Abstract
Stromatolites, organosedimentary structures formed by microbial activity, are found throughout the geological record and are important markers of biological history. More conspicuous in the past, stromatolites occur today in a few shallow marine environments, including Hamelin Pool in Shark Bay, Western Australia. Hamelin Pool stromatolites often have been considered contemporary analogs to ancient stromatolites, yet little is known about the microbial communities that build them. We used DNA-based molecular phylogenetic methods that do not require cultivation to study the microbial diversity of an irregular stromatolite and of the surface and interior of a domal stromatolite. To identify the constituents of the stromatolite communities, small subunit rRNA genes were amplified by PCR from community genomic DNA with universal primers, cloned, sequenced, and compared to known rRNA genes. The communities were highly diverse and novel. The average sequence identity of Hamelin Pool sequences compared to the >200,000 known rRNA sequences was only ∼92%. Clone libraries were ∼90% bacterial and ∼10% archaeal, and eucaryotic rRNA genes were not detected in the libraries. The most abundant sequences were representative of novel proteobacteria (∼28%), planctomycetes (∼17%), and actinobacteria (∼14%). Sequences representative of cyanobacteria, long considered to dominate these communities, comprised <5% of clones. Approximately 10% of the sequences were most closely related to those of α-proteobacterial anoxygenic phototrophs. These results provide a framework for understanding the kinds of organisms that build contemporary stromatolites, their ecology, and their relevance to stromatolites preserved in the geological record.
Fossil stromatolites are found throughout the geological record and are important biosignatures for the early Earth and in the search for extraterrestrial life. Stromatolites are defined as “organosedimentary structures predominantly accreted by sediment trapping, binding, and/or in situ precipitation as a result of the growth and metabolic activity of microorganisms” (52). Stromatolites are typically characterized by an irregular lamination, which can be concentric, wrinkled, or wavy and tend to develop convex upward structures in conical, columnar, domal, or upward widening shapes (8). Living stromatolites occur on Earth today in a few shallow marine environments, which includes Shark Bay in Western Australia and Exuma Sound in the Bahamas. Such living stromatolites have been interpreted as analogs of fossil stromatolites based on morphology. The oldest examples of preserved fossil stromatolites in the geological record are about 3.5 billion years old (Ga) and are found in Western Australia and South Africa (8, 10, 32, 53). Stromatolites in the geological record can provide insights into the nature of habitable environments on the early Earth, the antiquity of some microbial metabolisms and the evolution of biogeochemical cycles.
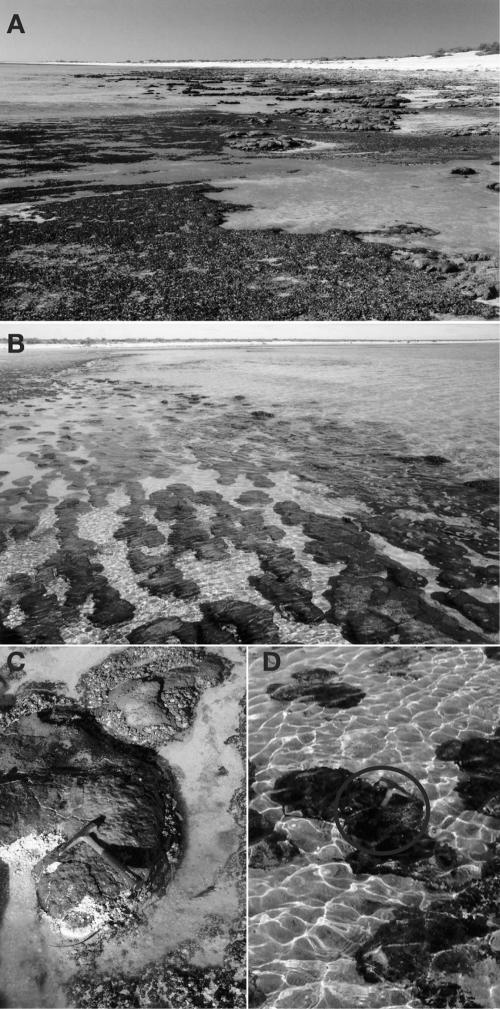
The stromatolites of Hamelin Pool, shown in Fig. 1, are examples of living stromatolites actively forming as a result of microbial activity. Stromatolites rise up to 0.5 m from the floor of this shallow hypersaline pool and display a variety of morphologies, including columnar, club, spheroidal, domal, nodular, or irregular shape (31). The stromatolites grow by accretion and precipitation of material to the outer surface layer, where the most conspicuous microbiology also occurs, commonly as a thinly stratified microbial mat (43, 49). Stromatolite morphology is often attributed to different kinds of microbial mat communities. Mat communities have been classified mostly by conspicuous cyanobacterial morphotypes and whether mats have a smooth or irregular appearance. Stromatolite communities also are structured vertically. Studies of Bahamian stromatolites, for instance, find that lithified interiors beneath surface mat communities are modified by endolithic microbes (44).
FIG. 1.
Pictures of the field site in the Hamelin Pool, Shark Bay. (A) Low-to-mid tide at the Hamelin Station exposing domal stromatolites and microbial mats. (B) Submerged irregular stromatolites at the Hamelin Station. (C) Sample HPDOM, at the tip of the hammer, collected from a domal stromatolite (26°23′32.4"S and 114°09′41.4′E). (D) Sample HPIRR, below the hammer (circled), from a submerged irregular stromatolite (∼20 m away from HPDOM).
Although Hamelin Pool stromatolites are among the most prominent examples of living geomicrobiological structures, we know little about the kinds of microorganisms that comprise these stromatolite-building communities. Previous studies have characterized growth rates of Hamelin Pool stromatolites by radiocarbon dating (12). Other studies have focused on the sedimentology (see, for example, reference 37), culture, and phylogeny of specific microorganisms (9, 36), biomolecular pigments (41), ecological relationships (3), and microscopy (5). Such studies have generally presumed or concluded that cyanobacteria, and in some cases photosynthetic eucaryotes, are the main constituents of Hamelin Pool stromatolite communities. However, assumptions that cyanobacteria dominate this ecosystem have not been based on strong evidence. Cyanobacteria are popularly associated with stromatolites mostly because they are easily observed microscopically as a consequence of their large and conspicuous morphologies.
The objective of the present study was to determine the molecular phylogenetic makeup of microbial communities that build Hamelin Pool stromatolites and perhaps to shed light on their relevance to Precambrian stromatolites. The composition of such microbial communities cannot be determined by methods that require cultivation because most organisms (>99%) from natural environments are not cultured by standard methods (40). Consequently, we determined the composition and diversity of communities by sequence analysis of small-subunit (SSU) rRNA genes amplified by PCR with universal primers (515F and 1391R) from genomic DNA extracted directly from stromatolite samples. The compositions of communities were compared by using phylogenetic statistics to assess the similarity of sequence collections in phylogenetic trees. With this information, we tested the following hypotheses: (i) the stromatolite communities are comprised primarily of cyanobacteria, (ii) endolithic communities of stromatolites are distinct from surface communities, and (iii) different stromatolite morphotypes are associated with different microbial communities. These analyses collectively provide new perspective on the structure and composition of the Hamelin Pool stromatolite communities.
MATERIALS AND METHODS
Sample location.
Hamelin Pool in Western Australia spans an area of ca. 1,220 km2 with an average tidal range of ca. 60 cm (31). Average water temperatures throughout the year are between 17 and 27°C, and annual evaporation is more than 2 m (31). These conditions create hypersaline environments in many embayments of Shark Bay, including Hamelin Pool, which has about twice the salinity of seawater.
