Abstract
One of the functions of RNA silencing in plants is antiviral defense. A hallmark of RNA silencing is spreading of the silenced state through the plant. Little is known about the nature of the systemic silencing signal and the proteins required for its production, transport, and reception in plant tissues. Here, we show that the RNA-dependent RNA polymerase RDR6 in Nicotiana benthamiana is involved in defense against potato virus X at the level of systemic spreading and in exclusion of the virus from the apical growing point. It has no effect on primary replication and cell-to-cell movement of the virus and does not contribute significantly to the formation of virus-derived small interfering (si) RNA in a fully established potato virus X infection. In grafting experiments, the RDR6 homolog was required for the ability of a cell to respond to, but not to produce or translocate, the systemic silencing signal. Taking these findings together, we suggest a model of virus defense in which RDR6 uses incoming silencing signal to generate double-stranded RNA precursors of secondary siRNA. According to this idea, the secondary siRNAs mediate RNA silencing as an immediate response that slows down the systemic spreading of the virus into the growing point and newly emerging leaves.
Endogenous RNA-dependent RNA polymerase (RDR) activity was demonstrated in plants more than 30 years ago (Astier-Manifacier and Cornuet, 1974), but its function remained unclear for many years. More recently, RDRs were proposed as part of an RNA silencing system (Lindbo et al., 1993) related to RNA interference in animals and quelling in fungi. Consistent with this idea, the SILENCING DEFECTIVE 1 (SDE1)/SUPPRESSOR OF GENE SILENCING 2(SGS2) locus was identified as a requirement for certain examples of transgene RNA silencing in Arabidopsis (Arabidopsis thaliana; Dalmay et al., 2000; Mourrain et al., 2000). Following a more recent nomenclature, we will henceforth refer to SDE1/SGS2 as RDR6 (Xie et al., 2004).
A likely biochemical role of RDRs in RNA silencing is to produce double-stranded (ds) RNA that is cleaved by RNase III-like enzymes called Dicer (DCR) in animals and Dicer-like (DCL) in plants (Bernstein et al., 2001; Golden et al., 2002; Carmell and Hannon, 2004). The resulting 21 to 24 nucleotide small interfering (si) RNAs are then recruited as the specificity determinants of silencing effector complexes that also include argonaute (AGO) proteins. The in vitro activity of RDR from Lycopersicum esculentum (Schiebel et al., 1993a, 1993b, 1998), Neurospora crassa (Makeyev and Bamford, 2002), and Schizosaccharomyces pombe (Motamedi et al., 2004) is consistent with a role in dsRNA synthesis.
There are several RNA silencing pathways (Baulcombe, 2004), and, correspondingly, there are multiple genes for RDR, DCR, DCL, and AGO proteins in many organisms. In some instances, when the initiator of silencing is a viral genome (Dalmay et al., 2000) or a gene that is transcribed into an RNA with ds regions (Beclin et al., 2002), there is no known requirement for an RDR. Similarly with micro (mi) RNAs, which are siRNA-like regulators of endogenous gene expression derived from partially dsRNA precursors, there is RDR-independent RNA silencing. The miRNA-mediated silencing is not affected by RDR mutations (Vaucheret et al., 2004) and the genomes of mammals and insects (Schwarz et al., 2002) do not encode RDRs, although a high proportion of the transcripts may be regulated by miRNAs (Brennecke et al., 2005; Lim et al., 2005). However, in plants, most of the non-miRNA silencing pathways have an RDR requirement. There are RDR6-dependent siRNAs, for example, that down-regulate Arabidopsis mRNAs (Peragine et al., 2004; Vazquez et al., 2004) and cis-acting siRNA mediators of heterochromatinization that require RDR2 (Chan et al., 2004; Xie et al., 2004; Herr et al., 2005). In addition, two plant RDR proteins have been implicated in virus defense (Dalmay et al., 2000; Mourrain et al., 2000; Xie et al., 2001). Arabidopsis RDR6 is implicated in defense against cucumber mosaic cucumovirus (CMV; Mourrain et al., 2000; Dalmay et al., 2001), and the tobacco ortholog of Arabidopsis RDR1 influences susceptibility to tobacco mosaic tobamovirus (TMV) and potato potexvirus X (PVX; Xie et al., 2001).
