Abstract
Geldanamycin and the closely related herbimycins A, B, and C were the first benzoquinone ansamycins to be extensively studied for their antitumor properties as small-molecule inhibitors of the Hsp90 protein chaperone complex. These compounds are produced by two different Streptomyces hygroscopicus strains and have the same modular polyketide synthase (PKS)-derived carbon skeleton but different substitution patterns at C-11, C-15, and C-17. To set the stage for structural modification by genetic engineering, we previously identified the gene cluster responsible for geldanamycin biosynthesis. We have now cloned and sequenced a 115-kb segment of the herbimycin biosynthetic gene cluster from S. hygroscopicus AM 3672, including the genes for the PKS and most of the post-PKS tailoring enzymes. The similarities and differences between the gene clusters and biosynthetic pathways for these closely related ansamycins are interpreted with support from the results of gene inactivation experiments. In addition, the organization and functions of genes involved in the biosynthesis of the 3-amino-5-hydroxybenzoic acid (AHBA) starter unit and the post-PKS modifications of progeldanamycin were assessed by inactivating the subclusters of AHBA biosynthetic genes and two oxygenase genes (gdmM and gdmL) that were proposed to be involved in formation of the geldanamycin benzoquinoid system. A resulting novel geldanamycin analog, KOS-1806, was isolated and characterized.
The herbimycins (19) and macbecins (18), belonging to the group of benzoquinone ansamycins (24), are closely related to geldanamycin (7) in structure and bioactivity. Streptomyces hygroscopicus NRRL 3602 produces geldanamycin, whereas S. hygroscopicus AM 3672 makes herbimycin A, a C-11,15 dimethoxy-17-desmethoxy analog of geldanamycin, as its major natural product.
Like geldanamycin, the herbimycins were discovered in the late 1970s and found to have weak antibacterial and antifungal properties and also to show plant growth-inhibitory effects (13, 19). Both compounds were later discovered to have potent antitumor activity at nanomolar concentrations (26, 33). Although initially identified as a tyrosine kinase inhibitor, geldanamycin was subsequently found to bind to members of the heat shock protein 90 (Hsp90) family. Interest in these compounds greatly increased when Neckers and coworkers (35) showed that Hsp90, an abundant protein chaperone in mammalian cells (23), is the primary cellular target of benzoquinone ansamycins and the basis of their antitumor properties. These findings led to a new therapeutic concept for anticancer research because Hsp90 plays a central role in various signal transduction pathways involved in tumorigenesis (23). Because of their potential to affect multiple targets simultaneously, benzoquinoid ansamycins are believed to be superior to agents, such as Herceptin and Gleevec, whose therapeutic value is limited to certain tumor types and can be compromised by the development of drug resistance.
Because of its nanomolar potency and apparent specificity for tumor cells, geldanamycin was the first small-molecule inhibitor of Hsp90 to undergo development as an anticancer drug. However, severe hepatotoxicity (30) led to its withdrawal from clinical trials in 1995. Nonetheless, during the 1990s, considerable interest in this class of anticancer compound led to the development of geldanamycin analogs with improved pharmacological profiles. 17-Allylamino-17-demethoxygeldanamycin and 17-(2′-dimethylamino) ethylamino-17-demethoxygeldanamycin are currently undergoing clinical trials (12, 27). Some pharmacological disadvantages of these compounds have yet to be overcome, and therefore more analogs of this class of compounds may be required if drug development is to succeed.
Both herbimycin and geldanamycin are produced by S. hygroscopicus strains and are assembled by modular polyketide synthases, large polypeptides composed of sets of active sites called modules. Each module contains the catalytic domains required for a single round of polyketide chain elongation by substrate condensation and various reductions or dehydrations; sequential action of the set of modules as a protein “assembly line” produces the nascent polyketide (9, 28). In most cases, dedicated post-PKS enzymes introduce additional modifications of the polyketide, such as glycosylation and oxidation, with important effects on biological activity.
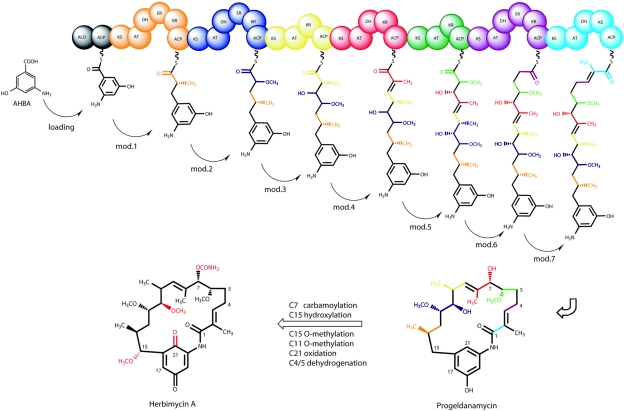
As is typical of all ansamycins, the carbon skeleton of geldanamycin is assembled from the 3-amino-5-hydroxybenzoic acid (AHBA) starter unit (2, 15) by successive condensation of two-carbon building blocks in seven chain-elongation steps that use one malonyl, four methylmalonyl, and two methoxymalonyl extender units (Fig. 1). We have proposed (21) that C-21 oxidation, C-17 oxidation/O-methylation, C-7 carbamoylation, and C-4,5 dehydrogenation occur after polyketide chain assembly. Because the polyketide backbone of herbimycin is identical to that of geldanamycin, PKS activity is expected to be very similar between the two pathways, with the structural differences in the products being introduced post-PKS.
FIG. 1.
Proposed pathway for herbimycin biosynthesis, starting from the ansamycin starter unit AHBA, followed by PKS processing through seven chain extension units (modules [mod.] 1 to 7) to give the hypothetical intermediate progeldanamycin, followed by formation of herbimycin A by PKS tailoring enzymes.
As geldanamycin does not show herbicidal effects such as those caused by herbimycin (13), we expect that these properties are acquired through methoxy substitution at C-11 and C-15. This raises the possibility that similar alterations of the geldanamycin moiety could result in derivatives with different bioactivity. To enable assessment of such structural alterations by genetic engineering, the corresponding herbimycin biosynthetic genes were needed.
We describe here the cloning and characterization of a major portion of the herbimycin (hbm) biosynthetic gene cluster containing the PKS genes and some other genes involved in post-PKS modifications, and we provide information about the AHBA biosynthesis genes that are essential for the formation of geldanamycin, as well as herbimycin.
MATERIALS AND METHODS
Media and growth conditions, bacterial strains, plasmids, and phages.
DNA manipulations were performed in Escherichia coli XL1-Blue (Stratagene) by standard procedures (25). For routine subcloning we used pLitmus 28 (New England Biolabs) and the pBluescript II plasmid vector series (Stratagene). The Streptomyces strains, phage, and plasmid vectors used in the present study are listed in Table 1. Lysogens were selected on solid R5 medium (14) by using a 2-ml overlay of 1 mg ml−1 neomycin (Sigma). Streptomyces lividans TK24, used as host strain for propagation of the φC31-phage derived vectors KC515 and pKOS-117A, was grown at 30°C on SFM solid medium (14) for sporulation and in liquid yeast extract-malt extract for preparation of protoplasts (14). Phage propagation was performed as described by Kieser et al. (14).
TABLE 1.
