Abstract
The gene (PH1074) encoding the NAD kinase of the hyperthermophilic archaeon Pyrococcus horikoshii was identified in the genome database, cloned, and functionally expressed in Escherichia coli. The recombinant enzyme was purified to homogeneity by heat treatment at 90°C for 20 min and one successive HiTrap affinity chromatography step. The purified enzyme was easily precipitated by dialysis against phosphate buffer without NaCl and imidazole and was usually stored in buffer containing 0.5 M NaCl and 0.5 M imidazole to avoid precipitation. The molecular mass of the active enzyme was determined to be 145 kDa by a gel filtration method, and the enzyme was composed of a tetramer of 37-kDa subunits. The archaeal enzyme utilized several nucleoside triphosphates, such as GTP, CTP, UTP, and ITP, as well as ATP and inorganic polyphosphates [poly(P)] as phosphoryl donors for NAD phosphorylation. The enzyme utilized poly(P)27 (the average length of the phosphoryl chain was 27) as the most active inorganic polyphosphate for NAD phosphorylation. Thus, this enzyme is categorized as an inorganic polyphosphate/ATP-dependent NAD kinase. The enzyme was the most thermostable NAD kinase found to date: its activity was not lost by incubation at 95°C for 10 min. The enzyme showed classical Michaelis-Menten-type kinetics for NAD and ATP, but not for poly(P)27. The Km values for NAD were determined to be 0.30 and 0.40 mM when poly(P)27 and ATP, respectively, were used as the phosphoryl donors. The Km value for ATP was 0.29 mM, and the concentration of poly(P)27 which gave half of the maximum enzyme activity was 0.59 mM. The enzyme required several metal cations, such as Mg2+, Mn2+, or Ni2+, for its activity. The deduced amino acid sequence showed a low level of identity to those of E. coli ATP-dependent NAD kinase (31%) and the inorganic polyphosphate/ATP-dependent NAD kinase of Mycobacterium tuberculosis (29%). This is the first description of the characteristics of a poly(P)/ATP-dependent NAD kinase from a hyperthermophilic archaeon.
NAD kinase (EC 2.7.1.23) catalyzes the formation of NADP through the phosphorylation of NAD. This is the only key enzyme that plays a crucial role in the regulation of the NADP level and of NADP-dependent anabolic/biosynthetic pathways in living organisms (18). In spite of the important physiological role of NADP, the purification and characterization of NAD kinases have been limited to those of several organisms, including vertebrates such as pigeons (the liver [2] and the heart [4]) and humans (16), yeasts such as Saccharomyces cerevisiae (12) and Candida utilis (5), and bacteria such as Mycobacterium tuberculosis H37Rv (10), Micrococcus flavus (10), Sphyngomonas sp. (20), and Bacillus subtilis (6). The NAD kinase genes from some bacteria, such as M. tuberculosis (10), M. flavus (11), and Escherichia coli (9), have been cloned and sequenced. Among them, NAD kinases from many organisms, except for a few bacteria, utilize ATP and some other nucleoside triphosphates, but not inorganic polyphosphate [poly(P)], as phosphate donors. On the other hand, the enzymes from M. tuberculosis, M. flavus, and B. subtilis have been reported to utilize inorganic polyphosphate [poly(P)] as well as ATP for NAD phosphorylation (6, 10). Poly(P) is a polymer of inorganic orthophosphate residues linked by high-energy phosphoanhydride bonds and is proposed to be a primitive energy source for living organisms (14). Although poly(P)/ATP-NAD kinases are postulated to be prototypes of ATP-dependent NAD kinases, there is no information so far about NAD kinases from archaea, which form the third and evolutionally oldest domain of life (25). Poly(P) could be used as a phosphoryl donor for enzymatic phosphorylation instead of ATP and ADP. NAD kinases from mesophiles have been used for the industrial production of NADP from NAD (8), and poly(P) is commercially available as a much less expensive phosphoryl donor than ATP. Stable poly(P)-dependent NAD kinase is more useful for enzymatic NADP production. The hyperthermophilic archaea have a high potential as a new source of much more stable enzymes than the counterparts of mesophiles. For this study, we have found a gene (open reading frame identification no. PH1074) encoding an NAD kinase homolog from a search of the genome sequence databases for an anaerobic hyperthermophilic archaeon, Pyrococcus horikoshii OT3 (13). We succeeded in the gene expression, purification, and characterization of the P. horikoshii NAD kinase. In this paper, we report the first biochemical characterization and primary structure of a hyperthermophilic NAD kinase.
MATERIALS AND METHODS
Materials.
