AGG AGT-3′) (1), U type (U16; 6-carboxytetramethylrhodamine [TAMRA]-5′-GTA GCA AGC TAC TCC CCG AT-3′) (34), and T type (T16; Cy5 labeled; 5′-CTC AGT AAA CTG CGC TCA CT-3′) (this study). The probes used for secondary symbionts were designed and tested for specificity and hybridization stringency by using previously described procedures (3, 14). Insect cell nuclei were simultaneously counterstained with 4′,6′-diamidino-2-phenylindole (DAPI) to aid cell localization and visualization. Insect cells were incubated overnight with 200 μl of a hybridization solution containing 20 mM Tris-HCl (pH 8.0), 0.9 M NaCl, 0.01% sodium dodecyl sulfate, 30% (vol/vol) formamide, each probe at a concentration of 70 pmol ml−1, and DAPI at a concentration of 140 pmol ml−1 in a dark humid chamber. They were then washed three times in PBS (5 min) and mounted in Citifluor AF1 (Citifluor Ltd., United Kingdom). The three control treatments used in all experiments were (i) a probe-free control for autofluorescence; (ii) an RNA-free control (treatment, prior to hybridization, with an RNase solution containing 150 mM NaCl, 10 mM Tris-HCl [pH 7.5], 1 mM EDTA, and 20 μg RNase [QIAGEN] ml−1 at 37°C for 30 min); and (iii) competitive suppression (addition of 4.2 nmol unlabeled probe to the hybridization solution) (data not shown) (3).
Confocal microscopy was performed with a Zeiss LSM 510 Meta attached to a Zeiss Axiovert 200 M fitted with a Plan-Apochromat ×63 oil immersion lens (N.A. 1.4). DAPI was excited with the 405-nm diode laser line, and emission was collected via a 420- to 428-nm band-pass filter. The fluorochromes TAMRA, Cy5, and FITC were excited with 543-nm HeNe, 633-nm HeNe, and 488-nm argon laser lines, respectively. Emission from TAMRA and Cy5 was collected via 560- and 650-nm long-pass filters, and emission from FITC was collected via a 505- to 570-nm band-pass filter. To prevent potential bleed-through problems under the experimental conditions, images were acquired sequentially.
Transmission electron microscopy.
A monolayer of A. albopictus C6/36 cells was grown in MMI containing 20% FBS to 90% confluence on plastic Thermanox coverslips (Nalge Nunc International, United States) in 24-well culture plates. The culture medium was then replaced with medium from a 10-day-old culture of cells infected with U type from M. rosae that had been filtered through a 5-μm filter to eliminate the insect cells. The inoculated coverslip cultures were incubated for 16 h at 26.5°C to allow the uptake of bacteria into the cells, washed with PBS, fixed in 3% glutaraldehyde, postfixed in 1% osmium tetroxide, dehydrated in acetone, and embedded in araldite resin. Sections (thickness, 60 nm) were stained using an LKB Ultrastainer with uranyl acetate and lead citrate and examined with a Philips CMI2 transmission electron microscope.
Nucleotide sequence accession numbers.
Consensus nucleotide sequences have been deposited in the National Center for Biotechnology Information GenBank database under accession numbers AY692358 to AY692365.
RESULTS
The first detailed inspection of C6/36 cells infected with bacterial preparations from aphids was conducted at the third passage (30 to 40 days postinfection). The secondary symbionts of aphids were predicted to be relatively slow growing (36); 10 to 20% of the 72 cultures from each aphid species had visible bacterial growth within 48 h after infection, and these cultures were excluded from the study.