Bacterial strains, plasmids, and phages used in this study
| Strain, plasmid, or phage | Properties or product | Source or reference |
|---|---|---|
| Strains | ||
| S. hygroscopicus NRRL 3602 | Wild type | NRRLa |
| S. hygroscopicus AM 3672 | Wild type | S. Omura, Kitasato Institute (Japan) |
| S. lividans TK24 | SLP1− SLP2−; str | 11 |
| S. hygroscopicus K390-76-1 | ahba-B::neo | This study |
| S. hygroscopicus K390-76-2 | ahba-N::neo | This study |
| S. hygroscopicus K313-115-1 | gdmA1::neo | This study |
| S. hygroscopicus K390-48-1 | hbmA1::neo | This study |
| S. hygroscopicus K390-13-1 | gdmM::neo | This study |
| S. hygroscopicus K390-33 | gdmL::neo | This study |
| Plasmid and phage vectors | ||
| pHP45 | aac(3)IV | 5 |
| KC1139 | Bifunctional oriT RK2 vector | 4 |
| pFDneoS | pUC19 containing aphII gene | 8 |
| KC515 | c+ Δatt-int::tsr::vph | 14 |
| pKOS305-117A | c+ Δatt-int::apr::vph | KC515 |
| pKOS256-166-2 | LDD PCR fragment of hbm | This study |
| pKOS279-78-17 | hbm PKS | This study |
| pKOS279-78-04 | hbm PKS | This study |
| pKOS205-110-12 | hbm PKS | This study |
| pKOS206-116-10 | ahba-B cluster | This study |
NRRL, Northern Regional Research Laboratory of the Agricultural Service.
YPD seed medium was prepared according to the manufacturer's instructions (Sigma-Aldrich, St. Louis, MO). Fermentation medium contained (per liter) 2.5 g of peptone (Difco), 2.5 g of tryptone (Difco), 2.5 g of yeast extract (Difco), 5 g of baby oatmeal (Gerber), and 10 g of beet molasses (Minn-dak). The medium was sterilized for 30 min at 121°C. For shake-flask fermentation, the geldanamycin- and herbimycin-producing strains, as well as their mutant derivatives, were grown under the conditions described previously (21). For fermentation of gdmM knockout mutant S. hygroscopicus K309-27-1 to produce compound KOS-1806, spores were seeded into 50 ml of YPD medium in a 250-ml flask. The culture was grown for 24 h at 30°C. An aliquot of 25 ml was transferred into 500 ml of YPD medium in a 2.8-liter Fernbach flask, and the culture was incubated at 30°C for 1 day and then used to inoculate a 20-liter bioreactor with a 12-liter working volume (New Brunswick Scientific BioFlo IV) at 4% (vol/vol) to produce KOS-1806. The temperature was maintained at 30°C. The pH was initially controlled at 6.5 with 2.5 N sulfuric acid or 2.5 N sodium hydroxide. Two days after inoculation, the pH was reduced to 6.0 for the rest of the process. Dissolved oxygen was controlled above 30% with agitation (200 to 400 rpm). The overall pressure in the bioreactors was manually set at 6 lb/in2, and airflow was controlled at 1 volume/volume per min (vvm). Foam was controlled by automatic addition of 50% (vol/vol) Antifoam B (JT Baker). The fermentations were harvested 4 days after inoculation.
Isolation and in vitro manipulation of DNA.
Manipulation and transformation of DNA in E. coli was performed according to standard procedures (25) and by using a QIAprep spin miniprep kit (QIAGEN) according to the supplier's protocols. Phage DNA was isolated by using a combination of the QIAGEN lambda minikit (QIAGEN) for polyethylene glycol-mediated DNA precipitation in combination with the ethanol precipitation method described by Kieser et al. (14). Restriction endonuclease digestions and ligations were performed by standard techniques (25). DNA fragments for labeling and subcloning were isolated with the QIAGEN gel extraction kit. The conditions for phage DNA transfection were as described by Kieser et al. (14). Genomic DNA preparation from Streptomyces for PCR and Southern blot hybridization was performed by using standard procedures (14).
For DNA amplification by PCR a standard protocol with initial sample denaturation at 94°C for 5 min was used. One amplification cycle consisted of sample denaturation (40 s), a primer variable melting temperature (Tm), and polymerization at 72°C. To estimate suitable polymerization intervals, we calculated 1 min/kb for Taq polymerase (Roche) and 1.5 min/kb for Pfu Ultra (Stratagene). The PCR mix was used as recommended by the manufacturer of the DNA polymerase, and amplification was performed with a thermocycler PCT-200 (MJ Research). For degenerate PCR, the optimal Tm was determined by Tm variation with a temperature gradient of 50 to 70°C.
For Southern blot hybridization analysis, Streptomyces chromosomal DNA was digested with suitable restriction enzymes overnight and electrophoresed in a 0.6% agarose gel for 8 h in TAE buffer (25). Gels were blotted on nitrocellulose membranes by using the neutral transfer method of the rapid transfer system (Schleicher & Schuell) as recommended by the manufacturer. For radioactive [32P]DNA probe labeling, we used [32P]dCTP (Amersham) with the random-primed labeling kit (Roche) to label 40 ng of probe DNA according to the manufacturer's instructions. Labeled probes were purified with the ProbeQuant G-50 spin column (Amersham). Hybridizations were performed by using the formamide hybridization protocol (25) under high-stringency conditions. For nonradioactive, colorimetric DIG-labeling we used the DIG Easy Hyb protocol (Roche) as recommended by the manufacturer.
DNA sequencing and analysis.
BAC and cosmid DNA was sequenced by using standard shotgun procedures (16). DNA sequencing was performed on an ABI 3730 capillary sequencer by using reagents and protocols provided by the manufacturer. DNA and corresponding protein sequences were analyzed with SEQUENCHER 4.2 (Gene Codes) and MAC VECTOR 7.1 (Accelerys) and compared to sequences in the public databases by using CLUSTAL W (32) and BLAST (1).
Isolation of the herbimycin PKS gene cluster.
A cosmid library was prepared by using a pSET152-based vector, pKOS97-64C (Z. Hu, unpublished data), with S. hygroscopicus AM 3672 total DNA partially digested with Sau3A that was ligated and introduced into E. coli by using the Gigapack III XL (Stratagene) in vitro packaging kit. DIG-labeled probes encompassing the PKS loading didomain and the carbamoyl transferase gene were used jointly in an approach similar to that described for detection of the geldanamycin PKS genes (21), using degenerate PCR to generate 100% matching probes from the genome. We amplified a 650-bp fragment for the carbamoyl transferase gene and a 690-bp fragment for the PKS loading didomain by using the primers listed in Table 2. The DNA fragments were subcloned with the pCR2.1-TOPO TA cloning kit (Invitrogen) to give pKOS256-166-2, the corresponding insert was confirmed by sequence analysis. The library was screened with this probe DNA by colony hybridization. Colonies that hybridized were further analyzed by DNA end-sequencing and restriction enzyme analysis, which revealed three overlapping clones, pKOS279-78-17, pKOS279-78-04 and pKOS205-110-12. These clones together cover a 115-kb region, including the herbimycin PKS genes.
TABLE 2.