The pET-15b vector was obtained from Novagen (Madison, WI). The E. coli strain BL21-CodonPlus-RIL(DE3) was purchased from Stratagene (La Jolla, CA). NAD and Proteus sp. glutamate dehydrogenase were purchased from Sigma (St. Louis, MO). Restriction enzymes and KOD DNA polymerase were purchased from New England Biolabs, Inc. (Beverly, MA) and Toyobo (Osaka, Japan), respectively. ATP, poly(P)n and poly(P)x (“x” indicates a mixture of polyphosphates with various chain lengths) were purchased from Wako Pure Chemicals (Osaka). All other chemicals were of reagent grade.
Enzyme assay and protein determination.
Poly(P)- and ATP-dependent NAD kinases were assayed by a modified two-step method as previously described (18). The reaction system was comprised, unless specified otherwise, of 100 mM bis-Tris-HCl (pH 6.8), 5.0 mM NAD, 5.0 mM MnCl2, phosphoryl donor [2.0 mg of poly(P)x, 2.0 mg of poly(P)n, or 5.0 mM ATP], and the enzyme in a total volume of 1.00 ml. After the reaction was carried out at 70°C for 30 min, ice-cold 30% HClO4 (40 μl) was added, and the mixture was kept on ice for 5 min. The mixture was centrifuged at 12,000 rpm for 10 min, and the supernatant was neutralized by the addition of 4 M KOH (40 μl). After the reaction mixture was clarified by centrifugation (12,000 rpm for 10 min), the NADP concentration of the supernatant solution was determined spectrophotometrically at 40°C using a reaction mixture (1.00 ml) containing 225 mM Tris-HCl (pH 7.8), 10 mM l-glutamate, 0.8 units Proteus sp. NADP-specific glutamate dehydrogenase, and an aliquot of the supernatant solution (50 μl). The absorbance of NADPH was monitored by the absorbance at 340 nm (extinction coefficient [ɛ] = 6.22 mM−1 cm−1). One unit of activity was defined as the amount of enzyme producing 1.0 μmol of NADP for 1 min at 70°C in a standard assay mixture containing poly(P)x as the phosphoryl donor, and the specific activity was expressed in units/mg of protein. NADH kinase activity was assayed by measuring NADPH formation from NADH. The assay mixture consisted of 100 mM Tris-HCl (pH 7.5), 5 mM NADH, 2 mg poly(P)x or 5 mM ATP, 5 mM MnCl2, and enzyme in a final volume of 1.0 ml. After incubation at 37°C for 60 min, ice-cold 30% HClO4 (40 μl) was added, and the mixture was kept on ice for 5 min. The mixture was centrifuged at 12,000 rpm for 10 min, and the supernatant was neutralized by the addition of 4 M KOH (40 μl). After the reaction mixture was clarified by centrifugation (12,000 rpm for 10 min), filtration (0.45-μm filter) of the supernatant was carried out, and an aliquot (5 μl) was used for NADPH determination by high-performance liquid chromatography (JASCO880-PU instrument with an Asahipak GS320HQ column). The peak of NADPH was detected by the absorbance at 340 nm, and the amount was determined by use of a calibration curve created from an NADPH standard. A reproducible assay for NADH kinase activity at higher temperatures such as 50 and 70°C was not achieved under these assay conditions. The protein concentration was determined by the Bradford method (3), with bovine serum albumin as the standard.
Cloning and expression of the gene encoding P. horikoshii NAD kinase.
The complete genome sequence of P. horikoshii OT3 has been reported by Kawarabayasi et al. (13). The amino acid sequence deduced from the gene (PH1074; accession number AB055976) of P. horikoshii encoding NAD kinase exhibited high homology with those of NAD kinases from other sources on the basis of a BLAST database search (1). The following set of oligonucleotide primers was used to amplify the gene fragment encoding NAD kinase by PCR: the first primer (5′-ATACCATATGAAGTTTGGTATAGTTGC-3′) was used to introduce a unique NdeI restriction site overlapping the 5′ initiation codon, and the other (5′-AGCCGGATCCCTAGTGTCTTTCCTTTATCC-3′) was used to introduce a unique BamHI restriction site in proximity to the 3′ end of the termination codon. The chromosomal P. horikoshii DNA was isolated as previously described (23) and used as the template. The amplified 0.8-kb fragment was ligated with a T-vector using a TA cloning kit (Invitrogen), and a hybrid plasmid, pTNADK, was constructed. Because an NdeI site (CATATG) is present in the gene PH1074, the site was silently mutated to CACATG using a QuikChange XL site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. pTNADK was used as the template, and the following set of oligonucleotide primers were used as the mutagenic primers: 5′-CGATCCTCAGGATAGAGCACATGACTAAAAAGGATATAC-3′ and 5′-GTATATCCTTTTTAGTCATGTGCTCTATCCTGAGGATCG-3′. After the nucleotidesequence was confirmed, the mutant plasmid was digested with NdeI and BamHI. The resulting 0.8-kb fragment was ligated into the expression vector pET15b, which was linearized with NdeI and BamHI, to generate pET15-NADK. E. coli BL21-CodonPlus-RIL(DE3) was transformed with pET15-NADK. The transformants were cultivated at 37°C in a medium (200 ml) containing 2.4 g of tryptone, 4.8 g of yeast extract, 1 ml of glycerol, 2.5 g of K2HPO4, 0.76 g of KH2PO4, and 10 mg of ampicillin until the absorbance at 600 nm reached 0.6. Induction was carried out by the addition of 1.0 mM isopropyl-β-d-thiogalactopyranoside to the medium, and the cultivation was continued for 12 h at 18°C. After cultivation, the cells were collected by centrifugation (8,000 × g for 10 min).