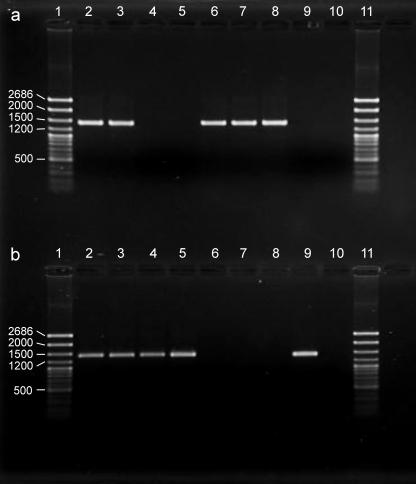
For the cultures examined at a magnification of × 600 at the third passage, rod-shaped bacteria were evident in 29% (19 of 65) and 42% (27 of 64) of the replicate cultures initiated from M. rosae and A. pisum, respectively, but no bacteria were detected in cultures initiated from A. fabae (n = 58). Samples from the cultures were subjected to diagnostic PCR for T type and U type (for M. rosae and A. pisum, the cultures selected contained bacterial rods). All nine cultures from A. pisum contained only U type; eight cultures from M. rosae contained U type, and seven of these cultures also contained T type; and six of the nine cultures from A. fabae were positive for only T type (Fig. 1). Thus, both T type and U type could be cultured in the C6/36 insect cells, and the estimated success rates were 26 to 67% for T type and 29 to 42% for U type (calculated from the data described above). Bacteria obtained from M. rosae and A. pisum were gram-negative rods and 2 to 9 μm long, characteristics that are consistent with the secondary symbiont U type (7). The cells of T type are generally <2 μm long (8), and they would not be readily detectable in association with living cells using an inverted microscope at a magnification of ×600.
FIG. 1.
Incidence of secondary symbionts in aphids and cell cultures. (a) T type. (b) U type. The aphid source material was M. rosae line ACD03/01 (lane 2), A. pisum line JF99/14 (lane 4), and A. fabae line CRT01/43 (lane 6). C6/36 cells were infected with bacterial preparations from M. rosae (lane 3), A. pisum (lane 5), and A. fabae (lane 7). Lanes 8 and 9 contained positive controls for T type and U type, respectively, and lane 10 contained the template-free negative control. The molecular ladder (lanes 1 and 11) was the BIOTC medium ladder (Geneflow, Fradley, United Kingdom). The bands were at (in descending order) bp 2686, 2000, 1500, 1200, 1000, 900, 800, 700, 500, 400, 300, 200, and 100. The analysis was conducted on a 1% agarose gel.
The persistence of secondary symbionts from the three aphid species was scored in insect cell cultures over multiple passages. U type from A. pisum and T type from A. fabae persisted for at least 20 passages (the maximum number of passages tested). The cultures bearing U type and T type from M. rosae tended to lose T type, so that by the 20th passage all of the cultures contained only U type. The loss of T type was not linked to the passaging procedure itself since one cell line bearing U type and T type maintained without passaging for 60 days (equivalent to six passages) yielded only U type (as determined by diagnostic PCR).
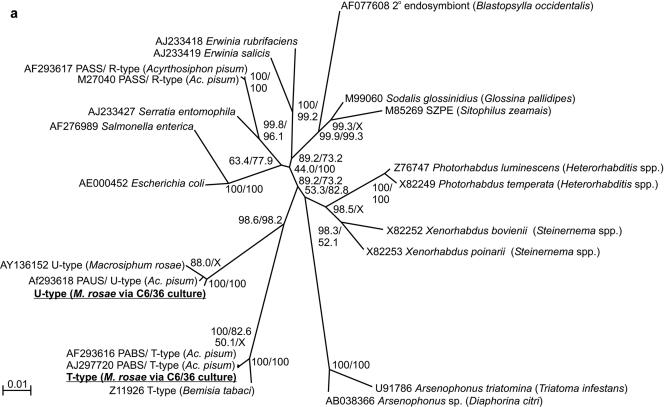
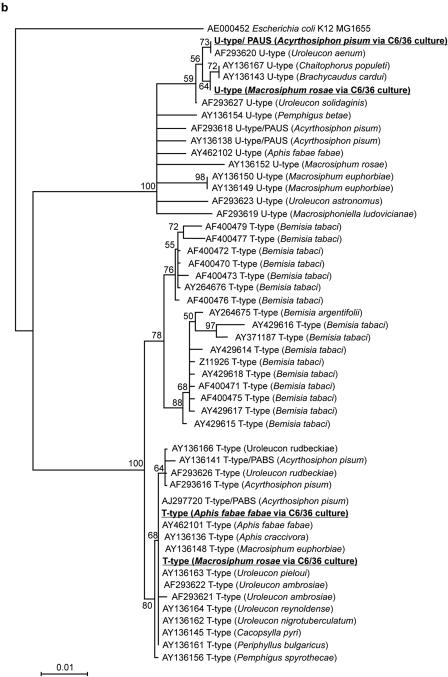
Bacterial identities within the cell cultures were confirmed using two complementary approaches. First, a 1,480-nt fragment of the 16S rRNA gene of the bacteria in the insect cells was amplified and sequenced. As shown by the unrooted neighbor-joining tree in Fig. 2a, the sequences could be assigned to the γ-proteobacteria and were distinct from the sequences of most other bacteria that have been found in insects, including the secondary symbiont known as R-type from aphids (27, 35). Consistent with the diagnostic PCR data (see above), the U-type sequences from M. rosae and