Design of gene disruption cassettes and oligonucleotides used
| Target | PCR primer (3′-5′)
|
Cassette design | |
|---|---|---|---|
| Name | Sequence | ||
| gdmA1 | LFfw | CGGGATCCGACGACCTGGGCCTGGACGAAAT | BamHI-LF-XhoI-neo-XbaI-RF-NsiI |
| LFbw | CCGCTGGAGGTGGTGGCCACCACGAACGTCGT | ||
| RFfw | GCTCTAGAAGTGATACGGCAGGCGCTGGCGAA | ||
| RFbw | TCGATGCATACTGCACGGTTTGGCGGAGGTGG | ||
| hbmA1 | LFfw | CGCGGATCCTGTCTGCTGGGCTACGGACACCAC | BamHI-LF-XhoI-neo-XbaI-RF-NsiI |
| LFbw | CCGCTCGAG AGTCCGTAGCATGCCGCCACACCT | ||
| RFfw | TGCTCTAGATTCGGCGTGGACTGATTCCCCGTC | ||
| RFbw | TGCATGCATCTTACCCGACAGCATCCACGGCAC | ||
| AHBA-B | LFfw | CGCGGATCCAGACCTCGACCACCGGTGTCTGGA | BamHI-LF-XhoI-neo-XbaI-RF-NsiI |
| LFbw | CCGCTCGAGCACGATTTCCAGCGCATGGCCCA | ||
| RFfw | TGCTCTAGACTCACCCGCTCGCCTTCGTCAT | ||
| RFbw | TGCATGCATTGAGCCACCACGGCGTGTGACA | ||
| AHBA-N | LFfw | CGCGGATCCTGCCAGTGGTGCATCTGCCGACCA | BamHI-LF-XhoI-neo-XbaI-RF-NsiI |
| LFbw | CCGCTCGAGTCACCCCGATGGGGTTGGCGAGTA | ||
| RFfw | TGCTCTAGATTGCCAGGCAGGGGCCGAATTGCT | ||
| RFbw | TGCATGCATCAGCGGCAGCTACAGAAGCGGTAC | ||
| gdmM | LFfw | CGGGATCCGACGACCTGGGCCTGGACGAAAT | BamHI-LF-XhoI-neo-XbaI-RF-NsiI |
| LFbw | ACGACGTTCGTGGTGGCCACCACCTCGAGCGG | ||
| RFfw | GCTCTAGAAGTGATACGGCAGGCGCTGGCGAA | ||
| RFbw | CAACCTCCGCCAAACCGTGCAGTATGCATGCA | ||
| gdmL | LFfw | CCCAAGCTTTCCGTCATCCTGCCGCAGCTCTGA | HindIII-LF-XbaI-neo-KpnI-RF-StuI |
| LFbw | GCTCTAGAGGTCGTGCTCTCCGTCAACACCGT | ||
| RFfw | GGGGTACCAGGGTGCCCTGAGCCAGGTGACAA | ||
| RFbw | TAGGCCTCGAACGCGATGAAGCCCTTCGGCA | ||
| LDD | LDfw | GAYGASCCSGCSTGGATGYTSTA | |
| LDbw | CCRTCSGTSCKGTACCASCCRTC | ||
Gene disruptions.
All gene disruptions were performed with a similar design with phages KC515 and pKOS305-117A (M. Piagentini, unpublished work) or plasmid pKC1139 (4) to deliver the corresponding neomycin resistance gene disruption cassettes. These cassettes consisted of two PCR-derived flanking regions (1 to 1.5 kb), where suitable restriction sites were introduced. The resulting DNA fragments were cloned, together with the neomycin resistance gene from pFDneoS (8). Detailed cloning information for each construct is summarized in Table 2.
For inactivation of the gdmA1 PKS and gdmM genes, the corresponding neomycin cassettes were cloned into phage KC515 at the BamHI-PstI sites (the thiostrepton resistance marker in KC515 [14] could not be used because both Streptomyces strains are resistant to this antibiotic). The resulting disruption mutants were selected on neomycin and confirmed by Southern blot hybridization. The cassettes for the disruption of hbmA1 and the ahba-B and ahba-N clusters were cloned into phage KOS305-117A carrying the apramycin resistance gene aac(3)IV from pHP45 (5) at the BamHI-BglII sites to replace the thiostrepton resistance gene. This phage construct was prepared to circumvent the thiostrepton resistance of S. hygroscopicus NRRL 3602 and the partial neomycin resistance we discovered for the herbimycin producer.
S. hygroscopicus AM 3672.
After transfection, the lysogens were grown under neomycin selection until sporulation occurred and screened for immediate double-crossover events, indicated either by loss of the second marker or by Southern analysis. Because of inappropriate restriction sites for gdmL, we used the temperature-sensitive plasmid pKC1139 (4) as a delivery system instead of phage KC515, using the same methodology as previously described for disruption of the gdmH gene (21). Genotypes of the disruption mutants were confirmed by Southern analysis with the neomycin resistance gene as the DNA probe. Phenotypic characterization was performed by metabolite production analysis.
General analytical procedures.
1H nuclear magnetic resonance (1H NMR) (400 MHz) and 13C NMR (100 MHz) were recorded in CDCl3 solution at 300 K with a Bruker DRX 400 spectrometer. Chemical shifts were referenced to δ 7.26 and 77.0 for 1H and 13C spectra, respectively. To determine the structure of KOS-1806, 1H, 13C, COSY, ctHMBC, and multiplicity-edited HSQC experiments were carried out. High-resolution electrospray ionization mass spectra were obtained by manual peak matching versus internal standards with an Applied Biosystems Mariner TOF spectrometer configured with a Turbo-Ionspray source in positive ion mode.
Detection, purification, and analysis of the novel geldanamycin analog KOS-1806.
After fermentation of the gdmM-null mutant strain, the culture broth was extracted with an equal volume of methanol and the supernatant was submitted for liquid chromatography-mass spectrometry analysis. The mass spectral fragmentation pattern indicated that the new peak (m/z 541) was a sodium adduct of a geldanamycin compound. For purification, the methanol was evaporated, the residue was extracted with ethyl acetate, and the extract was concentrated to solid material. The crude, oily product was dissolved in 21 ml of methanol and diluted to 60% (vol/vol) with water. The solution was loaded onto a C18 column (3 by 11 cm, 80 ml). The column was eluted with 70% (vol/vol) MeOH-H2O. Fractions (80 ml) were collected and analyzed by high-pressure liquid chromatography with 55% (vol/vol) MeOH-H2O as the mobile phase and an Inertsil column (25 by 4.6 cm). The fractions containing the target molecule were pooled and diluted to 25% (vol/vol) MeOH-H2O (350 ml). The solution was loaded onto a C18 column (3 by 11 cm, 80 ml) and eluted with 43% (vol/vol) MeOH-H2O. Fractions (40 ml) were collected and analyzed by high-pressure liquid chromatography. Fractions 15 to 22 containing the compound of interest were pooled and concentrated in vacuum to yield KOS-1806 (59 mg) as a white solid. High-resolution liquid chromatography-mass spectrometry data pointed to the sodium adduct of the previously detected geldanamycin analog. Electrospray ionization TOF mass spectroscopy indicated an m/z of 541.2896, compared to a calculated value for C28H42N2O7Na ([M + Na]+) of 541.2884. This assignment was confirmed by NMR. The structure of KOS-1806 was elucidated by interpretation of the two-dimensional NMR spectra and comparison of the proton and 13C NMR data with those of known geldanamycins. All spectral data of KOS-1806 were thus consistent with the structure shown in Fig. 3. 1H and 13C NMR data are shown in Table 3.
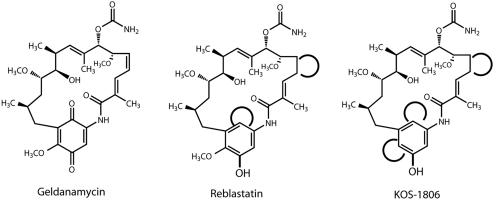
FIG. 3.