Purification of recombinant NAD kinase.
All procedures were carried out at room temperature. E. coli cells (about 6.5 g [wet weight]) harvested from a 0.50-liter culture were used as the starting material for the purification of NAD kinase. Unless otherwise stated, 20 mM Tris-HCl (pH 7.4) containing 10% glycerol was utilized as the standard buffer throughout the purification procedures used for this study. To prepare a crude extract, the cells were washed twice and suspended in the standard buffer. The cells were disrupted by sonication and centrifuged at 15,000 × g for 10 min. The resultant supernatant was used as the crude extract. Solid sodium sulfate was added to the crude extract at a final concentration of 0.2 M, the solution was heated at 90°C for 20 min, and denatured proteins were removed by centrifugation. The supernatant was dialyzed against the standard buffer and then applied to a HiTrap chelating column (5-ml volume; Pharmacia). Before the application of the enzyme, the column was equilibrated with 0.1 M NiSO4 (2.5 ml) and then with standard buffer containing 0.5 M NaCl (50 ml). After washing of the column with buffer containing 0.5 M NaCl and 0.05 M imidazole (50 ml), the enzyme was eluted with buffer containing 0.5 M NaCl and 0.5 M imidazole. The active fractions were pooled and used as the purified preparation. The purified enzyme was easily precipitated when NaCl and imidazole were removed from the buffer by dialysis. Thus, the buffer containing 0.5 M NaCl and 0.5 M imidazole was ordinarily used for the enzyme solution to avoid precipitation.
SDS-PAGE.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out as described by Laemmli (15) using 12.5% acrylamide and 0.1% SDS in a discontinuous Tris-glycine buffer system. maltose-binding protein--β-galactosidase (175 kDa), maltose-binding protein-paramyosin (83 kDa), glutamate dehydrogenase (62 kDa), aldolase (47.5 kDa), triosephosphate isomerase (32.5 kDa), β-lactoglobulin A (25 kDa), lysozyme (16.5 kDa), and aprotinin (6.5 kDa) were used as molecular mass standards (New England Biolabs). The protein sample was boiled at 100°C for 5 min in 60 mM Tris-HCl (pH 6.8) containing 2% SDS, 10% glycerol, 5% 2-mercaptoethanol, and 0.002% bromphenol blue.
Molecular mass determination.
The molecular mass of the purified enzyme was determined using an analytical Superose 6 (Amersham Bioscience) gel filtration column pre-equilibrated with 50 mM Tris-HCl buffer (pH 7.5) containing 0.2 M NaCl. Ferritin (440 kDa), catalase (232 kDa), aldolase (158 kDa), albumin (67 kDa), ovalbumin (43 kDa), chymotrypsinogen A (25 kDa), and RNase A (13.7 kDa) in gel filtration calibration kits (Amersham Bioscience) were used as molecular mass standards.
N-terminal amino acid sequencing.
Approximately 3 μg of the purified enzyme was subjected to SDS-PAGE as already described, followed by electroblotting onto a polyvinylidene difluoride membrane. The membrane was then stained with Ponceau S and destained. A protein band was excised and subjected to automated Edman degradation using a Shimadzu model PPSQ-10 protein sequencer.
Stability, pH optimum, and kinetic parameters.
To determine the thermostability of the enzyme, the enzyme solution (0.3 mg/ml) in 10 mM potassium phosphate buffer (pH 7.0) containing 0.5 M NaCl and 0.5 M imidazole was incubated at different temperatures, and the residual activity was determined by the standard assay method. To determine the pH stability, the enzyme (0.3 mg/ml) was incubated in buffers at various pH values (the pH was adjusted at 25°C) at 70°C for 60 min, and the remaining activity was then assayed by the standard two-step method. The buffers (100 mM) used were acetate buffer, morpholineethanesulfonic acid-NaOH, bis-Tris-HCl, Tris-HCl, and glycine-NaOH, for pH ranges of 3.5 to 5.0, 5.5 to 6.0, 6.0 to 7.0, 7.0 to 8.5, and 8.5 to 10.0, respectively. The same buffers were used to determine the optimal pH of the enzyme by running the standard assay method at 70°C. The Michaelis constants were determined from double-reciprocal-plot data obtained from the initial rate of NADP formation at 70°C.
