Abstract
The microbiota of the small intestine is poorly known because of difficulties in sampling. In this study, we examined whether the organisms cultured from the jejunum and feces resemble each other. Small-intestinal fluid samples were collected from 22 beagle dogs with a permanent jejunal fistula in parallel with fecal samples. In addition, corresponding samples from seven of the dogs were collected during a 4-week period (days 4, 10, 14, and 28) to examine the stability of the microbiota. In the jejunal samples, aerobic/facultative and anaerobic bacteria were equally represented, whereas anaerobes dominated in the fecal samples. Despite lower numbers of bacteria in the jejunum (range, 102 to 106 CFU/g) than in feces (range, 108 to 1011 CFU/g), some microbial groups were more prevalent in the small intestine: staphylococci, 64% versus 36%; nonfermentative gram-negative rods, 27% versus 9%; and yeasts, 27% versus 5%, respectively. In contrast, part of the fecal dominant microbiota (bile-resistant Bacteroides spp., Clostridium hiranonis-like organisms, and lactobacilli) was practically absent in the jejunum. Many species were seldom isolated simultaneously from both sample types, regardless of their overall prevalence. In conclusion, the small intestine contains a few bacterial species at a time with vastly fluctuating counts, opposite to the results obtained for the colon, where the major bacterial groups remain relatively constant over time. Qualitative and quantitative differences between the corresponding jejunal and fecal samples indicate the inability of fecal samples to represent the microbiotas present in the upper gut.
Although the intestinal microbiota is known to be of crucial importance to health, the significance of the relatively sparse microbiota in the small intestine has not been clarified due to major difficulties in investigating the intestinal microbiota in its natural habitat. Instead, fecal samples are widely used because of their easy accessibility. However, the microbiota present in feces does not necessarily reflect the specific features of that in the upper gastrointestinal tract, and different species may dominate in different parts of the gut (20, 36). Moreover, it is often difficult to characterize the bacterial diversity in fecal samples because of the poor or too high selectivity of selective agar media (13, 24), and the overgrowth of dominant flora can make it hard to identify predominant species, other than fecal ones. In contrast to the luminal microbiota of the colon, small-intestinal fluid seems to harbor a different microbial population, i.e., the ratios of aerobes and anaerobes, gram-negative and gram-positive bacteria, and total bacterial counts vary between these anatomical sites (7, 9, 16). There are also time-related and local variations in the composition and distribution of the microbiota in the upper gut (16, 20, 28). These characteristics have so far not been fully described, particularly because of the unavailability of suitable samples, obtained without anesthesia from living subjects, and the laborious nature of the analysis. The small intestine has been thought to have fewer species both in transit and attached to epithelia than to the distal colon (11). Still, very few studies have analyzed the small-intestinal microbiota of healthy subjects at species level, and hardly any data are thus available.
Recently, a dog fistula model was designed for an easy and painless access to jejunal chyme without disturbing the normal functions of the gut (12). The aim of the present study was to examine the similarity or dissimilarity of the microbial findings in small-intestinal fluid (jejunal chyme) and feces collected from 22 beagle dogs with such a fistula. In addition, the stability of the microbiota in the two sample types was followed in part of the dogs during a 4-week period.
(This work was presented in part at the 3rd World Congress on Anaerobic Bacteria and Infections, Glasgow, Scotland, May 2003, poster 6.026.)
MATERIALS AND METHODS
Study subjects and specimens.
Jejunal chyme and fecal samples were obtained from 22 healthy male laboratory beagle dogs (age, 1 to 3 years; body weight, 12 to 19 kg) with a permanent jejunal fistula (nipple valve) (12). The dogs were fed twice a day with commercial canned dog food (Pedigree, Waltham; Masterfoods, Helsinki, Finland). Additionally, four consecutive samples, collected at days 4, 10, 14, and 28, were obtained from seven of the dogs (the samples at day 14 were not available from two dogs). The experimental protocol was approved by the local ethics committee for animal experiment action in Helsinki, Finland, and was conducted according to the valid guidelines (www.hmso.gov.uk). The jejunal chyme samples were collected shortly after feeding (within 1.5 h), and fecal samples were collected shortly after defecation (within 1 h). Samples were immediately frozen as such at −70°C in compact tubes with no extra airspace and later quantitatively cultured in parallel per dog. The exposure time to air was minimized throughout the procedure with careful planning and preparations, including the use of prereduced dilution broth and agar plates kept under anaerobic conditions 18 h prior to their use. The small-intestine fluid and feces were thawed and homogenized, pH was measured (Benchtop 420 pH Meter, Orion), and the sample consistency, color, and odor were recorded.
Culture.