Structures of geldanamycin and monophenolic analogs. Distinctions from the geldanamycin structure are highlighted, and missing functions are indicated by open circles.
TABLE 3.
NMR assignments for KOS-1806a
| Carbon no. | δC | δH | COSY | HMBC |
|---|---|---|---|---|
| 25 | 12.6 | 1.33, 3H, s | H-9 | C-7,8,9 |
| 22 | 14.3 | 1.69, 3H, s | H-3 | C-1,2,3 |
| 26 | 17.9 | 0.89, 3H, d, J = 6 Hz | H-10 | C-9,10,11,23 |
| 28 | 18.7 | 0.69, 3H, d, J = 4.8 Hz | H-14 | C-13,14,15 |
| 4 | 19.2 | 2.02, 1H, m | H-4,5 | |
| 2.13, 1H, m | H-3,4,5 | |||
| 5 | 30.5 | 1.09, 1H, m | H-5,6 | |
| 1.21, 1H, m | ||||
| 14 | 31.2 | 1.78, 1H, m | H-13,28 | |
| 13 | 34.4 | 1.05, 1H, m | H-13,14 | |
| 1.49, 1H, m | H-13,15 | |||
| 10 | 35.1 | 2.30, 1H, m | H-9 | |
| 15 | 43.6 | 2.27, 1H, dd | H-15 | C-16,17 |
| 2.53, 1H, dd | H-14,15 | C-16,17,21 | ||
| 27 | 57.2 | 3.19, 3H, s | H-7 | C-12 |
| 23 | 59.3 | 3.29, 3H, s | C-6 | |
| 11 | 73.9 | 3.39, 1H, | H-10,11-OH | C-27 |
| 6 | 80.1 | 3.19, 1H, | H-5 | C-7,23 |
| 12 | 81.4 | 2.95, 1H, m | H-11,13,27 | C-14 |
| 7 | 81.8 | 4.81, 1H, d, J = 10 | H-6 | C-5,6,8,9,24,25 |
| 19 | 106.8 | 6.56, 1H, bs | C-17 | |
| 17 | 113.8 | 6.23, 1H, s | C-15,18,19,21 | |
| 21 | 117.2 | 6.16, 1H, s | C-17,19 | |
| 8 | 130.8 | |||
| 2 | 132.6 | |||
| 9 | 133.9 | 5.19, 1H, d, J = 8.8 | H-10,25 | C-7,25 |
| 3 | 134.9 | 5.65, 1H, s | H-4,22 | |
| 20 | 141.0 | |||
| 16 | 141.9 | |||
| 24 | 157.1 | |||
| 18 | 158.3 | |||
| 1 | 172.1 | |||
| 11-OH | 4.34, 1H, s | H-11 | ||
| 1-NH | 9.30, 1H, bs | C-2,17,19 |
COSY, correlation spectroscopy; HMBC, heteronuclear multiple bond correlation.
RESULTS
Nucleotide sequence of the hbm gene cluster.
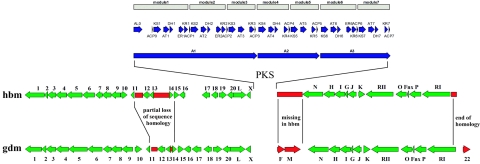
A cosmid library prepared from genomic DNA of S. hygroscopicus AM 3672 was screened by DNA hybridization by using labeled fragments of the hbm PKS loading domain gene segment and the hbmN carbamoyl transferase gene obtained by degenerate PCR on genomic DNA of S. hygroscopicus AM 3672. Three overlapping cosmids containing the hbm PKS gene cluster were isolated covering a 115-kb DNA segment. Sequencing revealed that the cloned DNA segment presumed to be involved in herbimycin biosynthesis contains 35 open reading frames (ORFs) with three large ORFs, A1, A2 and A3, coding for the PKS (Fig. 2; Table 4). The sequence data have been submitted to the DDBJ, EMBL, and GenBank databases under accession number AY947889.
FIG. 2.
Comparison of a 90-kb section of the herbimycin biosynthetic gene cluster with the genes responsible for geldanamycin production. PKS genes (A1, A2, and A3) and the PKS domains for both the hbm and gdm cluster are given in blue, with the modular organization indicated above each PKS gene by broad arrows. PKS tailoring genes are in green, and differences in gene organization or homology are indicated in red. “No homology” or “end of homology” indicates that these DNA regions are present but have <20% similarity; “missing in hbm” indicates that this region of DNA is absent in the herbimycin-producing gene cluster, whereas the comparable region contains the gdmF and gdmM genes in the geldanamycin producer.
The hbmA1-A3 ORFs encode the loading didomain and seven chain-extension modules of a multifunctional modular PKS that is essentially identical to the one described for geldanamycin (21). Each of the three predicted polypeptides is 95% similar to its gdm counterpart, supporting the expectation that the hbm polyketide product is identical to the hypothetical intermediate progeldanamycin and that differences between herbimycin and geldanamycin are introduced by post-PKS processing.
To probe the functions of the hbm and gdm PKS genes, each set was inactivated in its respective producer strain, which involved deletion of major parts of the A1ORF via insertion of a neomycin resistance gene. Delivery of the corresponding disruption cassettes was performed by homologous recombination using in the Streptomyces phages KC515 (gdmA1) and pKOS305-117A (hbmA1), respectively. The corresponding null mutants (gdmA1, K313-115-1) and (hbmA1, K390-48-1) were unable to produce herbimycin or geldanamycin when grown under standard fermentation conditions. Their genotypes were confirmed by Southern analysis with the neomycin resistance gene as probe. Detailed experimental procedures are described in the supplemental material.
Comparison of gdm and hbm tailoring genes.
As in the gdm biosynthetic gene cluster, the hbm PKS genes are flanked by sets of presumed biosynthetic genes (Fig. 2), some of which are believed to contribute to post-PKS modification by converting the hypothetical PKS intermediate, progeldanamycin (Fig. 1), into herbimycins A (Fig. 1), B, and C. The herbimycins and geldanamycin differ in the substitution pattern of their progeldanamycin backbone in three ways. First, the benzoquinoid system of all herbimycins lacks the methoxy group at the C-17 position that is present in geldanamycin. Second, an additional methoxy group is introduced at polyketide backbone position 15 in herbimycins A and C, where no such substitution is present in herbimycin B or has been reported for any geldanamycin analog. Third, the C-11 hydroxyl group of herbimycin A is O methylated, whereas herbimycin B and C remain hydroxylated at this position, like geldanamycin.
Comparison of a continuous 90-kb segment including the hbm PKS and tailoring genes with the corresponding segment of the gdm biosynthetic genes is shown in (Fig. 2), which is based on BLAST (1) analysis. The organization of genes flanking the hbm PKS is largely consistent with that observed for the gdm gene cluster. Pairwise comparisons of the corresponding ORFs revealed sequence similarities of 90 to 97%. However, there are two major differences, as highlighted in red. The first is a 4-kb segment upstream of the PKS genes between ORFs 11 and 14 where the homology between the clusters is partially lost. We could not deduce functions for ORFs 11 to 13 by sequence comparison with the databases. Only a part of ORF14 found in the hbm cluster is present in the gdm cluster. More interestingly, downstream of the PKS genes, the hbm cluster lacks homologs of gdmF and gdmM, and BLAST analysis of all ORFs identified in the 115-kb sequenced region failed to reveal a gdmF or gdmM homolog. Their absence was confirmed by a PCR analysis of total DNA from the herbimycin producer with nondegenerate primers to each end of the DNA flanking gdmF and gdmM in the region of the hbm gene cluster corresponding to the gdm genes (data not shown). These two genes are thought to be involved in post-PKS modifications of progeldanamycin. The lack of a gdmF homolog, an amide synthase believed to be responsible for cyclization of all proansamycin PKS products, raises the intriguing question of how cyclization of the nascent PKS product is achieved for herbimycin and whether a gdmF paralog is responsible. Furthermore, it should be noted that there are two structural features in both geldanamycin and herbimycin that are not consistent with the analyzed sequences. One is the formation of the C4/5 cis double bond, and another is the apparent absence of O-methyltransferases, which are required for formation of the methoxy residues at C-17 for geldanamycin and C-15 for herbimycin, respectively.