A. pisum were allied with, but not identical to, the U type previously reported for M. rosae; and the T type from A. fabae and M. rosae was very similar to a previously described T type found in A. pisum. The phylogenetic positions of the bacteria cultured in insect cells were investigated further by constructing neighbor-joining and maximum-likelihood trees rooted with E. coli (Fig. 2b). The tree topologies obtained by the two statistical methods were similar but not identical (data not shown); the basal clades obtained by both methods were the same, and the phylogenetic positions of the symbionts in cell culture were robust, but differences were observed in the apical branching patterns of the clades. These analyses clearly demonstrated the presence of two isolates of T type and U type with distinct 16S rRNA gene sequences, all within the known range of molecular diversity of these bacteria. At the level of the 16S rRNA sequence, no heterogeneity of either T type or U type was evident among the three sequences found in the aphid species.
FIG. 2.
Phylogenetic trees of 16S rRNA gene sequences of the bacteria cultured in insect cells in this study (underlined). (a) Phylogenetic position in the γ-proteobacteria (1,480-nt gene fragments); (b) sequence diversity relative to known U-type and T-type symbionts (635-nt gene fragments). The bootstrap percentages at the nodes were calculated by the neighbor-joining and maximum-likelihood methods. GenBank accession numbers, bacterial species or “informal” names, and (in parentheses) the relevant insect hosts (where relevant) are indicated. PABS, pea aphid Bemisia symbiont; PAUS, pea aphid U-type symbiont; PASS, pea aphid secondary symbiont.
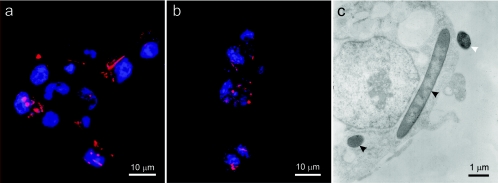
The second approach used to establish the identities of secondary symbionts in the cell cultures was a microscopic analysis using fluorescence in situ hybridization (FISH) of a DNA probe to 16S rRNA of the bacteria, a method that is independent of PCR amplification. Cell lines bearing secondary symbionts from M. rosae and A. pisum known to contain U type and cell lines bearing secondary symbionts from A. fabae known to contain T type (as determined by diagnostic PCR) were examined. Bacteria were detected in all inoculated cultures tested by FISH, and many of them appeared to be cytoplasmic. The number of bacteria per insect cell and the number of cells infected varied with passage and culture. In M. rosae cultures, rod-shaped bacteria were detected in 50% of the 100 insect cells examined, and each infected cell contained 1 to 20 bacteria that were up to 15 μm long and hybridized with both the general eubacterial probe and the U-type-specific probe (Fig. 3a) but not with the T-type-specific probe. For the insect cells in which T-type bacteria were detected by diagnostic PCR, about 25% of the 100 insect cells examined yielded a signal with the FISH probe specific for T type, and the readily detectable hybridizations were hybridizations of coccoid structures that were ca. 1.5 μm in diameter (Fig. 3b) and irregularly shaped structures whose dimensions were up to 3 μm. Transmission electron microscopy of cells bearing U type confirmed the presence of membrane-bound rod-shaped bacteria in the cytoplasm of the cells and also revealed extracellular bacteria (Fig. 3c).