Sample description.
Samples of two stromatolites were collected from the Hamelin Pool in the area of Hamelin Station in Shark Bay during the late afternoon of July 9, 2003. Samples of the upper ∼3 cm of stromatolites were collected from the intertidal zone, preserved in 70% ethanol in sterile 50-ml plastic tubes, and stored at 4°C.
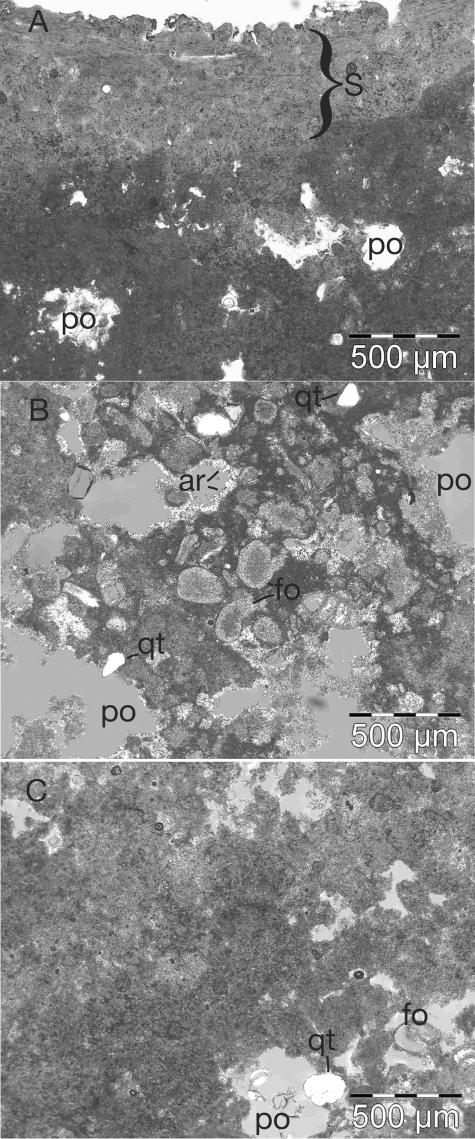
Sample HPDOM (for Hamelin Pool Domal) had a smooth, domal morphology (Fig. 1) with a thin (∼1-mm) indurated surface colored green, orange, and black. The shape of the HPDOM stromatolite was most similar to the “smooth surface mat” described by Reid et al. (43). It was collected when the stromatolite was directly exposed to air at low-to-mid tide. A chip of HPDOM was used to make a petrographic thin section for observation by optical microscopy. Figures 2A and B show representative images of the crust and interior of HPDOM, respectively. The beige-white porous interior consisted of submillimeter-sized carbonate sand, foraminifera, bivalve shells, shell fragments, and microcrystalline carbonate (micrite). Aragonite needles often coated or partially filled the pore spaces of HPDOM, and some detrital quartz grains and dolomite crystals (grown in situ) were observed. The microfabric of HPDOM most closely resembled the “unlaminated calcarenite” described by Reid et al. (43). HPDOM was subsampled by manual separation into a surface sample ca. 1 to 2 mm thick (HPDOM-S) and an interior sample taken from a vertical interval ca. 10 to 15 mm below the surface (HPDOM-I).
FIG. 2.
Light micrographs of Hamelin Pool stromatolites in thin section. (A) Image of sample HPDOM showing the surface (S) and the micritic interior close to the surface with pore spaces (po). (B) Representative image in crossed-polarized light of the interior of HPDOM showing an abundance of foraminifera (fo), detrital quartz grains (qt), and acicular aragonite (ar), which coat the interior of some pores (po). (C) Representative image of the interior of HPIRR showing the matrix dominated by micrite.
A second sample, HPIRR (for Hamelin Pool Irregular), was collected from a stromatolite with a dark green and black knobby surface and an irregular morphology. The surface of HPIRR resembled “knobby pustular” surfaces previously described (43). The stromatolite, shown in Fig. 1, was submerged at the time of collection. Microscopic observations in thin sections showed that HPIRR was different from HPDOM (Fig. 2C). HPIRR had a red-brown interior that was less porous and had fewer foraminifera and bivalve shells than HPDOM. The interior of HPIRR was mostly micritic (“massive micrite” [43]) with minor occurrences of detrital quartz grains, dolomite crystals, and shell fragments. A piece of the upper ∼5 mm of HPIRR was analyzed in bulk.
Laser scanning confocal microscopy (LSCM).
We imaged fluorescence associated with oxygenic phototrophs with a Leica TCS SP2 AOBS laser scanning confocal microscope. We imaged fluorescent emissions at 680 to 690 nm, which is due to chlorophyll molecules in reaction centers of photosystem II (PS II) of oxygenic phototrophs in vivo (6). Wavelengths that excite PS II fluorescence depend on associated accessory antenna pigments, which vary in different groups of oxygenic phototrophs. Thus, excitation profiles of PS II fluorescence are diagnostic of different kinds of oxygenic phototrophs (6). To determine excitation profiles, we measured the relative fluorescence intensity at 680 to 690 nm when samples were excited by wavelengths of 458, 476, 488, 496, 514, 543, and 633 nm. We also detected fluorescence of the antenna pigment phycoerythrin directly at 570 nm with an excitation wavelength of 543 nm (6).
Genomic DNA extraction.
We extracted duplicate community genomic DNA samples from each stromatolite sample (HPDOM-S, HPSOM-I, and HPIRR) and two negative controls with a bead-beating extraction protocol (16). A sample of each stromatolite (∼5 g) was crushed and homogenized in a flame-sterilized steel ore-crusher mortar and pestle (Fisher Scientific), and 0.25 to 0.35 g of crushed stromatolite was used in each DNA extraction. To prevent contamination, procedures were performed in a UV sterilized AirClean 600 PCR workstation (AirClean Systems). Community DNA was resuspended in 40 μl of TE (10 mM Tris, 1 mM EDTA). DNA concentrations were measured with the PicoGreen dsDNA quantitation kit (Molecular Probes) and a fluorometer. Average yields of extracted DNA were ∼5 μg/g of crushed rock.
PCR amplification of rRNA genes.
SSU rRNA genes were amplified by PCR with community DNA as a template and the universal primers 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 1391R (5′-GACGGGCGGTGWGTRCA-3′) (30). Each 50-μl PCR contained ∼50 ng of template DNA, 1.25 U of HotMaster Taq DNA Polymerase (Eppendorf), 1× HotMaster buffer (2.5 mM Mg2+), 1 mg of bovine serum albumin/ml, 50 μM concentrations of each deoxynucleoside triphosphate, and 0.2 μM concentrations of each primer. PCR conditions were 94°C for 2 min, followed by 30 cycles of 94°C for 15 s, 52°C for 15 s, and 65°C for 60 s, with a final 20- min extension at 65°C. PCR controls with no template or extraction control as a template were negative.
Clone library construction.
We constructed one clone library, 96 randomly selected rRNA clones, for each community DNA. PCR-amplified products from four independent PCRs were pooled to reduce the chances of PCR artifacts (28), purified by agarose gel electrophoresis with the Montage DNA gel extraction kit (Millipore), and cloned into the pCR4.0 vector by using the TOPO TA cloning kit and TOP10 chemically competent cells (Invitrogen). For each library, 96 colonies were picked, grown for 16 h, and processed into a quick plasmid preparation (1:4 dilution of overnight culture in Tris-EDTA, incubated at 85°C for 10 min and centrifuged).