RESULTS
Identification and expression of a hypothetical gene encoding NAD kinase in P. horikoshii.
In the genome database of P. horikoshii, we found a gene, PH1074, consisting of 834 bp at positions 978,541 to 979,374 of the entire genome. The predicted amino acid sequence (277 amino acids with a molecular mass of 31,414 Da) showed 31 and 29% identity with those of E. coli (9) and M. tuberculosis NAD kinases (10), respectively. We performed gene cloning, and the E. coli strain BL21-CodonPlus-RIL(DE3) was transformed with the expression vector pET15-NADK. In the cell extract of the transformant, NAD kinase activity was detected after incubation at 90°C for 20 min, suggesting the production of the hyperthermostable NAD kinase in E. coli.
Purification of the recombinant enzyme.
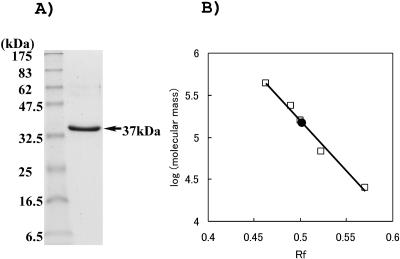
A typical result for purification of the recombinant enzyme from an extract of the E. coli transformant is summarized in Table 1. The enzyme was purified about 8.4-fold, with a 35% recovery rate, by heat treatment and a successive HiTrap affinity chromatography step. The purified enzyme was found to be homogeneous by SDS-PAGE (Fig. 1). About 22.5 mg of the purified enzyme was obtained from 0.5 liter of E. coli culture.
TABLE 1.
Summary of purification of P. horikoshii NAD kinase from recombinant E. coli cells
| Purification step | Total protein (mg) | Total activity (U) | Sp act (U/mg) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|
| Crude extract | 557 | 501 | 0.90 | 100 | 1.0 |
| Heat treatment at 90°C for 20 min | 59.9 | 421 | 7.0 | 84 | 7.8 |
| HiTrap chromatography | 22.5 | 174 | 7.6 | 35 | 8.4 |
FIG. 1.
Determination of molecular mass of purified P. horikoshii NAD kinase. (A) SDS-PAGE of recombinant P. horikoshii NAD kinase. Left lane, protein markers (New England Biolabs); right lane, purified P. horikoshii NAD kinase. (B) Molecular mass determination of native P. horikoshii NAD kinase by gel filtration chromatography. The molecular mass was plotted versus the elution volume/void volume. The void volume was determined with blue dextran 2000 (2,000 kDa). The elution position of the purified enzyme is indicated by a filled circle.
The N-terminal amino acid sequence of the purified enzyme was determined to be GSSHHHHHHSSGLVPRGSHMKFGIVARRD and was identical to the predicted sequence, including a histidine tag sequence in the N-terminal domain, deduced from the open reading frame.
Phosphoryl donor and acceptor specificities of P. horikoshii NAD kinase.
The enzyme used several nucleoside triphosphates, such as ATP, GTP, and UTP, as phosphoryl donors, but not nucleotide diphosphates such as ADP and nucleotide monophosphates such as AMP (Table 2). The activity of poly(P)x as the phosphoryl donor was examined and was higher than that of ATP. The circular polyphosphate meta(P)x was also very active as a donor. In addition, the donor activities of various chain lengths of poly(P) were examined. Commercially available poly(P) samples with various chain lengths were active as phosphoryl donors, except for orthophosphate and pyrophosphate, which were inert. Among the commercially available different chain lengths of poly(P), poly(P)27 exhibited the highest activity. Hereafter, we refer to the enzyme as poly(P)/ATP-NAD kinase.
TABLE 2.
Phosphoryl group donor specificitya
| Phosphoryl donorb | Relative activity (%) |
|---|---|
| Poly(P)x | 100 |
| AMP | 0 |
| ADP | 0 |
| ATP | 75 |
| GTP | 111 |
| CTP | 76 |
| TTP | 13 |
| UTP | 94 |
| ITP | 89 |
| Meta(P)x | 87 |
| Orthophosphate | 0 |
| Pyrophosphate | 0 |
| Poly(P)3 | 11 |
| Poly(P)5 | 22 |
| Poly(P)18 | 87 |
| Poly(P)27 | 101 |
| Poly(P)32 | 90 |
| Poly(P)46 | 67 |
| Poly(P)62 | 52 |
The specific activity obtained in an assay mixture composed of 100 mM bis-Tris-HCl (pH 6.8), 5 mM NAD, 5 mM MnCl2, and 2 mg/ml poly(P)x is represented as 100%.