The homogenates were serially diluted (10−1 to 10−7) in prereduced peptone-yeast extract broth at pH 7.0. Aliquots of 10 μl or 100 μl of the homogenates or appropriate dilutions were plated onto several nonselective and selective agar media and incubated at 35°C as appropriate (Table 1). A total of 33 agar plates were used for each fecal sample and 27 plates were used for each jejunal sample. Anaerobiosis was induced in anaerobic jars filled by the evacuation-replacement technique (Anoxomat WS8000; Mart B.V., Lichtenvoorde, The Netherlands) with a gas mixture (90% N2, 5% CO2, and 5% H2). NEYA and CCFA, the selective agars for clostridia, were plated both immediately and after 30-min alcohol shock for spore selection (15).
TABLE 1.
Nonselective and selective agar media used in the bacterial culture
| Agar medium | Enrichment or selection | Target bacteria | Atmosphere/incubation time in days | Reference |
|---|---|---|---|---|
| Brucella | Sheep blood, hemin, vitamin K1 | Total anaerobic counts | Anaerobic/5-7 | 15 |
| BBE | Bile, esculin, gentamycin | Bacteroides and Bilophila spp. | Anaerobic/5-7 | 15 |
| KVLB | Laked blood, kanamycin, vancomycin | Gram-negative anaerobes | Anaerobic/5-7 | 15 |
| BIF | Tomato juice, hemin, maltose | Bifidobacteria | Anaerobic/5-7 | 33 |
| MRS | Dextrose, ammonium citrate | Lactobacilli | Anaerobic/5-7 | 15 |
| CCFA | Fructose, egg yolk, cycloserine, cefoxitin | Clostridium difficile | Anaerobic/5-7 | 15 |
| NEYA | Egg yolk, neomycin | Clostridium spp. | Anaerobic/5-7 | 15 |
| Blood | Sheep blood | Total aerobic counts | 5% CO2/2-3 | 18 |
| CLED | Cystine, lactose; electrolyte deficient | Enterobacteriaceae, streptococci | Ambient air/2-3 | 18 |
| BE | Bile, esculin | Enterococci | Ambient air/2-3 | 18 |
| Sabouraud | Chloramphenicol | Yeasts | Ambient air/2-3 | 18 |
| RLB | Laked rabbit blood | Pigmenters, fluorescence | Anaerobic/4-7 | 15 |
| EYA | Egg yolk | Lipase and lecithinase producers | Anaerobic/4-7 | 15 |
Microbial identification.
The total counts and main groups of aerobic/facultative and anaerobic bacteria and yeasts were enumerated (detection limit, 102 CFU/g). To find all colonial morphotypes grown under the three different incubation conditions, a representative of each distinct colony type was recorded with a stereomicroscope, then isolated, and identified by established methods, including aerotolerance testing (blood agar in O2 and chocolate agar in CO for 5 days), gram staining, spot and screening tests (catalase, indole, nitrate, oxidase, lecithinase, and lipase), special antimicrobial potency disk patterns (vancomycin, kanamycin, colistin, sodium polyanethol sulfonate, penicillin, and oxgall), carbohydrate fermentation tests (prereduced anaerobically sterilized biochemicals), enzyme detection (diagnostic tablets; Rosco Diagnostica, Taastrup, Denmark), commercial test kits (Api system; bioMérieux, Marcy l'Etoile, France), and metabolic end product analysis by gas-liquid chromatography after peptone-yeast extract glucose broth culture (15, 22).
The presence of Clostridium difficile common antigen and toxin A in feces was determined with a commercial kit (Triage C. difficile Panel; Biosite, San Diego, CA) according to the manufacturer's instructions. For unclassified clostridia resembling C. difficile, genes encoding 16S rRNA were amplified and sequenced with the primers fD1 Mod and 533r by the method of Jalava et al. (14) and compared to the GenBank database.
Statistics.
Statistical analyses were performed using the SPSS 11.5 (SPSS, Chicago, IL, USA) and Epi Info 6 Statcalc (CDC, Atlanta, GAA) software for Windows. The pH values of the samples and number of isolates in the two sample types were compared by the paired-sample t test, and frequencies of different bacteria were compatred by Statcalc single-table tests. The differences between the bacterial counts were assessed by the Wilcoxon signed-rank test (nonparametric). A P value of <0.05 was considered statistically significant.
RESULTS
General features.
All jejunal samples were culture positive. The odor of the samples was mild and sometimes slightly sweet. A marked variation in viscosity, texture, and color was recorded in jejunal fluid, but differences in the visible features could not be connected to the culture results. The pH values varied very little within a sample category and were slightly higher in the jejunal samples than in the fecal samples: the pH in the jejunal samples was 6.8 g (range 6.4 to 7.5 g) versus a mean pH in the fecal samples of 6.6 (range, 6.1 to 7.4) (difference not significant).