FIG. 3.
Secondary symbionts in C3/36 cells. (a and b) Fluorescence in situ hybridization of Cy5-labeled probes (red) to U type isolated from M. rosae (a) and to T type isolated from A. fabae (b). (c) Transmission electron micrograph of U type. The black arrowheads indicate intracellular symbionts, and the white arrowhead indicates an extracellular symbiont.
In supplementary experiments we investigated whether U type and T type could be cultured in alternative insect cell lines, including the Drosophila S2 and Spodoptera SF9 lines. All recipient cell lines were positive for symbionts as determined by specific PCR assays after three passages. The lines infected with U type also contained rod-shaped bacteria that were detectable using an inverted microscope at a magnification of ×600. In separate experiments, both T type and U type derived from aphid homogenates were cultured successfully in Drosophila S2 cells (data not shown); the capacity of these bacteria from aphid homogenates to persist in SF9 cells was not investigated.
Rapidly growing microorganisms were detected within 48 h of inoculation of cells with aphid-derived material in a few of the cell cultures samples, including 10% (7 of 72) of the samples from M. rosae, 11% (8 of 72) of the samples from A. pisum, and 21% (15 of 72) of the samples from A. fabae. The incidence of these bacteria did not vary significantly with aphid species (χ2 [2 df] = 4.232, P > 0.05). When samples of all the cell cultures were tested for microorganisms cultivable on 5% sheep blood plates, only the cultures with the rapidly growing bacteria yielded bacterial colonies, including Micrococcus, Acinetobacter, and Staphylococcus colonies, as identified by sequencing of PCR-amplified 16S rRNA gene fragments, and colonies with the color and morphology of Serratia colonies (which were not studied further). All these genera have previously been described as genera that are associated with aphids, but they have not been described as symbiotic bacteria (15; C. Francois personal communication). No secondary symbionts were detected, and the insect cell cultures bearing these bacteria were not studied further.
DISCUSSION
Two aphid secondary bacteria informally known as T type and U type were isolated from three aphid species and cultured in cell lines of three insect species. The persistence of the bacteria in the cell cultures over extended periods, including multiple passages, demonstrated unambiguously that the bacterial cells are able to proliferate in cell culture. Symbionts were detected both in the cytoplasm of the cells and extracellularly, but the relative importance of the bacteria in these two locations to the persistence of the infections remains to be established. The intracellular symbionts appeared to be structurally intact and membrane bound, either singly or in groups (Fig. 3). Similar bacterial aggregations have not been reported for secondary symbionts in aphids, but they have been described for other intracellular bacteria, including pathogenic and symbiotic forms, including Salmonella, uropathogenic E. coli, Sodalis glossinidius, and Rickettsia spp. (6, 20, 21, 23). The aggregations may have arisen from either bacterial division within the cell or phagocytosis of multiple bacterial cells into a single vacuole.
The capacity of T type and U type to maintain persistent infections in all the insect cell lines tested (two dipteran lines and one lepidopteran line) demonstrates that there is potential compatibility with insect cells other than aphid cells. These bacteria have not been reported in dipterans or lepidopterans, and they are probably restricted to aphids and a few related phloem sap feeders (27). It is unlikely that their infective range is restricted by limited access to alternative host species for two reasons. First, there is strong phylogenetic and experimental evidence that the secondary symbionts are horizontally transmissible (5, 9, 27); and second, other insect-associated bacteria with mixed vertical and horizontal transmission patterns show broad host distributions (e.g., Wolbachia is estimated to occur in 20% of all insect species [38, 39]). Taxonomic restriction of the secondary symbionts to aphids may reflect specificity in interactions with particular cell types or physiological systems. For example, secondary symbionts may be able to invade the aphid across the gut wall (9) (during horizontal acquisition by ingestion of bacterial cells released from other aphids in honeydew) or the ovarial sheath (during vertical transmission) but may not be able to invade other insect taxa by these routes; or initial infections may be detected and eliminated by the humoral defense systems of insects other than aphids. Studies of the differences in the interactions of secondary symbionts with single cells and intact insects would promote our understanding of the molecular basis of infection and the invasiveness of bacteria in arthropods.