The final concentration used was 5 mM, except for poly(P)x and meta(P)x, for which the final concentration was 2 mg/ml.
As a phosphate acceptor of the enzyme, NADH was active, as was NAD. The poly(P)x (2 mg/ml)- and ATP (5 mM)-dependent NADH kinase activities of the enzyme were 0.035 μmol/min/mg and 0.79 μmol/min/mg, respectively, at 37°C.
Effects of pH and temperature on enzyme activity and stability.
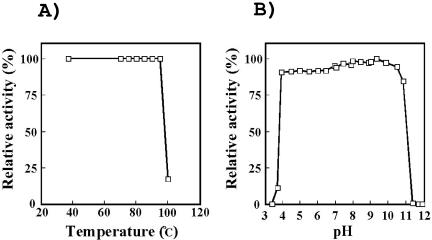
The effect of pH on NAD kinase activity was measured at various pHs at 70°C, and the optimum pH was about 6.8 (data not shown). The enzyme showed extremely high thermostability; upon heating at 95°C for 10 min, the activity was not lost, but about 80% of the activity was lost upon incubation at 100°C for 10 min (Fig. 2A). When the effect of pH on the stability of the enzyme was examined by incubation at 70°C for 60 min, the enzyme did not lose activity in a pH range of 4.0 to 10.5 (Fig. 2B). At a low temperature such as 4°C, the NAD kinase could be stored for more than 2 months without a loss of activity.
FIG. 2.
Effects of temperature and pH on stability of P. horikoshii NAD kinase. (A) Residual activity of the enzyme after treatment at various temperatures for 10 min. (B) The enzyme was incubated at 70°C for 60 min in buffer solutions with various pHs, and then the residual activity was measured.
Molecular mass and subunit structure.
The purified enzyme migrated as a single protein band corresponding to 37 kDa by SDS-PAGE (Fig. 1). The molecular mass of the P. horikoshii NAD kinase was estimated to be about 145 kDa by gel filtration, indicating that the enzyme is a tetramer consisting of four identical subunits.
Steady-state kinetic parameters.
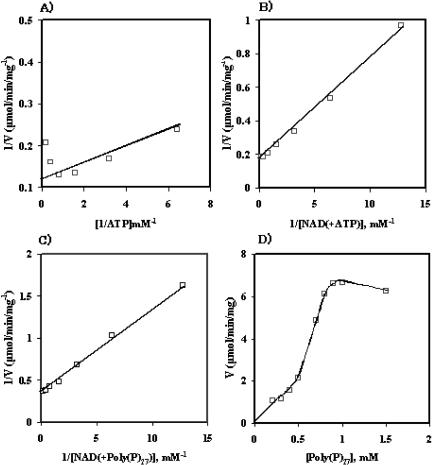
The P. horikoshii NAD kinase showed typical Michaelis-Menten kinetics for NAD and ATP (Fig. 3A to C) but did not show Michaelis-Menten kinetics for poly(P)27 (Fig. 3D). The Km values for NAD were 0.30 mM and 0.40 mM when poly(P)27 and ATP, respectively, were used as phosphoryl donors. The enzyme exhibited a higher affinity for NAD when poly(P)27 was used instead of ATP as the phosphoryl donor. On the other hand, the Km value for ATP was determined to be 0.29 mM, and the concentration of poly(P)27 which gave one-half the Vmax was 0.59 mM.
FIG. 3.
Initial-velocity analyses of NAD kinase. (A) Double-reciprocal plot of enzyme activity against ATP concentration in the presence of 5.0 mM NAD; (B) double-reciprocal plot of enzyme activity against NAD concentration in the presence of 5.0 mM ATP; (C) double-reciprocal plot of enzyme activity against NAD concentration in the presence of 2.0 mg poly(p)27; (D) dependence of the initial velocity on poly(P)27 concentration.
Effects of metal ions on NAD kinase activity.
The effect of metal ions was examined by assaying the activity after the addition of metal salts to the standard reaction mixture. The enzyme required divalent metals (chloride salts) for its activity (Table 3). In the case of poly(P)x, the addition of 5 mM MgCl2 was most effective, and several other metal ions, such as MnCl2 and NiCl2, gave high activities when poly(P)x was used as the phosphoryl donor. For ATP-dependent activity, CuCl2 gave the highest activity. In addition, the enzyme was slightly inhibited by 3 mM HgCl2 (35% inhibition) and p-mercuribenzoate (22% inhibition).
TABLE 3.
