Abstract
PCR-based molecular analyses can be hindered by the presence of unwanted or dominant DNA templates that reduce or eliminate detection of alternate templates. We describe here a reaction in which such templates can be exclusively digested by endonuclease restriction, leaving all other DNAs unmodified. After such a modification, the digested template is no longer available for PCR amplification, while nontarget DNAs remain intact and can be amplified. We demonstrate the application of this method and use denaturing gradient gel electrophoresis to ascertain the removal of target DNA templates and the subsequent enhanced amplification of nondigested DNAs. Specifically, plastid 16S rRNA genes were exclusively digested from environmental DNA extracted from plant roots. In addition, pure culture and environmental DNA extracts were spiked with various amounts of genomic DNA extracted from Streptomyces spp., and selective restriction of the Streptomyces 16S rRNA genes via the suicide polymerase endonuclease restriction PCR method was employed to remove the amended DNA.
In the field of molecular biology, the predominance of a DNA template within a single sample can bias or restrict molecular analyses. In environmental studies, for example, PCR-based analyses of cyanobacterial diversity in microbial mat systems have been limited by the presence of a dominant but known cyanobacterial population, Microcoleus chthonoplastes (2). Likewise, we found that DNA from the soil flagellate Heteromita globosa masked PCR-based rRNA gene analyses of fungal populations (4). In studies of bacterial populations associated with roots, plastid DNA, which is readily amplified with general bacterial primer sets, can easily predominate in total DNA extracts and subsequent PCR products (11, 13, 14). In all of these cases, the overwhelming presence of a single DNA template diminished the capacity of molecular tools to assess and detect diversity. In addition, the dominant DNA templates contained no mismatches with the general primers applied, and thus, more stringent PCR conditions would not have mitigated their impact. Such conditions are not exclusive to environmental microbiology. Hancock et al. (5) described a condition of heteroplasmic human mitochondrial DNA mutations where mutant and wild-type sequences coexist in the same individual. The mutant DNA is a small proportion of the wild type and difficult to detect at low concentrations. In that study, and elsewhere, those authors applied a methodology termed “PCR clamping” using peptide nucleic acids (PNAs) to suppress PCR amplification of wild-type or dominant sequences (3, 5, 12; for reviews, see references 10 and 15). PCR clamping using PNAs is one of several methods for overcoming the problem of dominant DNA templates. For example, oligonucleotide primers can be modified with a 3′ phosphoramidite spacer that inhibits PCR extension. Such primers, specific to Escherichia coli 16S rRNA genes, were used to inhibit amplification of E. coli rRNA genes from a bacterial artificial chromosome library (7). For specific reduction of the impact of plastid DNA, Nikolausz et al. (11) were forced to employ RNA extraction coupled with reverse transcriptase to avoid the predominance of plastid DNA and found the proportion of plastid cDNA much reduced. Elsewhere, Bebout et al. (2) employed freeze-thaw treatments of total DNA extracts to reduce the predominance of the cyanobacterium M. chthonoplastes in PCR-based analyses of cyanobacterial diversity.
We have developed a new technique to reduce or eliminate the impact of dominant DNAs on molecular analyses by target-specific endonuclease restriction. The method, termed suicide polymerase endonuclease restriction (SuPER), allows targeted DNA to be restricted exclusively while nontarget DNAs remain unmodified. To achieve this, three reactions are conducted simultaneously; these include primer annealing at stringent temperature, Taq DNA polymerase elongation, and endonuclease restriction. This technique can be applied in a similar manner to “PCR clamping” by PNAs or by application of primers modified to inhibit extension but does not require the synthesis of specialized primers. In addition, the method destroys the target DNA by endonuclease restriction and may be modified to assist in subsequent reactions that are not PCR based (i.e., generation of bacterial artificial chromosome libraries from genomic DNA). The SuPER method, in addition to effecting a near-complete removal of target DNA, is adaptable to any DNA template. This method does require that the target template be sufficiently different from nontarget templates to design a specific primer; however, only a single conserved location within a gene or target DNA sequence is required for the operation of this method. The method is adaptable, simple to optimize and perform, and inexpensive.