This study provides additional insight into the nature of interactions between the secondary symbionts and the insect host. Specifically, T type can be maintained in cell culture when it is derived from an aphid bearing T type as the sole secondary symbiont but not when it is derived from an aphid bearing both T type and U type. In the case of mixed infections of T type and U type, the T type was always eliminated. This is in contrast to the existence of T- and U-type coinfections in field aphids (17, 29) and in long-term laboratory aphid cultures (12). The basis for the difference between cell culture and the insect symbiosis is not known, but it may include limited overlap in the resources utilized by the T-type and U-type symbionts in aphids, with resource partitioning promoted by, for example, localization to different cells or tissues in the insect and access to a wider range of nutrients in the symbiosis than in cell culture. Additionally, the possibility that U type possesses traits (e.g., bacteriocins) which are directly antagonistic to T type expressed in cell culture but not in the symbiosis cannot be excluded.
The ability to culture single-taxon preparations in cell lines expands the research opportunities available for secondary symbionts of aphids. Previously, the only preparations available have been fractionated aphid homogenates with low yields often contaminated by other aphid-associated bacteria. Cell lines bearing secondary symbionts not only provide a new platform for the development of novel approaches and technologies to control insect pests but also have potential as a source of material for genomic analyses and short-term physiological studies of the bacteria in defined media (40). They make possible fundamental studies of the interactions between insect symbionts and animal cells, including the extent to which the establishment and persistence of intracellular symbiosis is dependent on traits also displayed by overt pathogens (37).
This study is the first demonstration that secondary symbionts of aphids can be cultured apart from the aphid-Buchnera symbiosis. To our knowledge, just one other secondary symbiont, S. glossinidius in tsetse flies, has been cultured previously (36). Our success with two secondary symbiont bacteria suggests that this approach may be generally applicable to many secondary symbionts in insects.
The secondary symbionts which we cultured in insect cells are known informally as T type (pea aphid Bemisia symbiont) and U type (pea aphid U-type symbiont). They have been described previously on the basis of their 16S rRNA gene sequences (8, 27), and their morphology has been identified by in situ hybridization (8, 34). We propose the provisional taxon names (30) “Candidatus Consessoris aphidicola” (from the Latin consessoris, meaning one who sits near, a neighbor) for T type and “Candidatus Adiaceo aphidicola” (from the Latin adiaceo, meaning to lie by the side of, be adjacent) for U type. These names were specifically chosen because their Latin origins suggest close physical proximity without implying function.
Acknowledgments
We thank P. O'Toole and T. Tsuchida for advice concerning the FISH analysis and J. Ferrari and C. R. Tosh for use of aphid lineages JF 99/14 and CRT01/43, respectively.
The Wellcome Trust (A.C.D.) and James Burgess Scholarship (S.M.C.) provided financial support.
REFERENCES
- 1.Amann, R., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buchner, P. 1965. Endosymbiosis of animals with plant microorganisms. Interscience Publishers, New York, N.Y.
- 3.Chandler, S. M. Host plants and the symbiotic bacteria of the black bean aphid (Aphid fabae Scopoli). Ph.D. thesis, submittted. The University of York, York, United Kingdom.
- 4.Chen, D. Q., B. C. Campbell, and A. H. Purcell. 1996. A new Rickettsia from a herbivorous insect, the pea aphid Acyrthosiphon pisum (Harris). Curr. Microbiol. 33:123-128. [DOI] [PubMed] [Google Scholar]
- 5.Chen, D. Q., C. B. Montllor, and A. H. Purcell. 2000. Fitness effects of two facultative endosymbiotic bacteria on the pea aphid, Acyrthosiphon pisum, and the blue alfalfa aphid, A. kondoi. Entomol. Exp. Appl. 95:315-323. [Google Scholar]
- 6.Dale, C., and I. Maudlin. 1999. Sodalis gen. nov. and Sodalis glossinidius sp. nov., a microaerophilic secondary endosymbiont of the tsetse fly Glossina morsitans morsitans. Int. J. Syst. Bacteriol. 49:267-275. [DOI] [PubMed] [Google Scholar]
- 7.Darby, A. C. 2002. The microbiota of the pea aphid Acyrthosiphon pisum. Ph.D. thesis. University of York, York, United Kingdom.