Inactivation of gdmM and gdmL.
We previously proposed that two oxidative post-PKS modifications, at positions C-17 and C-21, take place on the aromatic ring of geldanamycin (21). This hypothesis is supported by the discovery of reblastatin (Fig. 3), a nonbenzoquinoid geldanamycin analog with antitumor activity (29), isolated from a geldanamycin-producing strain (31). Reblastatin lacks the 21-hydroxyl (as well as the 4,5 double bond) and is therefore likely to be a shunt product of the normal biosynthetic pathway, resulting from the lack of aromatic ring oxidation and desaturation of the 4,5 position. In the gdm cluster we found two ORFs (ORF4 and gdmP) encoding proteins with sequence homology to CYP450 mono-oxygenases and two other ORFs flanking the PKS that also are candidates for the oxidations: gdmM, a homolog of rif19 (3), and gdmL. The deduced products of the latter two genes resemble FAD-dependent monooxygenases (21). Because the hbm cluster lacks a gdmM homolog in the sequenced segment and the C-17 position of herbimycin is not oxidized, we hypothesized that the monooxygenases encoded by gdmM and gdmL would play an important role in oxidation of the aromatic ring in progeldanamycin at positions C-17 or C-21 or both.
We were interested in engineering nonbenzoquinoid, reblastatin-like geldanamycin analogs to assess their effects on the pharmacological profile of this drug. Therefore, we attempted to inactivate the gdmM and gdmL genes to produce such analogs. Each gene was disrupted by insertion of a neomycin resistance gene and simultaneously deleting major parts of the corresponding ORFs. Interestingly, gdmL inactivation (mutant K390-33) had no discernible effect on the metabolite profile, even though its presence in the gdm and hbm clusters suggests that it could be involved in their biosynthesis. Disruption of gdmM (mutant K390-13-1), on the other hand, resulted in formation of a novel nonbenzoquinoid geldanamycin analog (KOS-1806) with a monophenolic structure (Fig. 3). As with reblastatin, post-PKS processing is interrupted and fails to form the C-4,5 cis double bond as well as the benzoquinone. However, like herbimycin, KOS-1806 remains unsubstituted at C-17. This result establishes that gdmM governs at least one of the post-PKS oxidation steps and also shows that gdmL is dispensable or that loss of its function is compensated for by a paralogous gene that is located outside of the sequenced gdm segment.
Inactivation of the genes required for AHBA formation.
We previously reported that neither the hbm nor the gdm gene cluster contains all of the genes required for formation of the ansamycin starter unit, AHBA (Table 5). A similar situation has been reported for the biosynthesis of ansamitocin (36), where the AHBA biosynthetic genes were identified in two subclusters separated by more than 30 kb. This contrasts with the situation for rifamycin where a complete ABHA biosynthetic gene set is found as part of the biosynthetic gene cluster (3).
TABLE 5.
ahba-B gene designations and functions
| ORF | Predicted function | Gene (reference) | Identity (%) | Size (aa)a |
|---|---|---|---|---|
| ahba-1a | Oxidoreductase (DAHP synthesis) | ansG (6) | 64 | 360 |
| ahba-1b | Phosphatase (DAHP synthesis) | ansH (6) | 73 | 231 |
| ahba-1c | Kinase (DAHP synthesis) | asm22 (36) | 63 | 265 |
| gdmO | Aminodehydroquinate synthase | asm47 (36) | 76 | 354 |
| ahba-3 | Aminodehydroquinate synthase | mitP (17) | 74 | 349 |
| ahba-4 | Aminodehydroquinate dehydratase | asm23 (36) | 75 | 149 |
| ahba-B | AHBA-synthase | ansF (6) | 79 | 388 |
aa, amino acids.
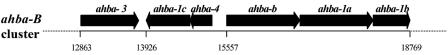
The geldanamycin producer has two different AHBA clusters (ahba-B and ahba-N) (21), whereas the herbimycin producer revealed only a ahba-B homolog (ahba-H) when its genomic DNA was analyzed by the same PCR method used for the NRRL 3062 DNA. Pairwise comparison of these deduced protein sequences to corresponding ones from other ansamycin gene clusters—ansamitocin (36), rifamycin (3), ansatrienin (6), and mitomycin C (17)—showed that the polypeptides encoded by the ahba-B and ahba-H loci are most similar to AHBA synthases involved in the biosynthesis of benzoquinone ansamycins and that the ahba-N proteins are most similar to those for the biosynthesis of naphthoquinone ansamycins. Because geldanamycin and herbimycin are benzoquinone ansamycins, we concluded that the product of ahba-B and not ahba-N was most likely to be involved in geldanamycin biosynthesis. Sequence analysis of a ahba-B-containing clone, pKOS256-116-10, which we detected in the initial screen for the gdm PKS genes (21), revealed six ORFs (Fig. 4) more than 30 kb apart from the gdm PKS cluster with sequence homology to other sets of AHBA biosynthetic genes reported for the previously mentioned ansamycins (Table 5). The sequence data have been submitted to the DDBJ, EMBL, and GenBank databases under accession number AY952143.
FIG. 4.
ahba-B biosynthetic gene cluster in S. hygroscopicus AM 3602.
Interestingly, a paralog of gdmO encoding a dehydroquinate synthase and the only ahba-related gene found close to the gdm PKS was also detected within the ahba-B group. As for the ABHA biosynthetic genes in the ansamitocin producer (36), the rifH homolog (amino-DHAP-synthase) required for early-stage AHBA biosynthesis (37) is missing from this group. We also identified a similar set of genes for the ahba-N cluster by end sequencing of various BAC clones; however, this cluster was not fully sequenced.
Two gene disruptions were carried out to test the essential role of the ahba-B cluster in geldanamycin biosynthesis. With the data we obtained for the ahba-B genes and end-sequencing data for the ahba-N cluster, we could use phage KC515 to delete the full set of either group of AHBA biosynthetic genes coincident with insertion of the neomycin resistance marker. The deletion mutants of the ahba-B (K390-76-1) genes consistently failed to produce geldanamycin, whereas deletion of the ahba-N group (K390-76-2) had no discernible effect on the production profile. Feeding of AHBA to a representative ahba-B disruption mutant (K390-76-1), fully restored geldanamycin production.
DISCUSSION
In this study we have shown that the structural similarities between herbimycin and geldanamycin are reflected in the organization of their corresponding gene clusters and hence in the predicted biosynthetic proteins. The strongest similarities were observed at the catalytic sites, with more variability seen around the interdomain linker regions. We propose that these two benzoquinone antibiotics are made from the same hypothetical polyketide intermediate, progeldanamycin, and that their structural differentiation therefore occurs through post-PKS biosynthetic steps.