Total bacterial counts.
The total bacterial counts in jejunal chyme were significantly lower (P < 0.001) than the counts in feces: median, 4 × 104 (range, 102 to 106) versus a median count of 3 × 1010 (range, 108 to 1011) CFU/g (wet weight), respectively. All growth in the jejunal samples was recorded and identified, while only bacterial morphotypes from dilutions of >105 to 106 were isolated from the fecal samples. The mean number of bacterial genera per species per sample isolated from the jejunum was significantly lower (P < 0.001) than that from feces: 7 (range, 1 to 11) versus 12 (range, 9 to 16), respectively. The anaerobe/aerobe ratio varied considerably both within and especially between the jejunal and fecal samples. In jejunal chyme, median anaerobic and aerobic growth was almost equally represented: 1 × 104 (range, 102 to 106) versus 7 × 103 (range, 102 to 106) CFU/g (difference not significant), respectively. In feces, anaerobic bacteria dominated over aerobic/facultative bacteria as follows: a median value of 3 × 1010 (range, 108 to 1011) versus 2 × 109 (range, 106 to 10 10) CFU/g (P = 0.010), respectively. Anaerobes covered a significantly lower proportion of the total jejunal bacterial concentration than their proportions in feces: 48% (range, 0.2 to 99.98%) versus 79% (range, 4.8 to 99.98%) (P = 0.004), respectively. Anaerobic species were consistently found in higher concentrations than aerobes in 54% of the jejunal samples and in 86% of the fecal samples. In the jejunal samples, the dominant aerobic/facultative bacterial group varied, while the abundant aerobic growth in feces comprised mostly streptococci. Those dogs that harbored high aerobic fecal counts had notably high aerobic counts in jejunal chyme as well.
Microbiota in jejunal chyme versus feces.
Figure 1 presents the overall prevalence of microbial groups and Table 2 shows the prevalence and mean counts of different anaerobic and aerobic bacteria and yeasts isolated from jejunal chyme and feces. The proportions of different bacteria of the total cultivable microbiota differed significantly between the jejunal and fecal samples (Fig. 2). In general, the high prevalence reflected the high proportion of the total count. The exceptions to this were aerobic gram-positive rods and bile-resistant Bacteroides spp., which were prevalent in feces but present at relatively low levels, and clostridia, which were the organisms most commonly found in the jejunum but comprised only 9% of the total jejunal count. On the other hand, coliforms and streptococci were present in 18% and 36% of the jejunal samples, respectively, but then covered 5% and 8% of the total jejunal count.
FIG. 1.
Prevalence of bacterial groups and yeasts in jejunal chyme and feces of 22 dogs at baseline. Statistical differences between the sample types were determined by the Epi Info 6 single-table test. *, P < 0.05; **, P < 0.001.
TABLE 2.
The prevalence and mean counts (CFU/g [wet weight]) of different anaerobic and aerobic bacteria and yeasts in jejunal chyme and feces of 22 beagle dogsa
| Bacterium or yeast | Jejunal chyme
|
Feces
|
P | ||
|---|---|---|---|---|---|
| n (%) | CFU | n (%) | CFU | ||
| Anaerobic | |||||
| Bile-resistant Bacteroides | 9 (41) | 2 × 102 | 21 (95) | 4 × 108 | ** |
| Fusobacteria | 11 (50) | 1 × 104 | 19 (86) | 1 × 109 | * |
| Other gram-negative rods | 8 (36) | 3 × 103 | 12 (55) | 3 × 107 | |
| Clostridia | 18 (82) | 3 × 104 | 22 (100) | 2 × 1010 | * |
| Bifidobacteria | 9 (41) | 4 × 103 | 14 (64) | 2 × 109 | |
| Lactobacilli | 1 (5) | 3 × 101 | 7 (32) | 7 × 106 | * |
| Other gram-positive rods | 7 (32) | 6 × 102 | 7 (32) | 1 × 108 | |
| Gram-positive cocci | 15 (68) | 3 × 104 | 22 (100) | 2 × 1010 | * |
| Gram-negative cocci | 4 (18) | 5 × 102 | 5 (23) | 5 × 107 | |
| Aerobic | |||||
| Coliforms | 4 (18) | 3 × 103 | 22 (100) | 1 × 109 | ** |
| Other gram-negative rods | 6 (27) | 1 × 103 | 2 (9) | 5 × 102 | |
| Gram-positive rods | 15 (68) | 5 × 104 | 21 (95) | 5 × 107 | * |
| Streptococci | 8 (36) | 6 × 103 | 22 (100) | 1 × 1010 | ** |
| Enterococci | 9 (41) | 6 × 103 | 18 (82) | 2 × 108 | * |
| Staphylococci | 14 (64) | 7 × 103 | 8 (36) | 4 × 105 | |
| Other gram-positive cocci | 10 (46) | 5 × 104 | 8 (36) | 5 × 107 | |
| Gram-negative cocci | 1 (5) | 2 × 101 | 1 (5) | 5 × 104 | |
| Yeasts | 6 (27) | 2 × 101 | 1 (5) | 2 × 103 | * |
Statistical differences in prevalence between the sample types were determined with the Epi Info 6 single-table test. *, P < 0.05; **, P < 0.001.