- 8.Darby, A. C., L. M. Birkle, S. L. Turner, and A. E. Douglas. 2001. An aphid-borne bacterium allied to the secondary symbionts of whitefly. FEMS Microbiol. Ecol. 36:43-50. [DOI] [PubMed] [Google Scholar]
- 9.Darby, A. C., and A. E. Douglas. 2003. Elucidation of the transmission patterns of an insect-borne bacterium. Appl. Environ. Microbiol. 69:4403-4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Douglas, A. E. 1998. Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu. Rev. Entomol. 43:17-37. [DOI] [PubMed] [Google Scholar]
- 11.Felsenstein, J. 1985. Confidence limits on phylogenies—an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 12.Ferrari, J., A. C. Darby, T. J. Daniell, H. C. J. Godfray, and A. E. Douglas. 2004. Linking the bacterial community in pea aphids with host-plant use and natural enemy resistance. Ecol. Entomol. 29:60-65. [Google Scholar]
- 13.Fukatsu, T., N. Nikoh, R. Kawai, and R. Koga. 2000. The secondary endosymbiotic bacterium of the pea aphid Acyrthosiphon pisum (Insecta: Homoptera). Appl. Environ. Microbiol. 66:2748-2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukatsu, T., K. Watanabe, and Y. Sekiguchi. 1998. Specific detection of intracellular symbiotic bacteria of aphids by oligonucleotide-probed in situ hybridization. Appl. Entomol. Zool. 33:461-472. [Google Scholar]
- 15.Grenier, A. M., C. Nardon, and Y. Rahbe. 1994. Observations on the microorganisms occurring in the gut of the pea aphid Acyrthosiphon pisum. Entomol. Exp. Appl. 70:91-96. [Google Scholar]
- 16.Hasegawa, M., H. Kishino, K. Hayasaka, and S. Horai. 1990. Mitochondrial-DNA evolution in primates—transition rate has been extremely low in the lemur. J. Mol. Evol. 31:113-121. [DOI] [PubMed] [Google Scholar]
- 17.Haynes, S., A. C. Darby, T. J. Daniell, G. Webster, F. J. F. van Veen, H. C. J. Godfray, J. I. Prosser, and A. E. Douglas. 2003. Diversity of bacteria associated with natural aphid populations. Appl. Environ. Microbiol. 69:7216-7223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hypsa, V., and C. Dale. 1997. In vitro culture and phylogenetic analysis of “Candidatus Arsenophonus triatominarum,” an intracellular bacterium from the triatomine bug, Triatoma infestans. Int. J. Syst. Bacteriol. 47:1140-1144. [DOI] [PubMed] [Google Scholar]
- 19.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 20.Labruna, M. B., T. Whitworth, M. C. Horta, D. H. Bouyer, J. W. McBride, A. Pinter, V. Popov, S. M. Gennari, and D. H. Walker. 2004. Rickettsia species infecting Amblyomma cooperi ticks from an area in the state of Sao Paulo, Brazil, where Brazilian spotted fever is endemic. J. Clin. Microbiol. 42:90-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyer-Bahlburg, A., J. Brinkhoff, V. Krenn, K. Trebesius, J. Heesemann, and H.-I. Huppertz. 2001. Infection of synovial fibroblasts in culture by Yersinia enterocolitica and Salmonella enterica serovar Enteritidis: ultrastructural investigation with respect to the pathogenesis of reactive arthritis. Infect. Immun. 69:7915-7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitsuhashi, J., and K. Maramorosch. 1964. Leafhopper tissue culture: embryonic, nymphal, and imaginal tissues from aseptic insects. Contrib. Boyce Thompson Inst. 22:435-460. [Google Scholar]