An additional manifestation of RNA silencing that is associated with virus defense in plants is a signal that spreads through the plasmodesmata and phloem (Palauqui et al., 1997; Voinnet and Baulcombe, 1997; Voinnet et al., 1998; Fagard and Vaucheret, 2000). The exact nature of the silencing signal is unknown, but siRNAs, especially those belonging to the 24 nucleotide class, are widely regarded as a strong candidate (Mlotshwa et al., 2002). A recent study showed that RDR6 is involved in long-range but not cell-to-cell signaling of RNA silencing (Himber et al., 2003).
In this paper, we describe an analysis of RDR-mediated defense against viruses in the virological model species Nicotiana benthamiana (Nb). We show that silencing of NbRDR6 causes plants to be hypersusceptible to PVX, potato potyvirus Y (PVY), and the Y satellite of CMV but not TMV, tobacco rattle tobravirus (TRV), turnip crinkle carmovirus (TCV), or CMV alone. The PVX hypersusceptibility was associated with the enhanced viral invasion of the growing point of the infected plant. We also show how NbRDR6 is implicated in systemic RNA silencing; it is not required for production or translocation of the silencing signal but it is required for cells to respond to received signal. By combining our findings about meristem invasion and systemic silencing, we derive a model of virus defense in which NbRDR6 recruits a PVX-derived silencing signal to trigger an immediate silencing response against virus as it enters the growing point and newly emerging leaves. This silencing signal-related mechanism could explain, at least in part, why PVX and possibly other viruses are not able to invade the meristem of infected plants.
RESULTS
Identification of the N. benthamiana Ortholog of AtRDR6
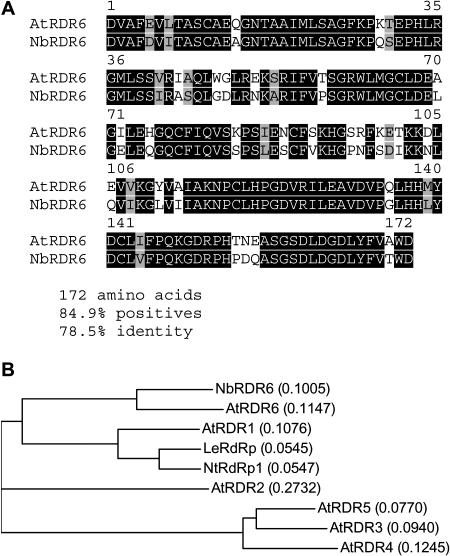
To investigate the role of RDR6 in antiviral defense, we generated an N. benthamiana line in which NbRDR6 was silenced. The silencing construct was based on a 516-bp fragment of N. benthamiana RDR6 cDNA that was PCR amplified using primers corresponding to highly conserved regions in RDR6 of Arabidopsis and other plants (Fig. 1A). The predicted translation product of the amplified DNA sequence is more similar (78.5% identical) to RDR6 than to any of the other Arabidopsis RDR proteins (42.3%, 27.3%, 25.0%, and 27.3% identity with RDR2, RDR3, RDR4, and RDR5, respectively; Fig. 1B), and we conclude that it represents a structural ortholog of Arabidopsis RDR6 henceforth referred to as NbRDR6.
Figure 1.
Alignment of the translated NbRDR6 sequence fragment to Arabidopsis RDR6 (A) and guide tree (B) of translated NbRDR6 aligned with all six Arabidopsis RDRs (AtRDR1–AtRDR6) and with RDR proteins from tomato and tobacco (LeRdRP and NtRdRP1, respectively). Calculated distance values according to the Neighbor Joining method (Saitou and Nei, 1987) are given in parentheses.