Restriction fragment length polymorphism analysis and sequencing.
rRNA gene clones were amplified by PCR from plasmid preparations with the vector primers T3 and T7. Amplified rRNA clones were initially screened by restriction fragment length polymorphism analysis to assess the diversity of clones prior to sequencing (25). The results showed that the libraries were sufficiently diverse to sequence all clones. PCR-amplified rRNA clones were prepared for DNA sequencing with the ExoSAP-it PCR cleanup kit (USB) and sequenced by using DYEnamic ET terminator cycle sequencing kit reaction mixtures on a MegaBACE 1000 DNA sequencer (Amersham Biosciences) with T3 and T7 vector primers. A total of 531 clones were analyzed in the present study.
Phylogenetic analysis and chimera detection.
Raw sequence data were analyzed and assembled into contiguous sequences with the PHRED and PHRAP software packages (17, 18). Sequences were compared to a current database of rRNA gene sequences from GenBank with BLAST (4). These steps were performed with the aid of XplorSeq, an in-house software package developed to automate and manage sequence analysis (D. N. Frank, unpublished software). Complete rRNA gene sequences were exported from XplorSeq and aligned to a database of >10,000 known SSU rRNA sequences (25) with the Arb software package (33). We used the method described by Hugenholtz et al. to identify chimeras (25). One chimera was identified and excluded from further analysis. Phylogenetic analysis was performed with the Arb and PAUP* 4.0 software packages (48) as previously described (23).
Richness and coverage estimators.
To estimate richness and coverage, sequences were collected into operational taxonomic units (OTUs) based on sequence identity at different fixed levels from 95 to 100% in 1% increments with a computer script written in the Python programming language that analyzes distance matrices exported from Arb. OTU richness and coverage estimators were calculated with the software program EstimateS version 6b1 (13) with default settings and 100 random sample repetitions.
Comparative phylogenetic methods.
We compared collections of community rRNA gene sequences with the phylogenetic test (p-test), which calculates differences between sequence sets in phylogenetic trees (34). This method calculates the probability that sequences in two sets compared are derived from the same sequence population with randomly permuted trees. The p-test tests whether there are more distinct lineages in a tree than expected by chance. P values were calculated as the ratio of 1,000 randomly permuted trees with fewer parsimony changes than the original tree. We considered P values of ≤0.05 as significant to reject the hypothesis that two sequence sets were derived from the same sequence population. The p-tests were determined with the Phylonode software module (C. Lozupone and R. Knight, unpublished software). We also used Phylonode to automatically identify internal nodes with significant differences. Each internal node was selected and tested to determine whether removal of that node significantly changed the result from the original tree. We adjusted P values for multiple tests with the Bonferroni correction.
Nucleotide sequence accession numbers.
Sequences of 478 rRNA gene clones determined are available under GenBank accession numbers AY851765 to AY852242. Clone names designate community (HPDOMI, HPDOMS, and HPIRR), replicate library number (1 or 2), and the clone number (e.g., A01).
RESULTS
We determined the composition of microbial communities from two morphologically distinct Hamelin Pool stromatolites with molecular phylogenetic methods. The first stromatolite, HPDOM, had a domal morphology and a smooth, ca. 1- to 2-mm-thick surface layer that was separated from the underlying interior layer in order to compare the microbial composition of the surface sample (HPDOM-S) to that of the interior (HPDOM-I). The second stromatolite, HPIRR, had an irregular morphology and a pustular surface layer that was not easily separated from the interior, and therefore the sample was analyzed in bulk (see Materials and Methods). For each sample, we constructed duplicate libraries of 96 randomly selected clones of SSU rRNA genes amplified by PCR with universal primers and determined their sequences.
Autosimilarity and sample representativeness.
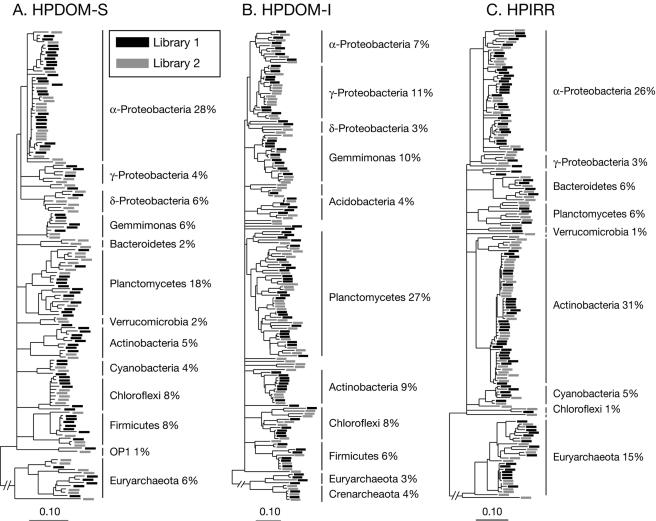
Microbial communities are highly complex, and we do not sample all unique sequences. Consequently, we tested whether the clone libraries were representative samples of rRNA genes amplified and cloned from community DNA by assessment of the autosimilarity (similarity among replicate samples randomly drawn from a population (11) of duplicate clone libraries from each stromatolite with the p-test (see Materials and Methods)). The results showed high autosimilarities with no significant differences in phylogenetic representation between duplicate libraries from the same stromatolite sample (P > 0.995). This indicates clone libraries were representative samples of at least the abundant phylogenetic diversity of sequences cloned from each community. The similarity of duplicate libraries is illustrated by the phylogenetic trees in Fig. 3, which show a relatively even distribution of sequences from duplicate libraries for each community. Based on these results, we combined duplicate libraries from each community and performed all additional analyses on combined sequence sets unless stated otherwise.
FIG. 3.
Phylogeny and representativeness of duplicate clone libraries. (A) HPDOM-S, (B) HPDOM-I, and (C) HPIRR. Each phylogenetic tree contains sequences from duplicate libraries, which, respectively, are represented by black and gray boxes. No significant differences were detected in statistical comparison of the phylogenetic composition of duplicate libraries. The phylogenetic lineages and relative proportion of sequences in the libraries are indicated for abundant sequences in the communities. Scale is 0.10 nucleotide changes per site.
Estimated richness and coverage.
In order to assess the richness (the actual diversity) and the coverage (how well a sample represents the population from which it was drawn) of clone libraries, we used the nonparametric estimators Chao1 and ACE for richness and Good's C and Chao's CACE for coverage (summarized in reference 29). These estimators use species-abundance data, so we collected sequences into OTUs, relatedness groups, based on sequence identity. Relationships between rRNA sequence variation and traditional taxonomic units such as species are not yet clear (47). Consequently, we defined OTUs at fixed thresholds from 95 to 100% sequence identity in 1% increments. Table 1 shows the estimated richness and coverage for OTU thresholds that range from 95 to 100% sequence identity. There was a sharp increase in estimated richness at the 100% threshold in each community, which reflects the large fraction of sequences (30 to 45%) with relatives that differed by <1% but are not identical. This phenomenon, dubbed “microdiversity,” is ubiquitous in environmental surveys, although its significance is not yet clear (1).
TABLE 1.