To demonstrate the concept and the effectiveness of the method, we conducted SuPER reactions with several environmental DNA extracts and these extracts spiked with various amounts of DNA from bacterial pure cultures. SuPER reactions were employed to digest exclusively plastid or Streptomyces 16S rRNA genes. To verify the digestion of the targeted DNA after the SuPER reaction, we subsequently conducted general bacterial PCR amplification and denaturing gradient gel electrophoresis (DGGE) analyses of the original and modified DNA samples. In addition, general guidelines for the SuPER reaction and subsequent PCR are presented.
MATERIALS AND METHODS
SuPER digest.
Primers for the SuPER reaction were designed using the probe design feature of the phylogenetic analysis software package ARB (8). Potential primers were evaluated by sequence analyses conducted using BLAST searches (1) and the ARB Probe Match feature. Primer sequences are listed in Table 1. Thermostable restriction enzymes were evaluated for use in the SuPER reaction by using Webcutter 2.0 (http://www.firstmarket.com/cutter/cut2.html) and the ARB phylogenetic package.
TABLE 1.
Primer sequences
| Primer | Sequence (5′→3′) | SSU rRNA gene location (positions)b | Target population | Reference or source |
|---|---|---|---|---|
| SuPER reaction | ||||
| Q491-F | GGG GAA TAA GCA TCG GCT AAC TCT G | 491-515 | Plastids | This study |
| Q491-R | CAG AGT TAG CCG ATG CTT ATT CCC C | 515-491 | Plastids | This study |
| S661-F | GTA GGG GAG ATC GGA ATT | 661-678 | Streptomyces | 6 |
| S661-R | AAT TCC GAT CTC CCC TAC | 678-661 | Streptomyces | 6 |
| Bacterial 16S rRNA gene amplification | ||||
| 341-Fa | CCT ACG GGA GGC AGC AG | 341-357 | Bacteria | 9 |
| 907-R | CCG TCA ATT CMT TTG AGT TT | 926-907 | Bacteria | 9 |
Contains the following 40-base GC clamp attached to the 5′ end to enhance separation in DGGE analyses: CGC CCG CCG CGC CCC GCG CCC GTC CCG CCG CCC CCG CCC G.
SSU, small subunit.
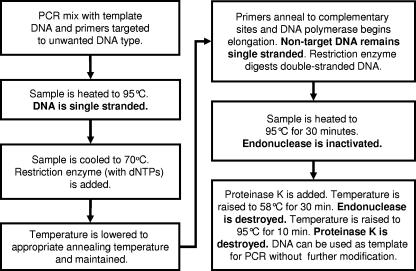
SuPER reactions were conducted according to the flow chart shown in Fig. 1. The theoretical operation of the reaction is shown in Fig. 2. Reaction mixtures for the SuPER stage were formulated as follows, in a final reaction volume of 25 μl: 0.75 U of Taq DNA polymerase (Red Taq; Sigma Chemical Co.), 1× Sigma PCR buffer, 3 μg bovine serum albumin (Roche Diagnostics, Mannheim, Germany), and 0.5 pmol of each primer. Final magnesium concentrations were adjusted by the addition of 25 mM MgCl2 to 4.0 mM in reactions employing the Streptomyces primers. SuPER reactions for the removal of plastid DNA were conducted with 1.5 mM MgCl2, already present in the PCR buffer.
FIG. 1.
Description and flow chart of the SuPER reaction prior to PCR analysis. dNTPs, deoxynucleoside triphosphates.
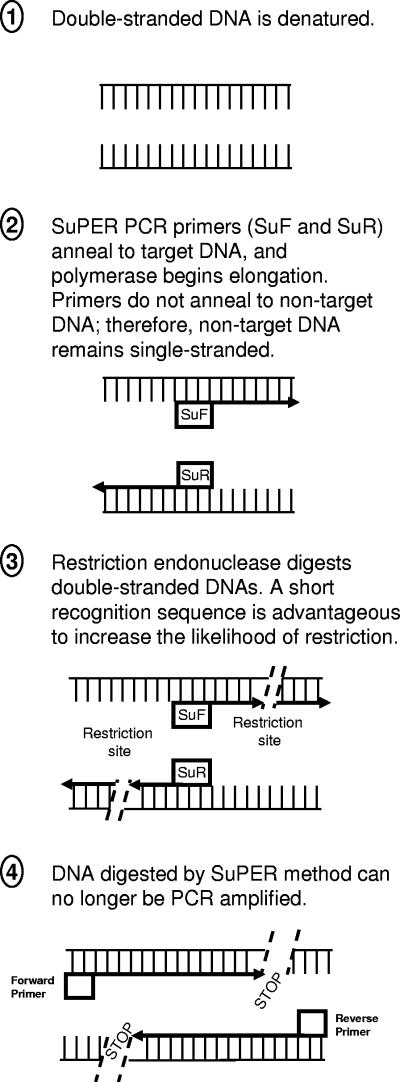
FIG. 2.