Effect of cations on NAD kinase activitya
| Cationb | Poly(P)x-dependent relative activity (%) | ATP-dependent relative activity (%) |
|---|---|---|
| None | 0 | 0 |
| MnCl2 | 100 | 62 |
| MgCl2 | 103 | 103 |
| NiCl2 | 80 | 36 |
| CaCl2 | 57 | 65 |
| ZnCl2 | 56 | 78 |
| CuCl2 | 100 | 158 |
The specific activity obtained in an assay mixture composed of 100 mM bis-Tris-HCl (pH 6.8), 5 mM NAD, 5 mM MnCl2, and 2 mg/ml poly(P)x is represented as 100%.
The concentration of cations used was 5 mM.
Sequence alignment of NAD kinases.
The amino acid sequence of P. horikoshii NAD kinase and those of enzymes from other sources were aligned based on sequences in the databases (Fig. 4). The P. horikoshii NAD kinase showed high sequence identities, of 90 and 82%, with the putative protein sequences from the Pyrococcus furiosus gene (PF1103) and the Pyrodictium abyssi gene (PAB1756), respectively, but only low identities with the Sulfolobus tokodaii putative protein ST2136 (39%), the Methanothermobacter thermautotrophicus putative protein MTH872 (42%), E. coli NAD kinase (31%) (9), and M. tuberculosis NAD kinase (29%) (10) according to the multiple program in CLUSTALW (24).
FIG. 4.
Alignment of the amino acid sequence of P. horikoshii NAD kinase with those of other NAD kinases based on sequences in the database using the CLUSTALW program (23). Asterisks, conserved residues. The two highly conserved regions, XXX-XGGDG-XL and DGXXX-TPTGSTXY, are boxed.
DISCUSSION
Although NAD kinase has been found in and purified from several organisms, including eukaryotes and mesophilic microorganisms, the enzyme has not been purified so far from either archaea or thermophiles. For this study, we found the gene homologue (PH1074) of NAD kinase in the hyperthermophilic archaeon P. horikoshii and ascertained that its gene product from a recombinant E. coli strain exhibited NAD kinase activity. This is the first report showing the presence of NAD kinase in a hyperthermophilic archaeon.
Two kinds of subunit molecular masses for microbial and animal NAD kinases have been reported to date. The subunit size of animal NAD kinases (about 49 kDa) is larger than that of microbial enzymes (30 to 35 kDa) because animal enzymes have an additional regulatory site in the C-terminal region (16). In addition, several different subunit structures have been found for NAD kinases from various organisms, including an octamer for the pigeon liver (2) and C. utilis (5) enzymes, a hexamer for the E. coli enzyme (9), a tetramer for the human (16), S. cerevisiae (12), and M. tuberculosis enzymes (10), and a dimer for the M. flavus enzyme (10). We found that the P. horikoshii NAD kinase consists of a tetramer of 37-kDa subunits (His-tagged protein). In this respect, the archaeal enzyme is similar to the human, S. cerevisiae, and M. tuberculosis enzymes, although the physiological, evolutional, and catalytically functional significances of the presence of different subunit structures are still unclear.
As expected, P. horikoshii NAD kinase was much more stable than its counterpart from mesophiles such as E. coli and M. tuberculosis; the E. coli enzyme loses one-half of its activity when incubated at 65°C for 10 min (9), but the P. horikoshii enzyme retains its full activity at temperatures up to 95°C. In addition, the P. horikoshii NAD kinase was highly stable over a wide range of pHs (pH 4 to 10.5) and could be stored for longer periods (for at least 2 months) at low temperatures. Highly stable P. horikoshii NAD kinase has much more potential usefulness for the production of NADP from NAD than its counterparts from mesophiles. Thermostable NAD kinase from Corynebacterium flaccumfaciens has been reported to be useful for enzymatic NADP production (17), but in this case, the enzyme is stable only up to 50°C. We could achieve the overproduction of stable NAD kinase in E. coli cells and a simple and large-scale preparation of the purified enzyme by only two procedures, heat treatment and affinity chromatography. In addition, the P. horikoshii enzyme utilizes inorganic poly(P)x as well as ATP as a phosphoryl donor. Inorganic poly(P)x is known to be much less expensive (about 1/1,000) than ATP and other triphosphonucleotides. This may prompt us to utilize the archaeal enzyme for NADP production.
The NAD kinases from M. tuberculosis, M. flavus, and B. subtilis have been known to utilize both poly(P) and ATP as phosphoryl donors (6, 10) and are part of a family of poly(P)/ATP-NAD kinases, which is different from the ATP-NAD kinase family. The archaeal NAD kinase may belong to the family of poly(P)/ATP-NAD kinases. While the NAD kinase of M. tuberculosis uses poly(P)4 as the most efficient phosphoryl donor, the P. horikoshii enzyme utilizes a much longer polyphosphate, poly(P)27. In addition, the P. horikoshii NAD kinase can also utilize meta(P)x, similar to the M. tuberculosis enzyme. Poly(P)/ATP-NAD kinases are thought to be prototypes of ATP-NAD kinases (14), and poly(P) is known to be easily produced under high-temperature conditions above 100°C (26). It seems reasonable that the P. horikoshii NAD kinase utilizes poly(P) as a successful phosphoryl donor because the optimum growth temperature of the hyperthermophilic archaeon is around 98°C.