Observed OTUs, estimated richness, and estimated coveragea
| OTU (%ID)b | Surface
|
Interior
|
Irregular
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Obsc | Chao1 | CACE | Obs | Chao1 | CACE | Obs | Chao1 | CACE | |
| 100 | 120 | 993 | 0.22 | 161 | 1309 | 0.11 | 148 | 1500 | 0.10 |
| 99 | 71 | 178 | 0.66 | 124 | 505 | 0.41 | 90 | 566 | 0.45 |
| 98 | 64 | 132 | 0.71 | 117 | 378 | 0.48 | 78 | 495 | 0.53 |
| 97 | 61 | 117 | 0.74 | 111 | 314 | 0.52 | 66 | 288 | 0.60 |
| 96 | 58 | 128 | 0.75 | 103 | 347 | 0.55 | 62 | 289 | 0.62 |
| 95 | 52 | 107 | 0.79 | 95 | 302 | 0.60 | 54 | 160 | 0.67 |
Estimates of ACE richness and Good's coverage were similar to Chao1 (total number of lineages predicted) and CACE (estimated fraction of lineages sampled), respectively (R2 > 0.98), and are not shown.
OTUs represent groups of sequences related by greater than or equal to the % sequence identity (%ID) indicated.
Obs, number of lineages observed.
We chose 97% sequence identity as a conservative cutoff for differentiation of “species” (21), although this likely underestimates actual species diversity. In order to test whether our libraries were sufficiently large to yield stable and unbiased estimates of richness at the 97% OTU threshold, we compared the estimated richness of each duplicate library to that of the duplicate libraries combined, as proposed by Kemp and Aller (29). We found no significant changes in the estimated richness at the 97% OTU threshold, which suggests the libraries were representative samples. This conclusion also was supported by the degree of coverage estimated for the libraries, which exceeded 50% for each stromatolite sample at the 97% OTU threshold (Table 1).
Comparison to known microbial diversity.
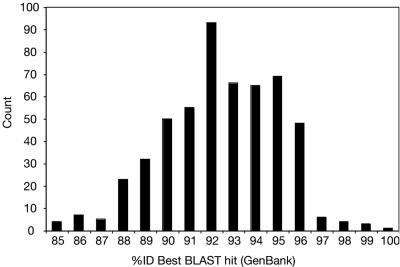
The relatedness of Hamelin Pool sequences to known rRNA gene sequences was determined by comparison to the >200,000 rRNA gene sequences currently in GenBank with BLAST to find the closest match for each Hamelin Pool sequence. Figure 4 shows the distribution of relatedness between Hamelin Pool sequences and their closest match in GenBank. Hamelin Pool sequences were related to known sequences by an average of only 92% sequence identity, which represents microbial diversity substantially different from what is currently known. Sequence relatedness ranged from 85 to 100% identity and varied by phylogenetic group. Table 2 shows the average relatedness of Hamelin Pool sequences to known sequences for each phylogenetic division represented. Only 3% of Hamelin Pool sequences were related to known sequences at ≥97% sequence identity, which suggests 97% of sequences represent new species (47).
FIG. 4.
Distribution of the relatedness of Hamelin Pool rRNA gene sequences to those currently available in GenBank.
TABLE 2.
Phylogenetic affiliation of Shark Bay community rRNA gene clones
| Phylogenetic divisiond | No. of clones (% of community)
|
Avg BLAST (%ID)b | Phylogenetic differencec
|
|||||
|---|---|---|---|---|---|---|---|---|
| HPDOM
|
HPIRR (all clones) | Both (all clones) | HPDOM/HPIRR | HPDOM-S/HPDOM-I | ||||
| Surface | Interior | Combineda | ||||||
| Bacteria | + | + | ||||||
| Acidobacteria | 0.6 | 4.3 | 2.5 | 1.7 | 92.9 | − | ||
| Actinobacteria | 4.7 | 9.2 | 7.0 | 31.8 | 15.3 | 94.5 | + | +/− |
| Bacteroides | 2.3 | 2.2 | 2.3 | 6.3 | 3.6 | 91.2 | + | − |
| Chlamydiaea | 0.5 | 0.3 | 1.1 | 0.6 | 91.0 | − | ||
| Chloroflexi | 7.6 | 7.6 | 7.6 | 1.1 | 5.5 | 89.9 | +/− | + |
| Cyanobacteria | 3.5 | 0.5 | 2.0 | 5.1 | 3.0 | 92.1 | +/− | − |
| Firmicutes | 7.6 | 6.0 | 6.8 | 4.5 | 94.5 | − | ||
| Gemmimonas | 5.3 | 9.8 | 7.5 | 1.7 | 5.6 | 92.8 | − | + |
| Nitrospira | 0.5 | 0.3 | 0.2 | 91.0 | ||||
| OP1* | 1.2 | 0.5 | 0.9 | 0.6 | 94.0 | |||
| OP5* | 1.1 | 0.5 | 0.4 | 87.7 | − | |||
| OS-K* | 0.5 | 0.3 | 0.2 | 93.0 | ||||
| Planctomycetes | 17.5 | 27.2 | 22.4 | 5.7 | 16.9 | 90.6 | +/− | +/− |
| Proteobacteria | 38.0 | 21.2 | 29.6 | 30.1 | 29.6 | + | + | |
| α-Proteobacteria | 28.1 | 7.1 | 17.6 | 26.7 | 20.3 | 93.4 | + | + |
| δ-Proteobacteria | 5.8 | 3.3 | 4.6 | 0.6 | 3.2 | 91.9 | +/− | − |
| γ-Proteobacteria | 4.1 | 10.9 | 7.5 | 2.8 | 6.0 | 92.6 | +/− | − |
| SBR1093* | 0.5 | 0.3 | 0.2 | 94.0 | ||||
| TM6* | 1.2 | 0.6 | 0.4 | 87.5 | ||||
| Verrucomicrobia | 2.3 | 0.5 | 1.4 | 1.1 | 1.3 | 91.9 | − | − |
| WS3* | 0.5 | 0.3 | 0.2 | 91.0 | ||||
| Archaea | + | − | ||||||
| Crenarchaeota | 1.8 | 4.3 | 3.1 | 0.6 | 2.3 | 95.3 | − | − |
| Eurvarchaeota | 6.4 | 2.7 | 4.6 | 15.3 | 8.1 | 90.4 | + | − |
| Total/avg (%) | 100 | 100 | 100 | 100 | 100 | 92.0 | ||
| No. of clones analyzed | 171 | 184 | 355 | 176 | 531 | 531 | ||
Relative proportion of HPDOM clones from surface and interior.
Average sequence identity of closest GenBank relatives for phylogenetic division.
Phylogenetic difference in the bacterial division between indicated communities as measured by “p-test.” P values were adjusted for multiple comparisons (Bonferroni). +, Significant difference with corrected P value; +/−, significant uncorrected, not significant corrected; −, no significant difference; blank, no comparison.
*, candidate division, known by sequence only.
Community composition and structure.
In order to determine phylogenetic relationships of organisms in the stromatolite communities, we aligned the Hamelin Pool sequences and calculated their positions in a phylogenetic tree with over 10,000 rRNA gene sequences representative of known bacterial and archaeal diversity (25) by using the Arb software package. The phylogenetic trees in Fig. 3 illustrate the relative topologies of sequences that represent each community. These trees show that some divisions contain clusters of microdiverse sequences, for example, actinobacterial sequences in HPIRR, whereas others are represented by a broader diversity of sequences, such as planctomycete sequences in all communities.