Diagram of the SuPER PCR method. Note that restriction enzymes may cut numerous times within the appropriate region depending upon DNA sequence and enzyme applied.
SuPER reaction mixtures containing DNA templates were initially heated to 95°C for 3 min, after which the temperature was dropped to 70°C. The restriction enzyme Tsp509I (New England Biolabs, Beverly, MA) combined with a deoxynucleoside triphosphate solution (200 μM final deoxynucleoside triphosphate concentration in the reaction mixture) was preheated to 70°C in the thermocycler block and then added to each reaction tube in amounts ranging from 0.1 to 5.0 U per 25 μl. For exclusive restriction of plastid 16S rRNA genes, the reactions were maintained at 70°C for 60 min, allowing simultaneous annealing of primers to plastid DNA, elongation of fragments, and digestion of double-stranded DNA. For Streptomyces reactions, in which shorter primers were employed, the incubation temperature was lowered to 60°C (for stringent digestion of Streptomyces 16S rRNA genes) and maintained for 60 min. Stringent reaction temperatures for SuPER reactions were determined using a temperature gradient thermocycler, and for each primer set, a series of reaction temperatures was tested and verified by DGGE analysis, as described below. All reaction mixtures were then heated to 95°C for 30 min to inactivate the restriction enzyme. After thermal inactivation, the samples were cooled to 4°C, and proteinase K (20-mg/ml solution; Amresco, Solon, OH) was added to each reaction mixture to a final concentration of 3.3 μg/μl. The reaction mixtures were then heated to 58°C for 30 min to digest polymerase and endonuclease and then heated to 95°C for 10 min to inactivate the proteinase K. All these reactions were conducted within the same tube incubated within a Tgradient thermal cycler (Whatman Biometra, Germany). After inactivation of proteinase K, the samples were used directly as a template for PCR amplification with bacterial primers as described below.
PCR amplification with general bacterial 16S rRNA gene primers.
Fragments of plastid and bacterial 16S rRNA genes were amplified from DNA templates using the primer set 341-F and 907-R (see Table 1 for primer sequences). Both primers are routinely applied for amplification of 16S rRNA genes from the bacterial domain (9) and match perfectly the 16S rRNA genes of plastids as well as most Streptomyces and Clavibacter spp. PCR mixes, per 50 μl, consisted of 1.5 U of Taq DNA polymerase (Red Taq; Sigma Chemical Co.), Sigma PCR buffer, 0.2 mM nucleotide mixture (Promega, Madison, WI), 6.25 μg bovine serum albumin (Roche Diagnostics, Mannheim, Germany), and 25 pmol of each primer. A final magnesium concentration of 1.5 mM or 4 mM was present. PCR mixes were initially denatured for 3 min at 95°C and then cycled 35 times through three steps: denaturation (94°C and 30 s), annealing (56°C and 30 s), and elongation (72°C and 30 s). A 2-min incubation at 72°C was added to the end of each PCR program. In some cases, weak PCR amplification after the SuPER reaction was overcome by increasing the magnesium concentration in the general bacterial PCR but retaining the same annealing temperature.
Verification of target template digestion.
DGGE was used to characterize PCR-amplified DNA in order to detect digestion of unwanted DNA templates. DGGE analyses were performed with a D-Gene system (Bio-Rad, CA) using the following ingredients and conditions: 1× TAE buffer (40 mM Tris HCl, 20 mM acetic acid, 1 mM EDTA [pH 8.3]) and 1-mm-thick polyacrylamide gels (6%). Gels contained a 20% to 60% denaturant gradient and were electrophoresed for 17 h at 100 V and 60°C. Gels were stained with GelStar nucleic acid stain (Biowhittaker Molecular Applications, Rockland, ME) and photographed on a UV transillumination table (302 nm) with a Kodak (Rochester, NY) digital camera.