We identified two separate sets of ABHA biosynthetic genes, ahba-B and ahba-N, in the geldanamycin producer, whereas only an ahba-B homolog (the ahba-H cluster) was found in the herbimycin producer but not completely characterized. Note that the AHBA genes are also found in two separate regions in the ansamitocin-producing organism (36). In the geldanamycin producer, we found that the ahba-N cluster is closely associated with a group of type I modular PKS genes (unpublished results), whereas the ahba-B cluster is not. Sequence analysis of the AHBA synthesis enzymes from both clusters was used to establish that the ahba-B set encodes proteins most similar to enzymes known to be involved in the biosynthesis of benzoquinone-type ansamycins, whereas the ahba-N set encodes enzymes that are more similar to those from producers of naphthoquinone-type ansamycins (data not shown). The ahba-B gene products are very similar to those reported for ansatrienin biosynthesis in S. collinus Tü 1892 (6). Inactivation of each ABHA biosynthetic gene set showed that only the ahba-B locus is required for formation of geldanamycin, unlike the case for ansamitocin, where both ABHA biosynthetic gene loci have genes that are essential for ansamitocin biosynthesis (36).
One feature of the biosynthesis of herbimycin and geldanamycin that cannot be explained by the gene sequence data is the formation of the C-4,5 cis double bond. PKS module 6, responsible for the corresponding chain-elongation step, contains an enoyl reductase (ER) domain and therefore should catalyze formation of a saturated C-4,5 carbon bond in progeldanamycin. Several observations support this idea. First, sequence comparison of ER6 with other functional ER domains in modular PKSs revealed no obvious active site mutations, and therefore there is no compelling evidence that ER6 is inactive in either PKS cluster. Second, given the possibility of an inactive ER6, recent findings about the stereoselectivity of ketoreductase (KR) domains (22) suggest that the nature of the corresponding hbm/gdm KR6 would not support the formation of a cis-unsaturated system at this position. Sequence comparisons of various KR domains have revealed that a particular Asp residue is present in all KRs that catalyze the formation of β-hydroxy products with the D absolute configuration. A group of KRs that support the formation of l-β-hydroxy products lack this residue, which in combination with a functional dehydratase domain, would result in a “cis” unsaturated polyketide product (22). This particular Asp residue is present in the gdm and hbm KR6 domain, which therefore qualifies it for the formation of a trans, not a cis, α,β-unsaturated system. More importantly, there are at least four geldanamycin analogs where the C-4,5 position remains saturated. One is reblastatin; another is the product of gdmM inactivation, KOS-1806, a reblastatin analog with a saturated 4,5 position; and a third is the C-4,5 saturated shunt product produced by a gdmA3 PKS acyltransferase 7 exchange mutant (20). Finally, Hong et al. (10) have recently reported that several post-PKS products with a saturated 4,5 double bond are present in a gdmN-null mutant. They show through bioconversion experiments that one of these compounds, 4,5-dihydro-7-descarbamoyl-7-hydroxygeldanamycin is converted to geldanamycin. A compound whose mass spectral data were consistent with the latter intermediate had previously been isolated from a gdmH-null mutant in which gdmN was partially inactivated (21). The accumulated data therefore support the belief that desaturation of the 4,5 position is the final step in geldanamycin biosynthesis (10).
Progeldanamycin therefore is very likely to have a C-4,5 saturated ansamycin system, which leads to the intriguing question of how the cis double bond is introduced. Catalysis by a fortuitous desaturase, utilizing a mechanism similar to that involved in the oxidative desaturation of fatty acids (34), is one possibility. However, this type of biochemical mechanism has not been reported for the biosynthesis of polyketide secondary metabolites to our knowledge. Oxidative processing by post-PKS enzymes that are unrelated to the typical fatty acid desaturases, such as the gdmP gene encoding a CYP450 enzyme, is a more plausible alternative.
Post-PKS processing steps distinguish the biosynthesis of herbimycin and geldanamycin. In contrast to the other ansamycin PKSs, the hbm PKS tailoring genes lack an amide synthase homolog in the region sequenced, raising the question of whether such a gene is located elsewhere in the genome or is not required for closure of the ansamycin ring system. If the putative hbmF gene is in fact absent, cyclization would have to be achieved through another mechanism. Overexpression of the gdmF gene in the herbimycin producer would be a way to address this question if that were to enhance the amount of herbimycin produced. Another intriguing observation is the apparent absence of O-methyltransferase genes (other than those required for methoxymalonate biosynthesis) in the gdm and hbm gene clusters, because formation of both of these benzoquinone antibiotics requires O methylation at C-17 for geldanamycin and at C-15 or C-11 for herbimycin.
We have not yet established the exact sequence of the post-PKS oxidations necessary to form the benzoquinone system of geldanamycin or herbimycin. However, the results obtained to date by us and others (10) provide the following insights. Inactivation of gdmH or gdmN does not interfere with the required post-PKS oxidations at C-17 and C-21, but C-4,5 desaturation is affected (10, 21), probably because the resulting intermediates are poor substrates for the putative C-4,5-desaturase. Inactivation of gdmM results in failure to perform any of the post-PKS oxidation steps and also blocks formation of the C-4,5 cis double bond. Similarly, a gdmA3 acyltransferase 7 swap mutant produces a compound that has not undergone the aromatic ring oxidations and C-4,5 desaturation. On the other hand, the work of Hong et al. (10) shows that C-21 oxidation to the benzoquinone and hydroxylation plus O methylation of C-17 can take place before 7-O carbamoylation and C-4,5 desaturation. On the basis of these observations, it seems that formation of the C-4,5 cis-unsaturated system is a late event in post-PKS processing that is preceded by C-21 oxidation and C-7 carbamoylation. The apparent absence of a gdmM homolog in the hbm gene cluster, together with the results from the corresponding gene disruption in the gdm gene cluster, suggest that gdmM is likely to be involved in C-17 oxidation. Furthermore, evidence from reblastatin, where C-17 is fully substituted but C-21 remains unoxidized, suggests that the C-17 and C-21 oxidations are two separate catalytic events. Yet neither C-21 oxidation nor substitution of C-17 are obligatory steps prior to 7-O carbamoylation. Because inactivation of gdmL did not affect the production of geldanamycin, other putative oxygenase genes in the gdm cluster, such as gdm04 and gdmP, are likely to be involved in formation of the benzoquinoid and 4,5 double bond found in geldanamycin and herbimycin.
Supplementary Material
TABLE 4.