Hamelin Pool stromatolite communities were comprised of rRNA genes representative of ca. 90% Bacteria and 10% Archaea. We detected no eucaryotic rRNA genes by PCR with universal primers in the present study. The stromatolite communities were complex, comprised of organisms with rRNA genes representative of 19 of the 52 identified divisions of Bacteria (42), as well as representatives of euryarchaeota and crenarchaeota. The most abundant rRNA gene sequences in the combined data set were representative of proteobacteria (∼29%), planctomycetes (∼17%), and actinobacteria (∼15%). In contrast, cyanobacterial sequences comprised <5% of the stromatolite communities. No sequences indicative of novel phylogenetic divisions were detected among the >500 clones analyzed.
Surface compared to endolithic communities.
In order to test the hypothesis that endolithic communities of stromatolites are distinct from surface communities, we compared rRNA gene sequences from HPDOM-I to those of HPDOM-S. The p-test showed a significant difference between the surface and interior communities (P < 0.001). Both communities were comprised of diverse sequences representative of Bacteria and Archaea. Sequences from the interior community represented 19 bacterial divisions. Sequences from the surface community represented 12 of the same bacterial divisions and one additional division, TM6.
With the p-test, we tested (i) whether there was a significant difference between the communities for each lineage in the tree and (ii) which lineages reduced the significant difference between the communities when removed from the tree. Lineages with significant differences are shown in Table 2. Five of twelve bacterial divisions with sequences from both communities showed significant differences (P < 0.05), as did the distribution of euryarchaeota and α-, δ-, and γ-proteobacteria. Except for the firmicutes, lineages with significant differences also contained most sequences. Removal of lineages did not reduce the significant difference between the communities, which suggests the difference between the communities is distributed throughout the phylogenetic tree and not concentrated in any particular lineage.
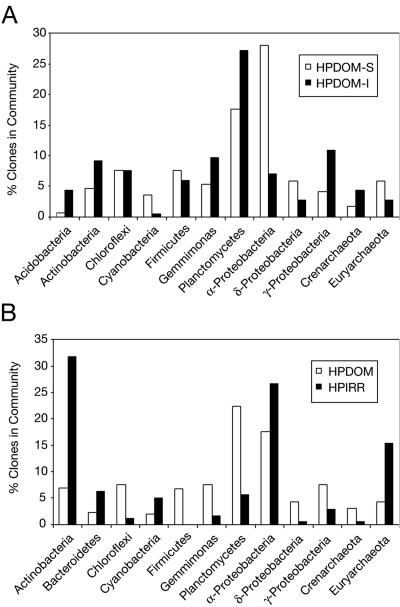
Figure 5A shows the relative distribution of clones from the surface and interior communities for the most abundant phylogenetic lineages. There were no identical sequences and few closely related sequences shared between the surface and interior communities. Only ca. 20% of sequences from the two communities were related at 97% or greater sequence identity, which suggests about 80% of each community differed from the others at a minimum of the species level. The most obvious differences in sequence composition between layers were in relative abundances of α-proteobacterial, planctomycete, and cyanobacterial sequences, as shown in Fig. 5A.
FIG. 5.
Comparison of the most abundant sequences by phylogenetic division for (A) HPDOM-S and HPDOM-I and (B) HPDOM and HPIRR.
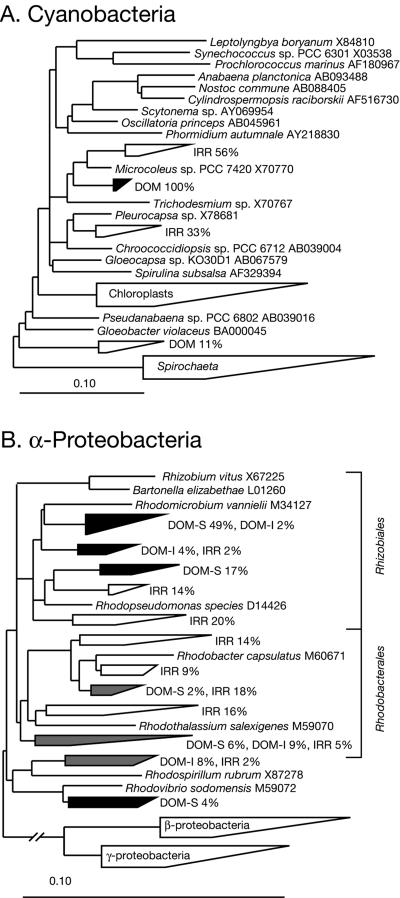
Cyanobacterial sequences were a remarkably small fraction of both communities, but were more abundant in the surface, which receives the most solar radiation. Phylogenetic analysis of the cyanobacterial sequences is summarized in Fig. 6A and showed that sequences from the surface and the interior were ≥98% identical. The HPDOM cyanobacterial sequences were most closely related to the filamentous Microcoleus spp., but by only ∼94% sequence identity, which indicates the most abundant cyanobacteria in HPDOM are relatively novel compared to known species. Sequences representative of α-proteobacteria also were more abundant in the surface community, and all were most closely related to cultured organisms known for anoxygenic photosynthesis, as shown in Fig. 6B. Planctomycete sequences were enriched in the interior community, and there were significant phylogenetic differences from exterior community, as shown in Table 2. Planctomycete sequences from both communities were substantially different from known sequences, with an average of ∼10% rRNA sequence divergence.
FIG. 6.
Diagramatic phylogenetic trees of Hamelin Pool stromatolite sequences and their cultivated relatives. Reference sequences of cultured representatives are shown in italics with associated GenBank accession numbers. Groups of related Hamelin Pool sequences are represented by wedges. The range of sequence divergence is represented by the lengths of the top and bottom edges. Open wedges contain sequences from HPIRR, black wedges contain sequences from HPDOM, gray wedges contain sequences from both, and wedges that contain reference sequences only are labeled inside the wedge. (A) Cyanobacterial sequences. Percentages indicate the fraction of total cyanobacterial sequences from HPDOM (DOM) or HPIRR (IRR). There were seven HPDOM sequences and nine HPIRR se-quences. (B) α-Proteobacterial sequences. Percentages indicate the fraction of total α-proteobacterial sequences from HPDOM (DOM) or HPIRR (IRR). The fraction of total HPDOM sequences from the surface (DOM-S) and interior (DOM-I) are shown. There were 53 HPDOM α-proteobacterial sequences and 45 HPIRR α-proteobacterial sequences.
Stromatolite communities and morphotypes.
In order to test the hypothesis that different microbial communities are associated with different stromatolite morphotypes, we combined sequences from the HPDOM-S and HPDOM-I samples (HPDOM) and compared them to HPIRR sequences. The p-test showed a significant phylogenetic difference between the communities (P < 0.001), which suggests that the communities are comprised of distinct microbial populations. Most sequences representative of particular phylogenetic groups in both communities contributed to this significant difference, as shown in Table 2, and no lineages reduced the significant difference between the communities when removed from the tree.
The communities were similar in their relatively low abundances of cyanobacterial sequences and high abundances of α-proteobacterial sequences, which were most closely related to known anoxygenic phototrophs (Fig. 6B). HPIRR had two distinct groups of cyanobacterial sequences, one most closely related to Microcoleus spp. sequences and the other to sequences of nonfilamentous Pleurocapsa spp. (Fig. 6A).
There were more differences than similarities in the composition of HPDOM and HPIRR at the bacterial phylogenetic division level, as shown in Table 2 and Fig. 5B. Only about 10% of sequences from the two communities were related at ≥97% sequence identity, and only about 15% of sequences were related at ≥95% sequence identity. The most obvious differences in composition of the two communities were in sequences representative of actinobacteria and euryarchaeota, which were more abundant in HPIRR, and planctomycetes, which were more abundant in HPDOM.