RESULTS
Primer design.
Using the ARB phylogenetic software package, primers targeting plastid 16S rRNA genes of some eudicotyledon plants, including cucumber and peanut, were developed. The plastid 16S rRNA gene primer applied to the SuPER system described here, located in the 16S rRNA from positions 491 to 515 (E. coli numbering), was synthesized in forward and reverse directions (primer Q491-F and its reverse complement, Q491-R, respectively). The Q491 primers are 25 bases long with a 52% GC content and have a high stringent annealing temperature. The primer S661-F was previously designed to target the 16S rRNA genes of bacteria from the genus Streptomyces (6). The S661 primers (S661-F and its reverse complement, S661-R) are 18 bases long with a 50% GC content. These primers were also chosen based on their location within the gene of interest (Fig. 2). Both plastid and Streptomyces primers were located between positions 341 and 926 (E. coli numbering) of the 16S rRNA genes (the region spanned by the general bacterial primers employed in the subsequent PCR). Cucumber plastid DNA has three Tsp509I restriction sites between positions 341 and 926 of the 16S rRNA gene (the region amplified by the 341-F and 907-R primers), one site between positions 341 and 491 and two sites between positions 491 and 926 (including one site within the 907-R primer). Bacteria from the genera Streptomyces and Clavibacter both have three Tsp509I restriction sites between positions 341 and 926 of the 16S rRNA gene; these include one site between positions 341 and 661, one position within the S661 primer, and one site within the 907-R primer.
Selective restriction of plastid 16S rRNA genes from environmental samples using the SuPER reaction.
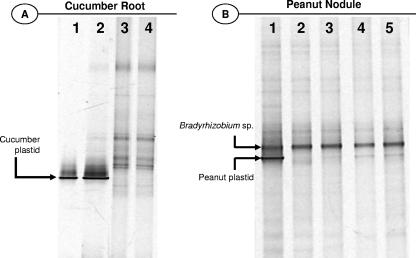
Samples of DNA extracted from soil-grown cucumber root and peanut nodules were subject to the SuPER reaction using the plastid primers (Q491-F and Q491-R) under optimized conditions (Fig. 3). To demonstrate the exclusive digestion of plastid DNA, bacterial PCR-DGGE analyses were conducted on pure cucumber DNA (Fig. 3A, lane 1), on DNA extracted from cucumber roots or peanut nodules grown in soil prior to the SuPER reaction (Fig. 3A, lane 2, and b, lane 1, respectively), and on the same DNAs after SuPER digestion of plastid DNA (Fig. 3A, lanes 3 and 4, and b, lanes 2 to 5). Bacterial 16S rRNA gene PCR-DGGE analysis of DNA extracted from cucumber roots and attached microorganisms demonstrated that the SuPER reaction digested cucumber plastid DNA almost entirely, resulting in the decreased amplification of plastid 16S rRNA genes and the increased amplification of nonplastid 16S rRNA gene templates. This reaction was equally successful in removing cucumber plastid DNA from SuPER reactions containing approximately 12 ng of total DNA (Fig. 3A, lane 3) and 36 ng of total DNA (Fig. 3A, lane 4). Likewise, plastid DNA was readily eliminated from analyses of peanut nodules (Fig. 3B). When decreased amounts of the restriction enzyme Tsp509I were used in the SuPER reaction (as low as 0.1 U per 25 μl), nearly total removal of plastid DNA still resulted (Fig. 3B, lanes 2 to 5). The application of 5 U was nonetheless the most effective in guaranteeing maximal digestion of the target template.
FIG. 3.