FIG. 2.
Mean proportions (percentage of the total bacterial count) of the most common bacterial groups in jejunal chyme and feces in 22 dogs. GPC, anaerobic gram-positive cocci; Clostr, clostridia; Strep, streptococci; Bifido, bifidobacteria; Fuso, fusobacteria; GPB, anaerobic gram-positive rods other than clostridia, lactobacilli, or bifidobacteria; Colif, coliforms; Lacto, lactobacilli; Bacter, bile-resistant Bacteroides spp.; Entc, enterococci; gpc, aerobic gram-positive cocci other than streptococci, enterococci or staphylococci; Staph, staphylococci; gpb, aerobic gram-positive rod. Statistical differences between the sample types were determined by the Epi Info 6 single-table test. *, P < 0.05; **, P < 0.001.
Several bacterial groups or species with an equal overall prevalence in the jejunal and fecal samples were seldom isolated simultaneously in the corresponding samples. Of the bacteria found in the jejunum, 25% were not detected in the corresponding fecal sample, nor were 45% of the fecal findings in the jejunum. Moreover, staphylococci, other catalase-producing cocci, nonfermentative gram-negative aerobic/facultative rods (other than coliforms), and yeasts were more frequent in jejunal chyme than in feces as part of the predominant flora, and some typically oropharyngeal species in the jejunal samples (neisseria and micrococci) were undetected in feces.
A group of unclassified clostridia, Clostridium hiranonis-like organisms, was rare in the jejunum but common in feces and present in 5% versus 82% of the samples (P < 0.001), respectively. These strains resembled C. difficile biochemically, i.e., they grew on CCFA agar medium, hydrolyzed proline, lacked the typical pungent odor of C. difficile, produced no or only traces of butyric and isocapronic acid, and remained unidentified and toxin negative by Triage C. difficile Panel (Biosite, Inc.). The amplified 16S rRNA sequence was 99% identical to C. hiranonis, but in contrast to C. hiranonis, the strains hydrolyzed gelatine and alkaline phosphatase and utilized mannitol. Bile-resistant Bacteroides spp., which were among the predominant bacterial findings in feces, were less frequent in jejunal chyme (Table 3). In the dogs positive for Bacteroides fragilis group organisms, Bacteroides fragilis was the dominant species in the jejunum, found in 8 of 9 (89%) dogs, whereas in feces B. fragilis was found in 9 of 21 (43%) dogs (P = 0.020), and Bacteroides vulgatus had the highest prevalence, found in 11 of 21 dogs (52%). Coliforms, which were present in all fecal samples, were isolated from only 18% of the jejunal samples (Table 2). Of the 166 fecal coliform isolates, 162 (98%) were identified as Escherichia coli, whereas among the nine jejunal isolates, six (67%) were identified as Citrobacter sp., two (22%) were identified as E. coli, and one (11%) was identified as Enterobacter aerogenes. In five dogs, the species with the highest counts were the same in the corresponding jejunal and fecal samples, whether anaerobic gram-positive cocci, Clostridium perfringens, or streptococci. The dominant species isolated from jejunal chyme varied between the dogs, since none of the bacterial groups dominated more often than any other, whereas those from feces were similar between the dogs (anaerobic gram-positive cocci or facultative streptococci dominated in 15 of 22 dogs). Although the bacterial composition of the jejunal samples varied from dog to dog, there was a coherence in the species variety, since only 3 of the 154 jejunal findings were isolated just once (anaerobic lactobacilli, C. hiranonis-like organism, or aerobic gram-negative cocci).
TABLE 3.
The prevalence and mean counts (CFU/g [wet weight]) of the bile-resistant Bacteroides species (B. fragilis group organisms and B. splanchnicus) in jejunal chyme and feces of 22 beagle dogsa
| Bacteria | Jejunal chyme
|
Feces
|
P | ||
|---|---|---|---|---|---|
| n (%) | CFU | n (%) | CFU | ||
| B. vulgatus | 2 (9) | 8 × 101 | 11 (50) | 4 × 108 | * |
| B. fragilis | 8 (36) | 2 × 102 | 9 (41) | 1 × 107 | |
| B. caccae | 0 (0) | 0 | 9 (41) | 6 × 107 | * |
| B. stercoris | 0 (0) | 0 | 3 (14) | 1 × 106 | |
| B. distasonis | 0 (0) | 0 | 1 (5) | 9 × 105 | |
| B. splanchnicus | 0 (0) | 0 | 7 (32) | 2 × 106 | * |
Statistical differences in prevalence between the sample types were determined with the Epi Info 6 single-table test. *, P < 0.05, **, P < 0.001.