Microscopy.
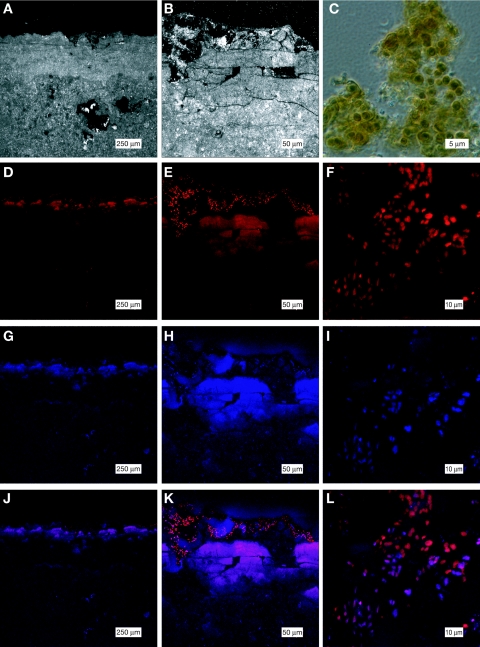
In order to determine the distribution of oxygenic phototrophs in Hamelin Pool stromatolite communities, we imaged fluorescence indicative of pigments associated with oxygenic photosynthesis in thin sections by LSCM. We also analyzed spectral signatures of fluorescent cells by LSCM; these signatures are diagnostic of photosynthetic pigments associated with different groups of oxygenic phototrophs (see Materials and Methods). Fluorescence was induced only at 633 nm and not other wavelengths (458, 476, 488, 496, 514, and 543 nm), which is indicative of cyanobacteria in vivo and contraindicative of cells that contain chloroplasts, which should be excited at 458 nm (6). Figure 7 shows the spatial distribution and spectral signature of fluorescent cells in HPDOM. All observable fluorescence was located in a layer ∼0.2 mm thick at the surface of the samples. Individual cells with diameters of 2 to 4 μm were observed at the surface. The underlying band of diffuse fluorescence was due to scattered fluorescence emitted by cells embedded in the mineral matrix. The excitation observed in cells at 633 nm and corresponding emission at 685 nm are diagnostic of phycocyanin. A subset of fluorescent cells shown in Fig. 7G to I also exhibited fluorescence diagnostic of phycoerythrin (excitation, 543 nm; emission, 590 nm) (6).
FIG. 7.
Cyanobacterial cells imaged in thin sections of the surface layer of HPDOM by LSCM shown at three magnifications. The top row shows reflected light images (A and B) and a phase-contrast image of cyanobacterial cells scraped from the surface of HPDOM (C). (D to F) Row 2 shows phycocyanin fluorescence (F 633/685). (G to I) Row 3 shows phycoerythrin fluorescence (F 543/590). (J to L) Row 4 shows the merge of images in rows 2 and 3.
DISCUSSION
Molecular methods.
Culture-independent molecular methods overcome many problems associated with the study of natural microbial communities by culture and microscopy. In principal, universal rRNA libraries provide a snapshot of the relative proportions of phylogenetic types in a community, and some properties of these individuals can be inferred from phylogenetic information. Organisms representative of a particular phylogenetic group are expected to have properties common to the group.
We acknowledge, however, that there are potential experimental artifacts, which can introduce biases in how well clone libraries represent the actual rRNA gene community in a sample. These include variable PCR amplification that results from primer selectivity and differential extraction of genomic DNA from environmental samples (28, 51). Different species may also have variable numbers of rRNA gene copies (2). Therefore, the relative abundance of rRNA genes surveyed from a sample with PCR-based methods may not correspond directly to the relative abundance of organisms in the original sample.
Nonetheless, studies show a significant correspondence between abundances measured with clone libraries and assessed with other methods, such as fluorescent in situ hybridization (27), rRNA hybridization (35), and clone libraries made by reverse transcription of rRNA (38). General correspondence is also observed between libraries made with different PCR primer combinations (20, 46). Collectively, the results show that, when used carefully, these methods accurately identify the most abundant organisms present in the original sample. Although we cannot rule out systematic biases in the present study, the high autosimilarity of our duplicate libraries indicates there was little random bias in the methods.
Sample representativeness.
The statistical treatment of our results suggests that the clone libraries were representative of the population of sequences amplified and cloned from the stromatolite communities. Sample collection is a complex and essential consideration in general ecology (11), which attracts attention in molecular microbial ecology as efforts to understand microbial diversity increase and the cost of DNA sequencing decreases (7, 19, 26). Natural microbial communities tend to be highly complex, and the in situ diversity is seldom entirely sampled with molecular methods. Consequently, for comparison and estimation of community compositions it is essential to assess how well samples represent communities (11, 34). We assessed sample representativeness with two methods that test different measures of autosimilarity. This approach has been proposed as a more objective and efficient means to assess sample representativeness than other methods currently used, such as estimation of rarefaction or coverage (11, 29).
Although results from comparative phylogenetic methods and rarefaction analysis suggested our samples were representative, we propose that measurement of the autosimilarity of sequence data by phylogenetic methods is more robust and objective than measurement by OTU-based methods because the latter approach imposes a taxonomic species concept on phylogenetic data. This can lead to questionable assumptions about the taxonomic relationships of organisms based on rRNA sequence variation. For example, evidence suggests the relationship between taxonomic units and SSU rRNA gene sequence varies by phylogenetic division (19), but the details have not been articulated. Stackebrandt and Goebel (47) studied this relationship in Bacteria and concluded that strains with SSU rRNA genes that are <97% identical are most likely different species, but strains with SSU rRNA genes that are >97% identical could be the same species or different species.
Phylogenetic composition.
The microbial communities of the Hamelin Pool stromatolites are comprised of ∼90% Bacteria and ∼10% Archaea, similar to results from other studies of different kinds of microbial mats (45). Much of the novel diversity we found was associated with bacterial phylogenetic divisions that are considered highly sampled, such as the α-proteobacteria, where we found sequences <90% identical to known sequences. This shows there is undiscovered novel diversity even in lineages represented by many sequences. We also obtained sequences characteristic of five candidate divisions of Bacteria, divisions with no cultured representatives that previously had been encountered in other environmental surveys (24, 42). These sequences constitute new and independent evidence that these candidate divisions in fact represent unique bacterial lineages.
We found sequences representative of a relatively low abundance of archaea, but both euryarchaeotes and crenarchaeotes were represented. Little is known about the metabolic diversity of archaea, and it is therefore not possible to assess their potential roles in the stromatolite communities. Euryarchaeal clones were most closely related to halophilic archaea, which is consistent with the hypersaline environment of the Hamelin Pool. Crenarchaeal rRNA sequences were not specifically associated with those of any cultured organisms. Absence of eucaryotic sequences suggests that if they were present in these stromatolites, they were relatively scarce, in contrast to previous observations (5). Our results corroborate the findings of Reid et al. (44), who also found that diatoms and other eucaryotes are scarce in Bahamian stromatolites.
Primary productivity, biomass, and community structure.
The emergent properties of stromatolite communities are governed by how community members contribute to the biomass, energy flow, and mineralization of the community. Our results provide a snapshot of the relative proportions of phylogenetic types in the stromatolite communities. The standing crop biomass of a community is a function of the relative abundance and size of its members. Community members are sustained by a flow of energy, which primary producers ultimately generate from an external source of energy, such as light or chemical energy. Both the relative size and the turnover rate of primary producers influences the number of individuals needed to sustain a community. A distinction between the overall biomass of the stromatolite communities and that of its primary producers is potentially important to understand the processes that form stromatolites.