Confirmation of removal of plastid DNA from DNA extracts of cucumber root and peanut nodule and concomitant enhanced detection of plant-associated bacteria. (A) Bacterial 16S rRNA gene PCR-DGGE analyses of cucumber and cucumber roots. Lane 1 contains PCR product amplified from pure cucumber DNA (plastid rRNA gene). Lane 2 contains PCR product amplified from total DNA extracted from cucumber root; the plastid 16S rRNA gene overwhelms the population analysis. Lanes 3 and 4 contain PCR product of the same DNA as that in lane 2 but modified by the SuPER reaction employing the plastid primers (Q491-F and Q491-R) starting with 12 and 36 ng of total DNA, respectively. The DGGE band representing plastid 16S rRNA is indicated. (B) Bacterial 16S rRNA gene PCR-DGGE analyses of DNA extracted from peanut nodules. Lane 1 contains PCR product amplified from peanut nodule DNA (and associated bacteria, primarily a symbiotic Bradyrhizobium sp.). Lanes 2 to 5 contain PCR product amplified from the same DNA as that in lane 1 but modified by the SuPER reaction employing the plastid primers. In lanes 2 to 5, 5, 1, 0.5 and 0.1 U of the restriction enzyme Tsp509I were applied to the SuPER reaction mixtures, respectively. The DGGE bands representing plastid and Bradyrhizobium sp. 16S rRNA genes are indicated.
Selective restriction of Streptomyces cattleya 16S rRNA genes from mixed samples with Clavibacter michiganensis DNA using the SuPER reaction.
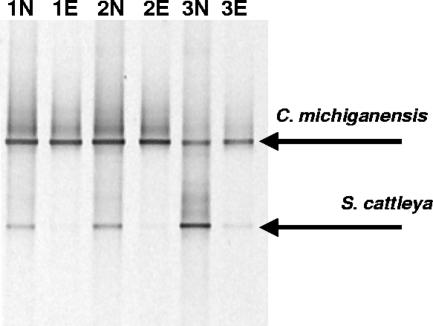
DNA of S. cattleya and C. michiganensis was mixed in various ratios and added as a template to SuPER reaction mixtures with the Streptomyces primers (S661-F and S661-R). Three DNA mixtures of S. cattleya and C. michiganensis with various total DNA levels and S. cattleya-C. michiganensis ratios were prepared. These contained 38, 103, and 123 ng of total DNA (per 25 μl), and this DNA was 57%, 21%, and 12% S. cattleya, respectively. The samples were then subjected to the stringent SuPER reaction with the Streptomyces primers. Control reactions without the Tsp509I enzyme were also conducted. In these control reactions, all other conditions and starting DNA concentrations were identical. After the SuPER reaction, the resulting templates were subjected to PCR with general bacterial primers 341-F and 907-R that anneal to 16S rRNA genes of S. cattleya and C. michiganensis, and the generated PCR products were analyzed by DGGE (Fig. 4). The PCR-DGGE analysis revealed that when the restriction enzyme was added, the SuPER reaction selectively restricted the target DNA (S. cattleya) while leaving unmodified DNA containing only a single mismatch to the primers employed (C. michiganensis). Weak PCR amplification of S. cattleya DNA was observed in samples that had contained the highest levels of S. cattleya DNA and concomitantly low levels of C. michiganensis DNA prior to the SuPER reaction. However, the level of PCR amplification of S. cattleya was greatly reduced relative to the no-enzyme control.
FIG. 4.
Confirmation of highly specific removal of a targeted DNA template from mixed target and nontarget (one mismatch) DNA samples. Mixtures of S. cattleya and C. michiganensis DNA were formulated with increasing Streptomyces DNA concentrations (lanes 1 to 3 with 12, 21, and 57% S. cattleya DNA and 123 ng, 103 ng, and 38 ng of total DNA, respectively), as described in the text. These samples were subjected to the SuPER reaction employing primers targeting 16S rRNA genes of bacteria from the genus Streptomyces (lanes 1E, 2E, and 3E). Control reactions, in which no enzyme was added, were also conducted (lanes 1N, 2N, and 3N). In SuPER reactions in which the Tsp509I enzyme was added, almost complete removal of S. cattleya DNA was observed. In control reactions without the Tsp509I enzyme, both S. cattleya and C. michiganensis DNA were strongly amplified by the bacterial 16S rRNA gene primers.
Selective restriction of Streptomyces coelicor 16S rRNA genes from spiked environmental samples using the SuPER reaction.