- 23.Mulvey, M. A., J. D. Schilling, and S. J. Hultgren. 2001. Establishment of a persistent Escherichia coli reservoir during the acute phase of a bladder infection. Infect. Immun. 69:4572-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muyzer, G., E. C. Dewaal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Neill, S. L., M. M. Pettigrew, S. P. Sinkins, H. R. Braig, T. G. Andreadis, and R. B. Tesh. 1997. In vitro cultivation of Wolbachia pipientis in an Aedes albopictus cell line. Insect Mol. Biol. 6:33-39. [DOI] [PubMed] [Google Scholar]
- 26.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 27.Russell, J. A., A. Latorre, B. Sabater-Munoz, A. Moya, and N. A. Moran. 2003. Side-stepping secondary symbionts: widespread horizontal transfer across and beyond the Aphidoidea. Mol. Ecol. 12:1061-1075. [DOI] [PubMed] [Google Scholar]
- 28.Sandstrom, J. P., J. A. Russell, J. P. White, and N. A. Moran. 2001. Independent origins and horizontal transfer of bacterial symbionts of aphids. Mol. Ecol. 10:217-228. [DOI] [PubMed] [Google Scholar]
- 29.Simon, J. C., S. Carre, M. Boutin, N. Prunier-Leterme, B. Sabater-Munoz, A. Latorre, and R. Bournoville. 2003. Host-based divergence in populations of the pea aphid: insights from nuclear markers and the prevalence of facultative symbionts. Proc. R. Soc. Lond. Ser. B Biol. Sci. 270:1703-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stackebrandt, E., W. Frederiksen, G. M. Garrity, P. Grimont, P. Kampfer, M. Maiden, X. Nesme, R. Rossello-Mora, J. Swings, H. G. Truper, L. Vauterin, A. C. Ward, and W. B. Whitman. 2002. Report of the Ad Hoc Committee for the Re-evaluation of the Species Definition in Bacteriology. Int. J. Syst. Evol. Bacteriol. 52:1043-1047. [DOI] [PubMed] [Google Scholar]
- 31.Swofford, D. L. 2003. PAUP*. Phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates, Sunderland, Mass.
- 32.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsuchida, T., R. Koga, and T. Fukatsu. 2004. Host plant specialization governed by facultative symbiont. Science 303:1989. [DOI] [PubMed] [Google Scholar]
- 34.Tsuchida, T., R. Koga, M. X. Y., T. Matsumoto, and T. Fukatsu. Characterization of a facultative endosymbiotic bacterium of the pea aphid Acyrthosiphon pisum. Microb. Ecol. 49:126-133. [DOI] [PubMed]
- 35.Unterman, B. M., P. Baumann, and D. L. McLean. 1989. Pea aphid symbiont relationships established by analysis of 16S rRNAs. J. Bacteriol. 171:2970-2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Welburn, S. C., I. Maudlin, and D. S. Ellis. 1987. In vitro cultivation of Rickettsia-like organisms from Glossina spp. Ann. Trop. Med. Parasitol. 81:331-335. [DOI] [PubMed] [Google Scholar]
- 37.Wernegreen, J. J. 2002. Genome evolution in bacterial endosymbionts of insects. Nat. Rev. Genet. 3:850-861. [DOI] [PubMed] [Google Scholar]
- 38.Werren, J. H., and D. M. Windsor. 2000. Wolbachia infection frequencies in insects: evidence of a global equilibrium? Proc. R. Soc. Lond. Ser. B Biol. Sci. 267:1277-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.West, S. A., J. M. Cook, J. H. Werren, and H. C. J. Godfray. 1998. Wolbachia in two insect host-parasitoid communities. Mol. Ecol. 7:1457-1465. [DOI] [PubMed] [Google Scholar]
- 40.Whitehead, L. F., and A. E. Douglas. 1993. A metabolic study of Buchnera, the intracellular bacterial symbionts of the pea aphid Acyrthosiphon pisum. J. Gen. Microbiol. 139:821-826. [Google Scholar]
- 41.Wilkinson, T. L., D. Adams, L. B. Minto, and A. E. Douglas. 2001. The impact of host plant on the abundance and function of symbiotic bacteria in an aphid. J. Exp. Biol. 204:3027-3038. [DOI] [PubMed] [Google Scholar]