The low abundance of cyanobacterial rRNA genes in the Hamelin Pool communities suggests that other kinds of organisms comprise much of the biomass. Surveys of hypersaline microbial-mat communities (45) and Bahamian stromatolite communities (L. K. Baumgartner, N. R. Pace, and P. T. Visscher, unpublished data) also find low relative proportions of cyanobacterial rRNA genes in universal libraries. Biogeochemical studies of the Bahamian stromatolites show most oxygenic photosynthesis occurs in a submillimeter layer near the surface (50). This suggests that the distribution of oxygenic phototrophs we observed in Hamelin Pool communities is similar to that of Bahamian communities.
If the main source of primary productivity in Hamelin Pool communities is oxygenic photosynthesis by cyanobacteria, then these stromatolites are supported by a relatively small fraction of the community (≤5% of rRNA genes). Such a scenario seems possible considering cyanobacteria can have very high turnover rates (39), and the cyanobacterial cells we observed microscopically had on average about twice the diameter of other cells. This observation implies cyanobacterial cells in the community have cell volumes about eight times larger than other organisms. In addition, there is no obvious external source of chemical energy in Hamelin Pool sufficient to drive new net primary production by metabolisms other than oxygenic photosynthesis.
Nonetheless, we find evidence that potential anoxygenic phototrophs are numerically abundant in the stromatolite community and thus may represent a considerable fraction of the standing biomass. Such organisms undoubtedly contribute to gross primary productivity in the community by photosynthesis, but anoxygenic phototrophs rely on a supply of electron donors such as sulfide, hydrogen, and reduced organics. Energy that typically produces such electron donors in comparable microbial systems originates from oxygenic phototrophs, cyanobacteria, and is transformed by a complex web of metabolisms (14). For example, sulfate reduction occurs in Bahamian stromatolites in an anoxic layer several millimeters below the photosynthetic zone (50). Such a sulfidic layer could support anoxygenic photosynthesis in stromatolite communities. Most other Hamelin Pool sequences were only distantly related to those of cultivated organisms, and so the metabolisms of the organisms they represent are not predictable.
Geological implications.
The significant differences we found between the HPDOM and HPIRR communities show that stromatolite morphology may be attributable to different microbial communities, although more comparisons are required to confirm this hypothesis. Ancient stromatolites have many morphologies (reviewed in reference 22), which may indicate that different microbial communities formed the basis for each unique structure. Variations in stromatolite morphology could result, for instance, from the nature of microbial extracellular materials, which might trap sediments differently or attract different community members.
Although cyanobacteria do not dominate Hamelin Pool stromatolite communities numerically, they likely play the main role in primary production. Our results suggest that novel and diverse microorganisms form much of the structure and fabric of the communities and, therefore, must contribute substantially to stromatolite formation. It is possible that microbial communities fueled by metabolisms other than oxygenic photosynthesis formed some ancient stromatolites. For example, stromatolites in Yellowstone hotsprings are built by communities in which the primary producers are considered anoxygenic photosynthetic chloroflexi fueled by electron donors derived from hydrothermal fluids (15). Therefore, we conclude that ancient stromatolites cannot be taken alone as evidence for oxygenic photosynthesis.
Acknowledgments
We thank members of the Pace Lab and in particular J. K. Harris, J. R. Spear, L. K. Baumgartner, and R. E. Ley for excellent discussions on the phylogeny of microbial communities. We also thank R. Knight and C. Lozupone for assistance with the Phylonode software package and valuable discussion. We acknowledge D. Stefoni and K. McNamara (CALM, Western Australia) for the permit to sample at the Shark Bay World Heritage site.
This study was supported by NSF grant DEB-0085490 to N.R.P. and the NASA Cooperative Agreement with the University of Colorado Center for Astrobiology to N.R.P. and S.J.M. D.P. thanks the Fonds de Recherche sur la Nature et les Technologies du Québec and the NAI for graduate support.
REFERENCES
- 1.Acinas, S. G., V. Klepac-Ceraj, D. E. Hunt, C. Pharino, I. Ceraj, D. L. Distel, and M. F. Polz. 2004. Fine-scale phylogenetic architecture of a complex bacterial community. Nature 430:551-554. [DOI] [PubMed] [Google Scholar]
- 2.Acinas, S. G., L. A. Marcelino, V. Klepac-Ceraj, and M. F. Polz. 2004. Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J. Bacteriol. 186:2629-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Qassab, S., W. J. Lee, S. Murray, A. G. B. Simpson, and D. J. Patterson. 2002. Flagellates from stromatolites and surrounding sediments in Shark Bay, Western Australia. Acta Protozool. 41:91-144. [Google Scholar]
- 4.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Awramik, S. M., and R. Riding. 1988. Role of algal Eukaryotes in subtidal columnar stromatolite formation. Proc. Natl. Acad. Sci. USA 85:1327-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beutler, M., K. H. Wiltshire, B. Meyer, C. Moldaenke, C. Luring, M. Meyerhofer, U. P. Hansen, and H. Dau. 2002. A fluorometric method for the differentiation of algal populations in vivo and in situ. Photosynth. Res. 72:39-53. [DOI] [PubMed] [Google Scholar]
- 7.Bohannan, B. J. M., and J. Hughes. 2003. New approaches to analyzing microbial biodiversity data. Curr. Opin. Microbiol. 6:282-287. [DOI] [PubMed] [Google Scholar]
- 8.Buick, R., J. S. R. Dunlop, and D. I. Groves. 1981. Stromatolite recognition in ancient rocks: an appraisal of irregularly laminated structures in an early Archean chert-barite unit from North-Pole, Western-Australia. Alcheringa 5:161-181. [Google Scholar]
- 9.Burns, B. P., F. Goh, M. Allen, and B. A. Neilan. 2004. Microbial diversity of extant stromatolites in the hypersaline marine environment of Shark Bay, Australia. Environ. Microbiol. 6:1096-1101. [DOI] [PubMed] [Google Scholar]
- 10.Byerly, G. R., D. R. Lowe, and M. M. Walsh. 1986. Stromatolites from the 3,300-3,500-Myr Swaziland Supergroup, Barberton Mountain Land, South-Africa. Nature 319:489-491. [Google Scholar]
- 11.Cao, Y., D. D. Williams, and D. P. Larsen. 2002. Comparison of ecological communities: the problem of sample representativeness. Ecol. Mono. 72:41-56. [Google Scholar]
- 12.Chivas, A. R., T. Torgersen, and H. A. Polach. 1990. Growth-rates and Holocene development of stromatolites from Shark Bay, Western-Australia. Aust. J. Earth Sci. 37:113-121. [Google Scholar]
- 13.Colwell, R. 1997. EstimateS: statistical estimation of species richness and shared species from samples. User's Guide, version 5, and application. [Online.] http://viceroy.eeb.uconn.edu/estimates.