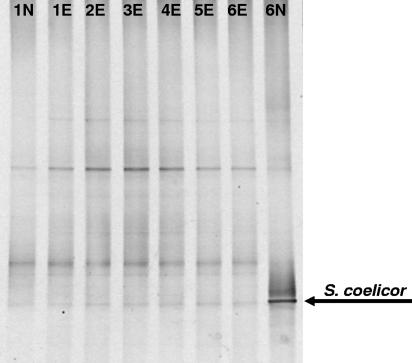
Total DNA extracted from an environmental sample (compost) and the same DNA spiked with various levels of S. coelicor DNA were subject to the SuPER reaction with the S661-F and S661-R primers and subsequent bacterial PCR-DGGE analysis (Fig. 5). All SuPER samples in this set of analyses contained 30 ng of environmental DNA per 25-μl volume. These reaction mixtures were then amended with S. coelicor DNA in amounts ranging from 0 to 165 ng (Fig. 5, lanes 1E to 6E). The reactions without Streptomyces DNA and with 165 ng of S. coelicor DNA were also subjected to no-enzyme control reactions (Fig. 5, lanes 1N and 6N, respectively). Selective restriction of Streptomyces DNA via the SuPER reaction restored the original environmental bacterial profile, regardless of the amount of S. coelicor DNA added. In the samples with 165 ng of S. coelicor DNA (lanes 6E and 6N), S. coelicor DNA comprised greater than 80% of the total DNA added to the reaction mixture and, in the no-enzyme control, dominated the DGGE profile (Fig. 5, lane 6N).
FIG. 5.
Selective removal of Streptomyces DNA in an environmental DNA sample (compost) spiked with different levels of S. coelicor DNA. All samples contained 30 ng of environmental DNA with 0 ng (lanes 1E and 1N), 33 ng (lane 2E), 66 ng (lane 3E), 99 ng (lane 4E), 132 ng (lane 5E), or 165 ng (lanes 6E and 6N) of amended Streptomyces DNA. Control reaction mixtures to which no restriction enzyme was added were also used (lanes 1N and 6N). After the SuPER reaction, all samples were subject to PCR-DGGE analyses with general bacterial primers. The compost bacterial population profile was restored in those samples spiked with Streptomyces DNA and subjected to the active (enzyme added) SuPER reaction.
DISCUSSION
Molecular analyses of some environmental samples are limited by the presence of an overwhelmingly dominant population. In such cases, the dominant and unwanted DNA template makes the detection and characterization of less-dominant sequences difficult and time-consuming. While specific PCR primers can be employed to circumvent the dominant template, ultimately, the problem requires a more general solution, particularly when attempting to detect unknown populations. To this end, we present here a rapid method for the targeted removal of DNA templates from mixed samples using a combination of DNA polymerase elongation and endonuclease restriction. Targeted DNA templates are exclusively restricted by creating a reaction environment in which stringent primer annealing, DNA polymerase fragment elongation, and thermostable endonuclease restriction operate at the same temperature. Under these reaction conditions, only target DNA templates become double stranded and are thus susceptible to digestion by the restriction enzyme. We have demonstrated here that there is at least a 10°C range (from 60 to 70°C) where these conditions can be met, and most likely, even a wider range of temperatures can be employed. However, at the lower end of temperatures, nonspecific DNA reannealing, and thus nonspecific DNA restriction, becomes a possibility. At higher temperatures, restriction enzymes may work less efficiently or be inactivated. Nonetheless, the range demonstrated here indicates that there is a great deal of flexibility for the design of appropriate primers for the SuPER reaction. Standard PCR primers, such as the 18-base S661 primers, can be employed for this method by increasing buffer magnesium concentrations, yielding stringent annealing at temperatures appropriate for the SuPER reaction. We also note that while we employed the SuPER reaction for digestion of 16S rRNA gene templates, there is no theoretical limitation to the application of the technique to other genes or DNA templates.