Comparison of geldanamycin and herbimycin ORFsa
| Gdm ORF
|
Proposed function | Hbm ORF
|
||
|---|---|---|---|---|
| Boundaries | Name | Name | Boundaries | |
| 1-1652b | ORF01 | Putative cation-transporting ATPase; homolog of S. coelicolor SC0860c and S. avermitillis SAV617 | ORF01 | 71-2359 |
| 1652-2083 | ORF02 | Putative secreted protein; homolog of S. coelicolor SC0861c and S. avermitilis SAV618 | ORF02 | 2359-2775 |
| 2070-3053 | ORF03 | Homolog of PvcA (P. aeruginosa PA2234) and of V. cholerae VC1949 | ORF03 | 2762-3745 |
| 3057-4313 | ORF04 | P450 | ORF04 | 3757-5013 |
| 4326-6152 | ORF05 | Asparagine synthase family | ORF05 | 5026-6852 |
| 6187-7617 | ORF06 | Transmembrane efflux protein | ORF06 | 6887-8317 |
| 7723-8526 | ORF07 | Permease (Fe); homolog of FtrE, S. coelicolor SC0998 | ORF07 | 8437-9240 |
| 8490-9572 | ORF08 | Homolog of FtrD, S. coelicolor SC0997 | ORF08 | 9204-10286 |
| 9572-10648 | ORF09 | Lipoprotein | ORF09 | 10286-11362 |
| 10649-15731 | ORF10-15 | Functions unclear | ORF10-15 | 11363-18359 |
| 15732-16415 | ORF16 | RhtB family transporter | ORF16 | 18360-19043 |
| 16502-17404 | ORF17 | Secreted protein | ORF17 | 21063-21965 |
| 17676-18467 | ORF18 | Hydrolase | ORF18 | 22155-22946 |
| 18621-19505 | ORF19 | Transcriptional regulator (AraC family) | ORF19 | 23100-23984 |
| 19555-20316 | ORF20 | Transcriptional regulator (TetR family) | ORF20 | 24036-24797 |
| 20357-21796 | GdmL | Flavin-dependent monooxygenase | HbmL | 24781-26277 |
| 21838-22308 | GdmX | Conserved JadX and MmyY homolog | HbmX | 26325-26795 |
| 22939-43464 | GdmAI | PKS modules 0 to 3 | HbmAI | 27677-48139 |
| Loading | AL0 X ACP0 | Loading | ||
| Module 1 | KS AT DH ER KR ACP | Module 1 | ||
| Module 2 | KS AT DH ER KR ACP | Module 2 | ||
| Module 3 | KS AT KR ACP | Module 3 | ||
| 43525-53829 | GdmAII | PKS modules 4 and 5 | HbmAII | 48197-58492 |
| Module 4 | KS AT DH KR ACP | Module 4 | ||
| Module 5 | KS AT KR ACP | Module 5 | ||
| 53859-65546 | GdmAIII | PKS modules 6 and 7 | HbmAIII | |
| Module 6 | KS AT DH ER KR ACP | Module 6 | 58519-70125 | |
| Module 7 | KS AT DH KR ACP | Module 7 | ||
| 6558-66331 | GdmF | Amide synthase | none | |
| 66328-67962 | GdmM | Flavin-dependent monooxygenase | none | |
| 68782-70791 | GdmN | Carbamoyltransferase | HbmN | 70662-72719 |
| 70853-71965 | GdmH | Methoxymalonyl-ACP biosynthesis pathway | HbmH | 72781-73893 |
| 71962-73074 | GdmI | Methoxymalonyl-ACP biosynthesis pathway | HbmI | 73890-75002 |
| 73071-73346 | GdmJ | ACP in methoxymalonyl-ACP biosynthesis pathway | HbmJ | 74999-75274 |
| 73343-74209 | GdmK | Methoxymalonyl-ACP biosynthesis pathway | HbmK | 75271-76137 |
| 74453-75019 | GdmG | O-Methyltransferase in methoxymalonyl-ACP biosynthesis | HbmG | 76381-77037 |
| 75234-78014 | GdmRII | LuxR-type transcriptional regulator | HbmRII | 77137-79917 |
| 78289-79353 | GdmO | Amino DHQ synthase | HbmO | 80193-81257 |
| GdmFdx | Ferredoxin | HbmFdx | 81334-81528 | |
| 79671-80864 | GdmP | P450 | HbmP | 81571-82764 |
| 81021-83909 | GdmRI | LuxR-type transcriptional regulator | HbmRI | 82921-86764 |
| 84662-85375 | ORF22 | Hydrolase | None | |
Gdm, geldanamycin; Hbm, herbimycin.
N terminus only.
Acknowledgments
We thank David Hopwood and Leonard Katz for helpful discussions and J. Carney and C. Tran for determining mass spectra. We also thank Rika Regentin for optimizing the fermentation procedures for the production of KOS-1806.
This study was supported in part by grants CA96262 and AL38947 from the National Institutes of Health.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Arakawa, K., R. Muller, T. Mahmud, T. W. Yu, and H. G. Floss. 2002. Characterization of the early stage aminoshikimate pathway in the formation of 3-amino-5-hydroxybenzoic acid: the RifN protein specifically converts kanosamine into kanosamine 6-phosphate. J. Am. Chem. Soc. 124:10644-10645. [DOI] [PubMed] [Google Scholar]
- 3.August, P. R., L. Tang, Y. J. Yoon, S. Ning, R. Muller, T. W. Yu, M. Taylor, D. Hoffmann, C. G. Kim, X. Zhang, C. R. Hutchinson, and H. G. Floss. 1998. Biosynthesis of the ansamycin antibiotic rifamycin: deductions from the molecular analysis of the rif biosynthetic gene cluster of Amycolatopsis mediterranei S699. Chem. Biol. 5:69-79. [DOI] [PubMed] [Google Scholar]
- 4.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. [DOI] [PubMed] [Google Scholar]
- 5.Blondelet-Rouault, M. H., J. Weiser, A. Lebrihi, P. Branny, and J. L. Pernodet. 1997. Antibiotic resistance gene cassettes derived from the omega interposon for use in Escherichia coli and Streptomyces. Gene 190:315-317. [DOI] [PubMed] [Google Scholar]
- 6.Chen, S., D. von Bamberg, V. Hale, M. Breuer, B. Hardt, R. Muller, H. G. Floss, K. A. Reynolds, and E. Leistner. 1999. Biosynthesis of ansatrienin (mycotrienin) and naphthomycin: identification and analysis of two separate biosynthetic gene clusters in Streptomyces collinus Tu 1892. Eur. J. Biochem. 261:98-107. [DOI] [PubMed] [Google Scholar]
- 7.DeBoer, C., P. A. Meulman, R. J. Wnuk, and D. H. Peterson. 1970. Geldanamycin, a new antibiotic. J. Antibiot. 23:442-447. [DOI] [PubMed] [Google Scholar]
- 8.Denis, F., and R. Brzezinski. 1991. An improved aminoglycoside resistance gene cassette for use in gram-negative bacteria and Streptomyces. FEMS Microbiol. Lett. 65:261-264. [DOI] [PubMed] [Google Scholar]
- 9.Donadio, S., M. J. Staver, J. B. McAlpine, S. J. Swanson, and L. Katz. 1991. Modular organization of genes required for complex polyketide biosynthesis. Science 252:675-679. [DOI] [PubMed] [Google Scholar]
- 10.Hong, Y. S., D. Lee, W. Kim, J. K. Jeong, C. G. Kim, J. K. Sohng, J. H. Lee, S. G. Paik, and J. J. Lee. 2004. Inactivation of the carbamoyltransferase gene refines post-polyketide synthase modification steps in the biosynthesis of the antitumor agent geldanamycin. J. Am. Chem. Soc. 126:11142-11143. [DOI] [PubMed] [Google Scholar]
- 11.Hopwood, D. A., T. Kieser, H. M. Wright, and M. J. Bibb. 1983. Plasmids, recombination and chromosome mapping in Streptomyces lividans 66. J. Gen. Microbiol. 129(Pt. 7):2257-2269. [DOI] [PubMed] [Google Scholar]