- 14.Des Marais, D. J. 2003. Biogeochemistry of hypersaline microbial mats illustrates the dynamics of modern microbial ecosystems and the early evolution of the biosphere. Biol. Bull. 204:160-167. [DOI] [PubMed] [Google Scholar]
- 15.Doemel, W. N., and T. D. Brock. 1974. Bacterial stromatolites: origin of laminations. Science 184:1083-1085. [DOI] [PubMed] [Google Scholar]
- 16.Dojka, M. A., P. Hugenholtz, S. K. Haack, and N. R. Pace. 1998. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl. Environ. Microbiol. 64:3869-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 18.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 19.Forney, L. J., X. Zhou, and C. J. Brown. 2004. Molecular microbial ecology: land of the one-eyed king. Curr. Opin. Microbiol. 7:210-220. [DOI] [PubMed] [Google Scholar]
- 20.Frank, D. N., G. B. Spiegelman, W. Davis, E. Wagner, E. Lyons, and N. R. Pace. 2003. Culture-independent molecular analysis of microbial constituents of the healthy human outer ear. J. Clin. Microbiol. 41:295-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goebel, B. M., and E. Stackebrandt. 1994. Cultural and phylogenetic analysis of mixed microbial populations found in natural and commercial bioleaching environments. Appl. Environ. Microbiol. 60:1614-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grotzinger, J. P., and A. H. Knoll. 1999. Stromatolites in Precambrian carbonates: evolutionary mileposts or environmental dipsticks? Annu. Rev. Earth Plan. Sci. 27:313-358. [DOI] [PubMed] [Google Scholar]
- 23.Harris, J. K., S. T. Kelley, and N. R. Pace. 2004. New perspective on uncultured bacterial phylogenetic division OP11. Appl. Environ. Microbiol. 70:845-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hugenholtz, P., C. Pitulle, K. L. Hershberger, and N. R. Pace. 1998. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180:366-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hughes, J. B., J. J. Hellmann, T. H. Ricketts, and B. J. M. Bohannan. 2001. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 67:4399-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Juretschko, S., A. Loy, A. Lehner, and M. Wagner. 2002. The microbial community composition of a nitrifying-denitrifying activated sludge from an industrial sewage treatment plant analyzed by the full-cycle rRNA approach. Syst. Appl. Microbiol. 25:84-99. [DOI] [PubMed] [Google Scholar]
- 28.Kanagawa, T. 2003. Bias and artifacts in multitemplate polymerase chain reactions (PCR). J. Biosci. Eng. 96:317-323. [DOI] [PubMed] [Google Scholar]
- 29.Kemp, P. F., and J. Y. Aller. 2004. Bacterial diversity in aquatic and other environments: what 16S rDNA libraries can tell us. FEMS Microbiol. Ecol. 47:161-177. [DOI] [PubMed] [Google Scholar]
- 30.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Inc., New York, N.Y.
- 31.Logan, B. W., and D. E. Cebulski. 1970. Sedimentary environments of Shark Bay, Western Australia, p. 1-37. In B. W. Logan, G. R. Davies, J. F. Read, and D. E. Cebulski (ed.), Carbonate sedimentation and environments, Shark Bay, Western Australia., vol. memoir 13. American Association of Petroleum Geologists, Tulsa, Okla. [Google Scholar]
- 32.Lowe, D. R. 1980. Stromatolites 3,400-Myr old from the Archean of Western-Australia. Nature 284:441-443. [Google Scholar]
- 33.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin, A. P. 2002. Phylogenetic approaches for describing and comparing the diversity of microbial communities. Appl. Environ. Microbiol. 68:3673-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Massana, R., A. E. Murray, C. M. Preston, and E. F. DeLong. 1997. Vertical distribution and phylogenetic characterization of marine planktonic Archaea in the Santa Barbara Channel. Appl. Environ. Microbiol. 63:50-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neilan, B. A., B. P. Burns, D. A. Relman, and D. R. Lowe. 2002. Molecular identification of cyanobacteria associated with stromatolites from distinct geographical locations. Astrobiology 2:271-280. [DOI] [PubMed] [Google Scholar]
- 37.Noffke, N., G. Gerdes, and T. Klenke. 2003. Benthic cyanobacteria and their influence on the sedimentary dynamics of peritidal depositional systems (siliciclastic, evaporitic salty, and evaporitic carbonatic). Earth-Sci. Rev. 62:163-176. [Google Scholar]
- 38.Nogales, B., E. R. B. Moore, E. Llobet-Brossa, R. Rossello-Mora, R. Amann, and K. N. Timmis. 2001. Combined use of 16S ribosomal DNA and 16S rRNA to study the bacterial community of polychlorinated biphenyl-polluted soil. Appl. Environ. Microbiol. 67:1874-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Overmann, J., and F. Garcia-Pichel. 2001. The phototrophic way of life, p. 1. In M. Dworkin, A. Balows, H. G. Truper, W. Harder, and K. Schleifer (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed. Springer-Verlag, New York, N.Y.
- 40.Pace, N. R. 1997. A molecular view of microbial diversity and the biosphere. Science 276:734-740. [DOI] [PubMed] [Google Scholar]
- 41.Palmisano, A. C., R. E. Summons, S. E. Cronin, and D. J. Desmarais. 1989. Lipophilic pigments from cyanobacterial (blue-green-algal) and diatom mats in Hamelin Pool, Shark Bay, Western-Australia. J. Phycol. 25:655-661. [DOI] [PubMed] [Google Scholar]
- 42.Rappe, M. S., and S. J. Giovannoni. 2003. The uncultured microbial majority. Annu. Rev. Microbiol. 57:369-394. [DOI] [PubMed] [Google Scholar]
- 43.Reid, R. P., N. P. James, I. G. Macintyre, C. P. Dupraz, and R. V. Burne. 2003. Shark Bay stromatolites: microfabrics and reinterpretation of origins. Facies 49:299-324. [Google Scholar]
- 44.Reid, R. P., P. T. Visscher, A. W. Decho, J. F. Stolz, B. M. Bebout, C. Dupraz, L. G. Macintyre, H. W. Paerl, J. L. Pinckney, L. Prufert-Bebout, T. F. Steppe, and D. J. Des Marais. 2000. The role of microbes in accretion, lamination and early lithification of modern marine stromatolites. Nature 406:989-992. [DOI] [PubMed] [Google Scholar]
- 45.Spear, J. R., R. E. Ley, A. B. Berger, and N. R. Pace. 2003. Complexity in natural microbial ecosystems: the Guerrero Negro experience. Biol. Bull. 204:168-173. [DOI] [PubMed] [Google Scholar]
- 46.Spear, J. R., J. J. Walker, T. M. McCollom, and N. R. Pace. 2005. Hydrogen and bioenergetics in the Yellowstone geothermal ecosystem. Proc. Natl. Acad. Sci. USA 102:2555-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 48.Swofford, D. L. 2002. PAUP*: phylogenetic analysis using parsimony (*and other methods), command reference version 4.0. Sinauer Associates, Sunderland, Mass.
- 49.Visscher, P. T., R. P. Reid, and B. M. Bebout. 2000. Microscale observations of sulfate reduction: correlation of microbial activity with lithified micritic laminae in modern marine stromatolites. Geology 28:919-922. [Google Scholar]
- 50.Visscher, P. T., R. P. Reid, B. M. Bebout, S. E. Hoeft, I. G. Macintyre, and J. A. Thompson. 1998. Formation of lithified micritic laminae in modern marine stromatolites (Bahamas): the role of sulfur cycling. Am. Miner. 83:1482-1493. [Google Scholar]
- 51.von Wintzingerode, F., U. B. Gobel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 52.Walter, M. R. 1976. Introduction, p. 1-3. In M. R. Walter (ed.), Stromatolites. Elsevier, Amsterdam, The Netherlands.
- 53.Walter, M. R., R. Buick, and J. S. R. Dunlop. 1980. Stromatolites 3,400-3,500 Myr old from the North-Pole area, Western-Australia. Nature 284:443-445. [Google Scholar]