To employ this methodology, the sequence of the dominant and unwanted DNA must be known. However, we note that the sequences of such DNAs are almost always known or easy to acquire precisely because of their dominance. Once the sequence of the unwanted template is known, the SuPER reaction can be employed, provided that a specific primer can be designed. An increasing number of mismatches between target and nontarget templates at primer locations will theoretically decrease the likelihood of primer annealing to nontarget templates. We have shown that under certain conditions, even a single mismatch between target and nontarget templates can be discriminated using the SuPER method. When operated under stringent temperatures, the SuPER reaction did not digest nontarget DNA templates, as demonstrated in this study using the S661 Streptomyces primers. In particular, we note that the discrimination against the mismatch template (C. michiganensis) was a result of the mismatch between the primers and the target DNA, not the absence of appropriate restriction sites for the Tsp509I enzyme. C. michiganensis, in addition to containing a Tsp509I restriction site within the primer position (i.e., 16S rRNA gene position 675 [E. coli numbering]), also contained Tsp509I restriction sites at 16S rRNA gene positions 559 and 918. These sites would have been restricted by the Tsp509I enzyme had the primers annealed, and fragment elongation proceeded.
The restriction enzyme Tsp509I was chosen for the SuPER reaction since (i) it operated at high temperatures similar to those of the stringent annealing temperature of the applied primers, (ii) it operated under a variety of buffer conditions, including PCR buffer, and (iii) its short recognition sequence (AATT) increased the potential number of restriction sites. It should be possible to use other thermostable restriction enzymes independently or simultaneously, provided they operate adequately under reaction buffer conditions that also support DNA polymerase activity. While the restriction enzyme Tsp509I operated efficiently in the PCR buffer, heat-inactivated Tsp509I enzyme, although unable to digest DNA, inhibited subsequent PCRs. To circumvent this inhibition, samples were treated with proteinase K prior to PCR amplification. Following this treatment, the proteinase K was inactivated at 95°C, and thereafter, the samples were used directly as a template for subsequent PCRs. In addition to heat-inactivated Tsp509I enzyme, primers used for the SuPER reaction were found to be inhibitory to the subsequent PCR. This inhibition was avoided by diluting these primers in the initial SuPER reaction to a concentration of roughly 1/20 of that of standard PCR primer concentrations.
Near-complete removal of target DNA could be achieved using the SuPER reaction. In those reactions with relatively high levels of nontarget DNAs (i.e., DNA unmodified by the SuPER reaction), target DNAs were essentially undetectable by PCR-DGGE after the SuPER reaction. To achieve maximum digestion of the target template when using a single primer location (e.g., Q491-F and Q491-R), it is critical that the enzyme cuts on both sides of the SuPER primer location so as to digest both strands of DNA (Fig. 2). To achieve a reduction in the subsequent PCR amplification of the targeted template, these cuts must be located within the region amplified by the subsequent PCR (Fig. 2). If, as in the case of the S661 primers, the primers contain a recognition sequence for the restriction enzyme employed, the addition of Taq DNA polymerase to the SuPER reaction mixture is not strictly required for effective digestion of the target template (data not shown). In any case, however, both forward and reverse primers are required for optimal removal of unwanted DNA. The use of only a single primer will theoretically result in half of the total target DNA being digested (i.e., one of two strands from the target template). Such a “half-digestion” nonetheless yielded a partial decrease in amplification of the unwanted template (data not shown).
The application of inverse and complementary primers to anneal to a single position on opposite strands of the target DNA could potentially result in dimer formation, thus reducing the pool of primer available for annealing to target locations. Although we did not observe any difficulty in digesting target templates using the inverse and complementary primers, it should be possible to use two (or more) different primers and achieve restriction of the target template, provided that these different primers have similar stringent annealing temperatures. However, we were most successful in eliminating target templates using inverse and complementary primers.
Although there are several techniques available for diminishing the impact of a DNA template from molecular analyses, the SuPER reaction has several advantages. First, the method does not require the synthesis of modified or specialized primers, and due to the low concentration of primer that is required, even a small-scale synthesis is sufficient for a large number of reactions. Second, unlike the other techniques, the SuPER reaction destroys the target DNA template by endonuclease restriction. This may be particularly advantageous for removing DNA templates prior to the generation of environmental clone libraries, and by using only a single primer, the method may be employed to remove a single strand of DNA, if so desired. Although there is a possibility for chimeric sequences forming during subsequent PCR (16), none were observed in the systems we examined. Multiple restriction enzymes might be employed to further digest target templates and reduce the possibility of chimera formation. This method can be applied to medical, forensic, and microbial ecology fields or any other field where an undesired DNA template inhibits molecular studies. While the general conditions for the SuPER reaction have been detailed here, new primers will have to be optimized for appropriate magnesium concentrations and annealing temperatures. However, these optimizations are no more cumbersome than those with normal PCR primer sets.