Stability.
Out of the seven dogs with five consecutive samples collected during a 4-week period, two dogs repeatedly had a low proportion of aerobes/facultatives in every jejunal and fecal sample, and one dog had a high proportion of these organisms in every jejunal sample. In the four remaining dogs, the proportion of aerobes of the total count fluctuated widely between different sampling occasions (jejunal chyme, <0.1 to 98%; feces, 0.1 to 78%). All seven dogs were negative for yeasts at day 0, but two dogs later harbored yeasts in two samples, and one dog had yeast in one sample.
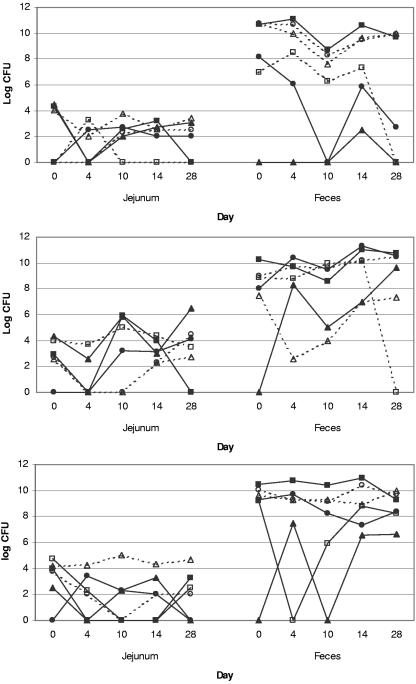
After a positive finding at baseline (day 0), the same species was repeatedly found in 42% of the consecutive jejunal samples and in 60% of the consecutive fecal samples of the same dog. The composition of the jejunal microbiota was less stable than the fecal microbiota; in jejunal samples, the predominant species varied between the dogs and remained seldom the same throughout the study. Figure 3 presents the shifts in the counts of the predominant findings in the jejunal and fecal samples in three selected dogs during the 28-day follow-up.
FIG. 3.
The counts of the predominant findings in the consecutive jejunal (CFU per milliliter) and fecal (CFU per gram) samples of three dogs (top, middle, and bottom panels). Solid squares (▪), anaerobic gram-positive cocci; solid circles (•), fusobacteria; solid triagles (▴), anaerobic gram-negative rods (other than fusobacteria or Bacteroides spp.); open squares (□), bifidobacteria; open circles (○), clostridia; open triangles (Δ), streptococci and enterococci.
DISCUSSION
This is the first study where microbial differences between the canine small-intestinal fluid and feces have been described in detail. The culture-based analysis of the whole jejunal microbiota in parallel with the corresponding fecal culture revealed considerable differences in the relative quantities of predominant bacterial groups in the corresponding samples. In this homologous group of beagle dogs, the composition of the jejunal flora was unique for every individual and changed over time, being fairly simple with only a few bacterial species at a time with vastly fluctuating counts. This is opposite from findings for the colon, since the fecal microbiota was more similar between the dogs and the major bacterial groups remained relatively constant over time.
The simultaneous processing of the samples from each subject was performed in parallel to make the comparison between the two sample types and consecutive samples as feasible as possible. The validity of the culture was monitored by plating an extensive series of dilutions together with the undiluted sample. Often the view on the plates of the parallel jejunal and fecal samples seemed fairly similar, although at different dilution levels and with fewer colony morphotypes; real differences could only be detected after careful analysis of the plates or further identification of the isolates. The selection criterion for picking isolates was due to differing morphologies of the colonies present, not the assumed relative importance of a finding or the proportion of the isolate on the agar.
Marteau et al. (20) reported facultative bacteria, including the lactobacillus-enterococcus group and E. coli species, to be part of the dominant flora in the human cecum and present in almost equal quantities as in feces, while obligate anaerobes were significantly more numerous in feces. Though with lower counts and fewer E. coli, we found a similar difference in the ratio of aerobes to anaerobes between the jejunum and feces. In contrast to the recent studies of beagle dogs (4) and labrador dogs (10), where no bifidobacteria but numerous Lactobacillus organisms were reported, we found bifidobacteria in 64% but lactobacilli only in 32% of the dogs. There are reports of C. difficile findings in healthy puppies and adult dogs (5, 19), whereas Struble et al. (30) and we failed to isolate C. difficile in dogs without diarrhea. Instead, we isolated unidentified clostridia resembling C. difficile (C. hiranonis-like organisms), which were dominant in the fecal clostridial populations. It is possible that previously reported nontoxigenic C. difficile findings with selective agar have included unclassified clostridia similar to those found in the present study. Unlike in feces, B. fragilis is the most frequent isolate of the B. fragilis group from gut mucosal surfaces (23). Accordingly, we found it dominant in the small intestine but not in feces. Yeasts were isolated more often from the jejunum than feces (27% versus 5%). There are contradictory opinions on the role of yeasts in the gut. Yeasts may have a reservoir in the upper gut, as part of the normal microbiota, but act as opportunistic pathogens under favorable conditions. There is no agreement on whether yeasts are enriched in the colon only when other species are suppressed and whether high counts are a result from rather than a cause of intestinal disturbance (1, 3, 17).