A BLAST search in the gene databases showed that the P. horikoshii enzyme exhibits high homology to hypothetical proteins from P. furiosus (PF1103; 90% identity), P. abyssi (PAB1756; 82% identity), and Methanobacterium thermautotrophicus (MTH872; 42% identity) (Fig. 4). This indicates that these archaeal strains may exhibit poly(P)/ATP-NAD kinases similar to that of P. horikoshii. An alignment of the amino acid sequences showed that the primary structure of P. horikoshii NAD kinase contains two highly conserved regions, XXX-XGGDG-XL and DGXXX-TPTGSTXY, which are found in E. coli NAD kinase (9), where “X” and “-” represent a hydrophobic amino acid residue and any amino acid residue, respectively. The NAD kinases of P. horikoshii and M. tuberculosis can utilize poly(P), but the E. coli enzyme cannot. Recently, the crystal structure of the M. tuberculosis enzyme was solved, and a novel fold was observed in the molecule (7). However, the poly(P) binding sites of the enzyme have not yet been clarified because the structure was determined for an apoform (7) and a complex with NAD (19). At present, we cannot identify which residue(s) is responsible for the interaction with poly(P) from this information. A comparison of the three-dimensional structures of the poly(P)/ATP-NAD kinase of P. horikoshii and the ATP-NAD kinase of E. coli may elucidate which amino acid(s) is responsible for the phosphoryl donor specificity and thermostability of the former enzyme and may provide some clues about the evolution of poly(P)-specific NAD kinases to ATP-specific NAD kinases.
In this study, we identified the NAD kinase gene of P. horikoshii and characterized the encoded enzyme. For the de novo synthesis pathway of NADP from l-aspartate and dihydroxyacetone phosphate, we have already found six enzyme homologs (genes PH0011, -0013, -0015, -0182, -0464, and -1074) which are responsible for the NADP biosynthesis pathway in P. horikoshii based on genome information. Among the six enzymes, the NAD kinase is the third enzyme which we have characterized, in addition to l-aspartate oxidase (22) and nicotinamide mononucleotide adenylyltransferase (21). We are now attempting the gene expression and characterization of the other three enzymes. These studies offer us much information for a better understanding of the total system of NAD and NADP biosynthesis pathways in archaea.
Acknowledgments
This study was funded by the National Project on Protein Structural and Functional Analyses (Protein 3000 project) promoted by the Ministry of Education, Science, Sports, Culture, and Technology of Japan and was supported in part by the Asahi Glass Foundation, Tokyo, Japan.
We thank I. Kanazawa and M. W. Bhuiya of the Department of Bioscience and Technology, Faculty of Engineering, University of Tokushima, for their kind technical assistance.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Apps, D. K. 1975. Pigeon-liver NAD kinase. The structural and kinetic basis of regulation of NADPH. Eur. J. Biochem. 55:475-483. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Bulygina, E. R., and V. I. Telepneva. 1980. Isolation of NAD-kinase from pigeon heart. Biokhimia 45:2019-2027. [PubMed] [Google Scholar]
- 5.Butler, J. R., and E. T. McGuinness. 1982. Candida utilis NAD+ kinase: purification, properties and affinity gel studies. Int. J. Biochem. 14:839-844. [DOI] [PubMed] [Google Scholar]
- 6.Garavaglia, S., A. Galizzi, and M. Rizzi. 2003. Allosteric regulation of Bacillus subtilis NAD kinase by quinolinic acid. J. Bacteriol. 185:4844-4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garavaglia, S., N. Raffaelli, L. Finaurini, G. Magni, and M. Rizzi. 2004. A novel fold revealed by Mycobacterium tuberculosis NAD kinase, a key allosteric enzyme in NADP biosynthesis. J. Biol. Chem. 279:40980-40986. [DOI] [PubMed] [Google Scholar]
- 8.Kawai, S., S. Mori, T. Mukai, H. Matsukawa, and K. Murata. 2001. Establishment of a mass-production system for NADP using bacterial inorganic polyphosphate/ATP-NAD kinase. J. Biosci. Bioeng. 92:447-452. [DOI] [PubMed] [Google Scholar]