Acknowledgments
We thank Joy Schochet, Jonathan Green, Oded Beja, and Eduard Jurkevitch for useful comments on the manuscript.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bebout, B. M., S. P. Carpenter, D. J. Des Marais, M. Discipulo, T. Embaye, F. Garcia-Pichel, T. M. Hoehler, M. Hogan, L. L. Jahnke, R. M. Keller, S. R. Miller, L. E. Prufert-Bebout, C. Raleigh, M. Rothrock, and K. Turk. 2002. Long-term manipulations of intact microbial mat communities in a greenhouse collaboratory: simulating earth's present and past field environments. Astrobiology 2:383-402. [DOI] [PubMed] [Google Scholar]
- 3.Behn, M., C. Thiede, A. Neubauer, W. Pankow, and M. Schuermann. 2000. Facilitated detection of oncogene mutations from exfoliated tissue material by a PNA-mediated ‘enriched PCR’ protocol. J. Pathol. 190:69-75. [DOI] [PubMed] [Google Scholar]
- 4.Green, S. J., S. Freeman, Y. Hadar, and D. Minz. 2004. Molecular tools for isolate and community studies of Pyrenomycete fungi. Mycologia 96:439-451. [PubMed] [Google Scholar]
- 5.Hancock, D. K., F. P. Schwarz, F. Song, L. J. Wong, and B. C. Levin. 2002. Design and use of a peptide nucleic acid for detection of the heteroplasmic low-frequency mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes (MELAS) mutation in human mitochondrial DNA. Clin. Chem. 48:2155-2163. [PubMed] [Google Scholar]
- 6.Inbar, E., S. J. Green, Y. Hadar, and D. Minz. Competing factors of compost concentration and proximity to root affect the distribution of Streptomycetes. Microb. Ecol., in press. [DOI] [PubMed]
- 7.Liles, M. R., B. F. Manske, S. B. Bintrim, J. Handelsman, and R. M. Goodman. 2003. A census of rRNA genes and linked genomic sequences within a soil metagenomic library. Appl. Environ. Microbiol. 69:2684-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nielsen, P. E. 2001. Peptide nucleic acid: a versatile tool in genetic diagnostics and molecular biology. Curr. Opin. Biotechnol. 12:16-20. [DOI] [PubMed] [Google Scholar]
- 11.Nikolausz, M., K. Marialigeti, and G. Kovacs. 2004. Comparison of RNA- and DNA-based species diversity investigations in rhizoplane bacteriology with respect to chloroplast sequence exclusion. J. Microbiol. Methods 56:365-373. [DOI] [PubMed] [Google Scholar]
- 12.Orum, H., P. E. Nielsen, M. Egholm, R. H. Berg, O. Buchardt, and C. Stanley. 1993. Single base pair mutation analysis by PNA directed PCR clamping. Nucleic Acids Res. 21:5332-5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seghers, D., L. Wittebolle, E. M. Top, W. Verstraete, and S. D. Siciliano. 2004. Impact of agricultural practices on the Zea mays L. endophytic community. Appl. Environ. Microbiol. 70:1475-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sessitsch, A., B. Reiter, U. Pfeifer, and E. Wilhelm. 2002. Cultivation-independent population analysis of bacterial endophytes in three potato varieties based on eubacterial and Actinomycetes-specific PCR of 16S rRNA genes. FEMS Microbiol. Ecol. 39:23-32. [DOI] [PubMed] [Google Scholar]
- 15.von Wintzingerode, F., O. Landt, A. Ehrlich, and U. B. Gobel. 2000. Peptide nucleic acid-mediated PCR clamping as a useful supplement in the determination of microbial diversity. Appl. Environ. Microbiol. 66:549-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang, G. C.-Y., and Y. Wang. 1997. Frequency of formation of chimeric molecules as a consequence of PCR coamplification of 16S rRNA genes from mixed bacterial genomes. Appl. Environ. Microbiol. 63:4645-4650. [DOI] [PMC free article] [PubMed] [Google Scholar]