In comparison to the human fecal studies using 16 rRNA hybridization with slightly varying sets of group-specific probes (8, 27, 21), we found a significantly lower proportion of Bacteroides spp. (20 to 39% versus 0.3%, respectively) in canine feces by culture and a comparable proportion of bifidobacteria (1 to 7.3% versus 3.6%) and enterobacteria (1% versus 1%). Further, compared to findings for the Clostridium coccoides phylogenetic group, we found a significantly higher proportion of anaerobic gram-positive cocci combined with clostridia (16 to 29% versus 67%) and, compared to the low-G+C-content group, a slightly higher proportion of streptococci (1 to 12% versus 19%). Notably, the variations between individuals were vast, partly due to the methodological features of culture and to normal microbial fluctuation above and under the detection limit. Although a substantial part of the large bacterial community of the gut is not detectable by culture (31, 35), each technique, including molecular biological procedures (34) and the community cellular fatty acid analysis (25), has inherent limitations. Selective culture media promote recovery and presumptive identification to genus level, but the identification needs to be confirmed by other methods. In the present study, a wide array of microbes was covered with different levels of identification. All isolates were identified to the genus/group level, and selected bacteria were identified further (coliforms, Bacteroides spp., and C. difficile candidates). All individual identification results were not reported because of the large amount of scattered data, i.e., the findings that were relatively rare (e.g., Prevotella and Porhyromonas) or too laborious to identify further (e.g., Eubacterium-like organisms and gram-positive aerobic rods) were collected in groups. This limited repertoire of the species level identification, combined with the genus/group level identification, was able to detect differences between the sample types, which was the main objective of the study.
The ileum has been thought to be the transition zone for increasing bacterial counts (105 to 109), moving toward the colon (26); however, in the present study we detected bacterial growth of 106 CFU/g, including coliforms and anaerobes, already in the jejunum of some healthy beagles. The count crossed the level regarded as the limit for small-intestinal overgrowth (6, 29). The permanent jejunal fistula allowed postfeeding sampling and may therefore have provided exceptionally productive samples, yielding high bacterial counts. The observation suggests that the current limit may be underestimated and questions the feasibility of such a definitive limit. It remains to be confirmed whether the limit for small-intestinal overgrowth is arguable for humans as well. Small-intestinal overgrowth can be found in subjects with no predisposing conditions, and bacterial concentrations may be similar in jejunal biopsies of healthy human volunteers and patients with gastrointestinal symptoms (6, 29, 32). Speculation on overgrowth of small-intestinal microbes leading to opportunistic infections should be reported with caution, and the early steps in infections may not be explained solely by microbial growth exceeding 105 CFU/g.
In conclusion, the jejunal microbiota was found to be distinctive in species distribution, proportions of main bacterial groups, and variability, representing both qualitative and quantitative differences in the corresponding fecal samples. The present analysis was performed at a more detailed level than in previous studies in human subjects that described the features of the microbiota in the different parts of the upper gut (2, 9, 16, 20). In addition to the effects brought by differences in distribution of dominant species, the metabolic activities and microbial effects of the highly adaptive microbiota can vary in different niches of the body. A better conception of the local microbiota gives an insight to host-microbe interactions in the different parts of the gut. A limited species composition and thus possibly pronounced effects of single species, like clostridia or staphylococci, should not be overlooked, since specific microbial features of different niches may have clinical relevance in various disorders and diseases of the gastrointestinal tract.
Acknowledgments
We thank the late Hannele Jousimies-Somer for her contribution to the study and Terhi Virtanen and Arja Kanervo-Nordström for technical assistance.
The study was financed collaboratively by the National Public Health Institute (KTL), Helsinki, Finland, and Ipsat Therapies, Ltd., Espoo, Finland.