- 9.Kawai, S., S. Mori, T. Mukai, W. Hashimoto, and K. Murata. 2001. Molecular characterization of Escherichia coli NAD kinase. Eur. J. Biochem. 268:4359-4365. [DOI] [PubMed] [Google Scholar]
- 10.Kawai, S., S. Mori, T. Mukai, S. Suzuki, T. Yamada, W. Hashimoto, and K. Murata. 2000. Inorganic polyphosphate/ATP-NAD kinase of Micrococcus flavus and Mycobacterium tuberculosis H37Rv. Biochem. Biophys. Res. Commun. 276:57-63. [DOI] [PubMed] [Google Scholar]
- 11.Kawai, S., S. Mori, and K. Murata. 2003. Primary structure of inorganic polyphosphate/ATP-NAD kinase from Micrococcus flavus, and occurrence of substrate inorganic polyphosphate for the enzyme. Biosci. Biotechnol. Biochem. 67:1751-1760. [DOI] [PubMed] [Google Scholar]
- 12.Kawai, S., S. Suzuki, S. Mori, and K. Murata. 2001. Molecular cloning and identification of UTR1 of a yeast Saccharomyces cerevisiae as a gene encoding an NAD kinase. FEMS Microbiol. Lett. 200:181-184. [DOI] [PubMed] [Google Scholar]
- 13.Kawarabayasi, Y., M. Sawada, H. Horikawa, Y. Haikawa, Y. Hino, S. Yamamoto, M. Sekine, S. Baba, H. Kosugi, A. Hosoyama, Y. Nagai, M. Sakai, K. Ogura, R. Otsuka, H. Nakazawa, M. Takamiya, Y. Ohfuku, T. Funahashi, T. Tanaka, Y. Kudoh, J. Yamazaki, N. Kushida, A. Oguchi, K. Aoki, T. Yoshizawa, Y. Nakamura, F. T. Robb, K. Horikoshi, Y. Masuchi, and H. Kikuchi. 1998. Complete sequence and gene organization of the genome of a hyperthermophilic archaebacterium, Pyrococcus horikoshii OT-3. DNA Res. 5:55-76. [DOI] [PubMed] [Google Scholar]
- 14.Kornberg, A., N. N. Rao, and D. Ault-Riche. 1999. Inorganic polyphosphate: a molecule of many functions. Annu. Rev. Biochem. 68:89-125. [DOI] [PubMed] [Google Scholar]
- 15.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 16.Lerner, F., M. Niere, A. Ludwig, and M. Ziegler. 2001. Structural and functional characterization of human NAD kinase. Biochem. Biophys. Res. Commun. 288:69-74. [DOI] [PubMed] [Google Scholar]
- 17.Matsushita, H., S. Yokoyama, and A. Obayashi. 1986. NADP+ production using thermostable NAD+ kinase of Corynebacterium flaccumfaciens AHU-1622. Can. J. Microbiol. 32:585-590. [DOI] [PubMed] [Google Scholar]
- 18.Moat, A. G., and J. W. Foster. 1987. Biosynthesis and salvage pathways of pyridine nucleotides, p. 1-24. In D. Dolphin, R. Poulson, and A. Avramovic (ed.), Pyridine nucleotide coenzymes, part B. John Wiley & Sons Inc., New York, N.Y.
- 19.Mori, S., M. Yamasaki, Y. Maruyama, K. Momma, S. Kawai, W. Hashimoto, B. Mikami, and K. Murata. 2004. Crystallographic studies of Mycobacterium tuberculosis polyphosphate/ATP-NAD kinase complexed with NAD. J. Biosci. Bioeng. 98:391-393. [DOI] [PubMed] [Google Scholar]
- 20.Ochiai, A., S. Mori, S. Kawai, and K. Murata. 2004. Overexpression, purification, and characterization of ATP-NAD kinase of Sphingomonas sp. A1. Protein Expr. Purif. 36:124-130. [DOI] [PubMed] [Google Scholar]
- 21.Sakuraba, H., K. Kanai, S. Goda, Y. Kawarabayasi, and T. Ohshima. 2003. A nicotinamide mononucleotide adenylyltransferase with unique adenylyl group donor specificity from a hyperthermophilic archaeon, Pyrococcus horikoshii OT3. J. Mol. Catalysis B 23:273-279. [Google Scholar]
- 22.Sakuraba, H., T. Satomura, R. Kawakami, S. Yamamoto, Y. Kawarabayasi, H. Kikuchi, and T. Ohshima. 2002. l-Aspartate oxidase is present in the anaerobic hyperthermophilic archaeon Pyrococcus horikoshii OT-3: characteristics and role in the de novo biosynthesis of nicotinamide adenine dinucleotide proposed by genome sequencing. Extremophiles 6:275-281. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., p. 9.14-9.23. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 24.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woese, C. R., O. Kandler, and M. L. Wheelis. 1990. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. USA 87:4576-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamagata, Y., H. Watanabe, M. Saitoh, and T. Namba. 1991. Volcanic production of polyphosphates and its relevance to prebiotic evolution. Nature 352:516-519. [DOI] [PubMed] [Google Scholar]