- 12.Ivy, P. S., and M. Schoenfeldt. 2004. Clinical trials referral resource: current clinical trials of 17-AG and 17-DMAG. Oncology 615:619-620. [PubMed] [Google Scholar]
- 13.Iwai, Y., A. Nakagawa, N. Sadakane, S. Omura, H. Oiwa, S. Matsumoto, M. Takahashi, T. Ikai, and Y. Ochiai. 1980. Herbimycin B, a new benzoquinonoid ansamycin with anti-TMV and herbicidal activities. J. Antibiot. 33:1114-1119. [DOI] [PubMed] [Google Scholar]
- 14.Kieser, T., J. Bibb, M., M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 15.Kim, C. G., T. W. Yu, C. B. Fryhle, S. Handa, and H. G. Floss. 1998. 3-Amino-5-hydroxybenzoic acid synthase, the terminal enzyme in the formation of the precursor of mC7N units in rifamycin and related antibiotics. J. Biol. Chem. 273:6030-6040. [DOI] [PubMed] [Google Scholar]
- 16.Lin, X., S. Kaul, S. Rounsley, T. P. Shea, M. I. Benito, C. D. Town, C. Y. Fujii, T. Mason, C. L. Bowman, M. Barnstead, T. V. Feldblyum, C. R. Buell, K. A. Ketchum, J. Lee, C. M. Ronning, H. L. Koo, K. S. Moffat, L. A. Cronin, M. Shen, G. Pai, S. Van Aken, L. Umayam, L. J. Tallon, J. E. Gill, J. C. Venter, et al. 1999. Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature 402:761-768. [DOI] [PubMed] [Google Scholar]
- 17.Mao, Y., M. Varoglu, and D. H. Sherman. 1999. Molecular characterization and analysis of the biosynthetic gene cluster for the antitumor antibiotic mitomycin C from Streptomyces lavendulae NRRL 2564. Chem. Biol. 6:251-263. [DOI] [PubMed] [Google Scholar]
- 18.Muroi, M., M. Izawa, Y. Kosai, and M. Asai. 1980. Macbecins I and II, new antitumor antibiotics. II. Isolation and characterization. J. Antibiot. 33:205-212. [DOI] [PubMed] [Google Scholar]
- 19.Omura, S., Y. Iwai, Y. Takahashi, N. Sadakane, A. Nakagawa, H. Oiwa, Y. Hasegawa, and T. Ikai. 1979. Herbimycin, a new antibiotic produced by a strain of Streptomyces. J. Antibiot. 32:255-261. [DOI] [PubMed] [Google Scholar]
- 20.Patel, K., M. Piagentini, A. Rascher, Z. Q. Tian, G. O. Buchanan, R. Regentin, Z. Hu, C. R. Hutchinson, and R. McDaniel. 2004. Engineered biosynthesis of geldanamycin analogs for hsp90 inhibition. Chem. Biol. 11:1625-1633. [DOI] [PubMed]
- 21.Rascher, A., Z. Hu, N. Viswanathan, A. Schirmer, R. Reid, W. C. Nierman, M. Lewis, and C. R. Hutchinson. 2003. Cloning and characterization of a gene cluster for geldanamycin production in Streptomyces hygroscopicus NRRL 3602. FEMS Microbiol. Lett. 218:223-230. [DOI] [PubMed] [Google Scholar]
- 22.Reid, R., M. Piagentini, E. Rodriguez, G. Ashley, N. Viswanathan, J. Carney, D. V. Santi, C. R. Hutchinson, and R. McDaniel. 2003. A model of structure and catalysis for ketoreductase domains in modular polyketide synthases. Biochemistry 42:72-79. [DOI] [PubMed] [Google Scholar]
- 23.Richter, K., and J. Buchner. 2001. Hsp90: chaperoning signal transduction. J. Cell Physiol. 188:281-290. [DOI] [PubMed] [Google Scholar]
- 24.Rinehart, K. L., Jr., and L. S. Shield. 1976. Chemistry of the ansamycin antibiotics. Fortschr. Chem. Org. Naturst. 33:231-307. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook, J., and D. W. Russel. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor, Cold Spring Harbor, N.Y.
- 26.Sasaki, K., H. Yasuda, and K. Onodera. 1979. Growth inhibition of virus transformed cells in vitro and antitumor activity in vivo of geldanamycin and its derivatives. J. Antibiot. 32:849-851. [DOI] [PubMed] [Google Scholar]
- 27.Schnur, R. C., M. L. Corman, R. J. Gallaschun, B. A. Cooper, M. F. Dee, J. L. Doty, M. L. Muzzi, J. D. Moyer, C. I. DiOrio, E. G. Barbacci, et al. 1995. Inhibition of the oncogene product p185erbB-2 in vitro and in vivo by geldanamycin and dihydrogel danamycin derivatives. J. Med. Chem. 38:3806-3812. [DOI] [PubMed] [Google Scholar]
- 28.Staunton, J., and K. J. Weissman. 2001. Polyketide biosynthesis: a millennium review. Nat. Prod. Rep. 18:380-416. [DOI] [PubMed] [Google Scholar]
- 29.Stead, P., S. Latif, A. P. Blackaby, P. J. Sidebottom, A. Deakin, N. L. Taylor, P. Life, J. Spaull, F. Burrell, R. Jones, J. Lewis, I. Davidson, and T. Mander. 2000. Discovery of novel ansamycins possessing potent inhibitory activity in a cell-based oncostatin M signalling assay. J. Antibiot. 53:657-663. [DOI] [PubMed] [Google Scholar]
- 30.Supko, J. G., R. L. Hickman, M. R. Grever, and L. Malspeis. 1995. Preclinical pharmacologic evaluation of geldanamycin as an antitumor agent. Cancer Chemother. Pharmacol. 36:305-315. [DOI] [PubMed] [Google Scholar]
- 31.Takatsu, T., M. Ohtsuki, A. Muramatsu, R. Enokita, and S. I. Kurakata. 2000. Reblastatin, a novel benzenoid ansamycin-type cell cycle inhibitor. J. Antibiot. 53:1310-1312. [DOI] [PubMed] [Google Scholar]
- 32.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uehara, Y., Y. Murakami, K. Suzukake-Tsuchiya, Y. Moriya, H. Sano, K. Shibata, and S. Omura. 1988. Effects of herbimycin derivatives on src oncogene function in relation to antitumor activity. J. Antibiot. 41:831-834. [DOI] [PubMed] [Google Scholar]
- 34.Villorbina, G., L. Roura, F. Camps, J. Joglar, and G. Fabrias. 2003. Enzymatic desaturation of fatty acids: delta11 desaturase activity on cyclopropane acid probes. J. Org. Chem. 68:2820-2829. [DOI] [PubMed] [Google Scholar]
- 35.Whitesell, L., E. G. Mimnaugh, B. De Costa, C. E. Myers, and L. M. Neckers. 1994. Inhibition of heat shock protein HSP90-pp60v-src heteroprotein complex formation by benzoquinone ansamycins: essential role for stress proteins in oncogenic transformation. Proc. Natl. Acad. Sci. USA 91:8324-8328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu, T. W., L. Bai, D. Clade, D. Hoffmann, S. Toelzer, K. Q. Trinh, J. Xu, S. J. Moss, E. Leistner, and H. G. Floss. 2002. The biosynthetic gene cluster of the maytansinoid antitumor agent ansamitocin from Actinosynnema pretiosum. Proc. Natl. Acad. Sci. USA 99:7968-7973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu, T. W., R. Muller, M. Muller, X. Zhang, G. Draeger, C. G. Kim, E. Leistner, and H. G. Floss. 2001. Mutational analysis and reconstituted expression of the biosynthetic genes involved in the formation of 3-amino-5-hydroxybenzoic acid, the starter unit of rifamycin biosynthesis in Amycolatopsis mediterranei S699. J. Biol. Chem. 276:12546-12555. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.