REFERENCES
- 1.Aldemir, M., O. F. Kokoglu, M. F. Geyik, and H. Buyukbayram. 2002. Effects of octreotide acetate and Saccharomyces boulardii on bacterial translocation in an experimental intestinal loop obstruction model of rats. Tohoku J. Exp. Med. 198:1-9. [DOI] [PubMed] [Google Scholar]
- 2.Bentley, D. W., R. L. Nichols, R. E. Condon, and S. L. Gorbach. 1972. The microflora of the human ileum and intrabdominal colon: results of direct needle aspiration at surgery and evaluation of the technique. J. Lab. Clin. Med. 79:421-429. [PubMed] [Google Scholar]
- 3.Bernhardt, H., and M. Knoke. 1997. Mycological aspects of gastrointestinal microflora. Scand. J. Gastroenterol. Suppl. 222:102-106. [DOI] [PubMed] [Google Scholar]
- 4.Buddington, R. K. 2003. Postnatal changes in bacterial populations in the gastrointestinal tract of dogs. Am. J. Vet. Res. 64:646-651. [DOI] [PubMed] [Google Scholar]
- 5.Buogo, C., A. P. Burnens, J. Perrin, and J. Nicolet. 1995. Presence of Campylobacter spp., Clostridium difficile, C. perfringens and salmonellae in litters of puppies and in adult dogs in a shelter. Schweiz. Arch. Tierheilkd. 137:165-171. [PubMed] [Google Scholar]
- 6.Corazza, G. R., M. G. Menozzi, A. Strocchi, L. Rasciti, D. Vaira, R. Lecchini, P. Avanzini, C. Chezzi, and G. Gasbarrini. 1990. The diagnosis of small bowel bacterial overgrowth. Reliability of jejunal culture and inadequacy of breath hydrogen testing. Gastroenterology 98:302-309. [DOI] [PubMed] [Google Scholar]
- 7.Finegold, S. M., V. S. Sutter, and G. E. Mathisen. 1983. Normal indigenous intestinal flora, p. 3-31. In D. J. Hentges (ed.), Human intestinal microflora in health and disease. Academic Press, New York, N.Y.
- 8.Franks, A. H., H. J. Harmsen, G. C. Raangs, G. J. Jansen, F. Schut, and G. W. Welling. 1998. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64:3336-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorbach, S. L., A. G. Plaut, L. Nahas, L. Weinstein, G. Spanknebel, and R. Levitan. 1967. Studies of intestinal microflora. II. Microorganisms of the small intestine and their relations to oral and fecal flora. Gastroenterology 53:856-867. [PubMed] [Google Scholar]
- 10.Greetham, H. L., C. Giffard, R. A. Hutson, M. D. Collins, and G. R. Gibson. 2002. Bacteriology of the Labrador gut: a cultural and genotypic approach. J. Appl. Microbiol. 93:640-646. [DOI] [PubMed] [Google Scholar]
- 11.Guarner, F., and J.-R. Malagelada. 2003. Gut flora in health and disease. Lancet 361:512-519. [DOI] [PubMed] [Google Scholar]
- 12.Harmoinen, J., J. Mättö, M. L. Rinkinen, M. Wilsson-Rahmberg, and E. Westermarck. 2001. Permanent jejunal fistula: promising method for obtaining small intestinal chyme without disturbing intestinal function. Comp. Med. 51:252-256. [PubMed] [Google Scholar]
- 13.Hartemink, R., and F. M. Rombouts. 1999. Comparison of media for the detection of bifidobacteria, lactobacilli and total anaerobes from faecal samples. J. Microbiol. Methods 36:181-192. [DOI] [PubMed] [Google Scholar]
- 14.Jalava, J., P. Kotilainen, S. Nikkari, M. Skurnik, E. Vänttinen, O. P. Lehtonen, E. Eerola, and P. Toivanen. 1995. Use of the polymerase chain reaction and DNA sequencing for detection of Bartonella quintana in the aortic valve of a patient with culture-negative infective endocarditis. Clin. Infect. Dis. 21:891-896. [DOI] [PubMed] [Google Scholar]
- 15.Jousimies-Somer, H., P. Summanen, D. M. Citron, E. J. Baron, H. M. Wexler, and S. M. Finegold. 2002. Wadsworth-KTL anaerobic bacteriology manual, 6th ed. Star Publishing, Belmont, CA.
- 16.Justesen, T., O. H. Nielsen, I. E. Jacobsen, J. Lave, and S. N. Rasmussen. 1984. The normal cultivable microflora in upper jejunal fluid in healthy adults. Scand. J. Gastroenterol. 19:279-282. [PubMed] [Google Scholar]
- 17.Krause, R., E. Schwab, D. Bachhiesl, F. Daxbock, C. Wenisch, G. J. Krejs, and E. C. Reisinger. 2001. Role of Candida in antibiotic-associated diarrhea. J. Infect. Dis. 184:1065-1069. [DOI] [PubMed] [Google Scholar]
- 18.MacFaddin, J. F. 1985. Media for isolation, cultivation, identification, maintenance of medical bacteria, vol. 1. Williams & Wilkins, Baltimore, MD.
- 19.Marks, S. L., E. J. Kather, P. H. Kass, and A. C. Melli. 2002. Genotypic and phenotypic characterization of Clostridium perfringens and Clostridium difficile in diarrheic and healthy dogs. J. Vet. Intern. Med. 16:533-540. [DOI] [PubMed] [Google Scholar]
- 20.Marteau, P., P. Rochart, J. Dore, C. Bera-Maillet, A. Bernalier, and G. Corthier. 2001. Comparative study of bacterial groups within the human cecal and fecal microbiota. Appl. Environ. Microbiol. 67:4939-4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuki, T., K. Watanabe, J. Fujimoto, Y. Miyamoto, T. Takada, K. Matsumoto, H. Oyaizu, and R. Tanaka. 2002. Development of 16S rRNA-gene-targeted group-specific primers for the detection and identification of predominant bacteria in human feces. Appl. Environ. Microbiol. 68:5445-5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murray, P. R., E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.). 2003. Manual of clinical microbiology, 8th ed. ASM Press, Washington, D.C.
- 23.Namavar, F., E. B. Theunissen, A. M. Verweij-Van Vught, P. G. Peerbooms, M. Bal, H. F. Hoitsma, and D. M. MacLaren. 1989. Epidemiology of the Bacteroides fragilis group in the colonic flora in 10 patients with colonic cancer. J. Med. Microbiol. 29:171-176. [DOI] [PubMed] [Google Scholar]
- 24.Nelson, G. M., and S. E. George. 1995. Comparison of media for selection and enumeration of mouse fecal flora populations. J. Microbiol. Methods 22:293-300. [Google Scholar]
- 25.Peltonen, R., W. H. Ling, O. Hänninen, and E. Eerola. 1992. An uncooked vegan diet shifts the profile of human fecal microflora: computerized analysis of direct stool sample gas-liquid chromatography profiles of bacterial cellular fatty acids. Appl. Environ. Microbiol. 58:3660-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saltzman, J. R., and R. M. Russell. 1994. Nutritional consequences of intestinal bacterial overgrowth. Compr. Ther. 20:523-530. [PubMed] [Google Scholar]
- 27.Sghir, A., G. Gramet, A. Suau, V. Rochet, P. Pochart, and J. Dore. 2000. Quantification of bacterial groups within human fecal flora by oligonucleotide probe hybridization. Appl. Environ. Microbiol. 66:2263-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simon, G. L., and S. L. Gorbach. 1986. The human intestinal microflora. Dig. Dis. Sci. 31(Suppl.):147-162. [DOI] [PubMed] [Google Scholar]
- 29.Stotzer, P. O., A. Brandberg, and A. F. Kilander. 1998. Diagnosis of small intestinal bacterial overgrowth in clinical praxis: a comparison of the culture of small bowel aspirate, duodenal biopsies and gastric aspirate. Hepatogastroenterology 45:1018-1022. [PubMed] [Google Scholar]
- 30.Struble, A. L., Y. J. Tang, P. H. Kass, P. H. Gumerlock, B. R. Madewell, and J. Silva, Jr. 1994. Fecal shedding of Clostridium difficile in dogs: a period prevalence survey in a veterinary medical teaching hospital. J. Vet. Diagn. Investig. 6:342-347. [DOI] [PubMed] [Google Scholar]
- 31.Suau, A., R. Bonnet, M. Sutren, J. J. Godon, G. R. Gibson, M. D. Collins, and J. Dore. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sullivan, Å., H. Törnblom, G. Lindberg, B. Hammarlund, A.-C. Palmgren, C. Einarsson, and C. E. Nord. 2003. The micro-flora of the small bowel in health and disease. Anaerobe 9:11-14. [DOI] [PubMed] [Google Scholar]
- 33.Sutter, V. R., D. M., Citron, M. A. C., Edelstein, S. M., Finegold. 1985. Wadsworth anaerobic bacteriology manual, 4th ed. Star Publishing, Belmont, CA.
- 34.Tannock, G. W. 2001. Molecular assessment of intestinal microflora. Am. J. Clin. Nutr. 73(Suppl.):410S-414S. [DOI] [PubMed] [Google Scholar]
- 35.Wilson, K. H., and R. B. Blitchington. 1996. Human colonic biota studied by ribosomal DNA sequence analysis. Appl. Environ. Microbiol. 62:2273-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zoetendal, E. G., A. von Wright, T. Vilpponen-Salmela, K. Ben-Amor, A. D. Akkermans, and W. M. de Vos. 2002. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl. Environ. Microbiol. 68:3401-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]