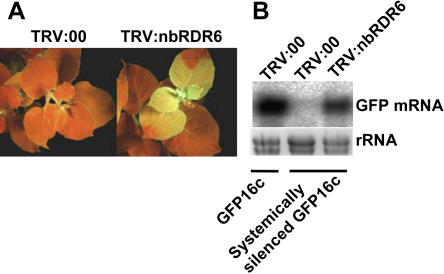
To confirm that NbRDR6 is functionally similar to RDR6, we cloned a 243-bp portion of the original 516-bp NbRDR6 cDNA into a TRV silencing vector. This construct (TRV:RDR6) was inoculated to N. benthamiana in which a green fluorescent protein (GFP) transgene was posttranscriptionally silenced, and, after 4 to 6 weeks, these plants exhibited a breakdown of GFP silencing, similar to the phenotype of RDR6 mutants in Arabidopsis (Dalmay et al., 2000). The loss of silencing was manifested as an increase in GFP fluorescence and RNA levels (Fig. 2, A and B). No breakdown of GFP silencing occurred in plants inoculated with the empty TRV vector, showing that this phenotype is caused specifically by the virus-induced silencing of NbRDR6 in TRV:RDR6-infected tissue.
Figure 2.
Virus-induced gene silencing of NbRDR6 in N. benthamiana. Two-week-old GFP16c plants were systemically silenced for GFP by transient expression of a GFPi. Once silencing was established, TRV VIGS vectors either containing the NbRDR6 fragment (TRV:RDR6) or empty (TRV:00) were inoculated. Five to six weeks later, TRV:RDR6-inoculated but not TRV:00-inoculated plants showed a breakdown of maintenance of the GFP silencing, visible as green fluorescence under UV light (A) and reestablished expression of the GFP mRNA (B) in the virus-infected tissues.
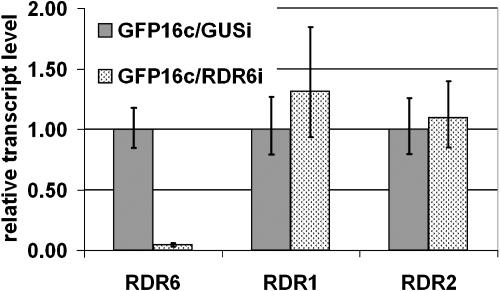
For analysis of NbRDR6 without the complication of TRV infection we generated the N. benthamiana line RDR6i, in which NbRDR6 is constitutively silenced by an RNAi hairpin construct. The GFP16c/RDR6i line additionally expresses GFP under the control of the cauliflower mosaic virus 35S promoter. In quantitative real-time reverse transcription (RT)-PCR analyses the NbRDR6 transcript levels were reduced to 4% of the level in a line carrying an unrelated RNAi construct, targeting the β-glucuronidase (GUS) gene (GFP16c/GUSi), whereas expression of the N. benthamiana orthologs of RDR1 and RDR2 was not affected significantly (Fig. 3). The GFP16c/RDR6i plants phenocopied the Arabidopsis rdr6 mutants; silencing of GFP by a sense GFP construct was impaired, whereas an inverted repeat GFP construct (GFPi) induced local silencing of GFP and formation of GFP-derived siRNA (Supplemental Fig. 1, A and B). GFP silencing by GFPi was equally efficient in GFP16c and GFP16c/RDR6i plants, indicating that expression of the RDR6i construct did not overload and inhibit the silencing machinery (Supplemental Fig. 1C).
Figure 3.
Quantitative real-time RT-PCR for RDR6, RDR1, and RDR2 transcript levels in N. benthamiana lines carrying the RDR6i construct or an unrelated RNAi construct (GUSi). Mean values are based on four pools of five plants each. Error bars represent ses of the mean. Relative transcript levels were calculated using the ΔΔC(t) method (Livak and Schmittgen, 2001) with GAPDH transcripts serving as an internal standard.
Reduced Expression of NbRDR6 Results in Hypersusceptibility to Some Viruses
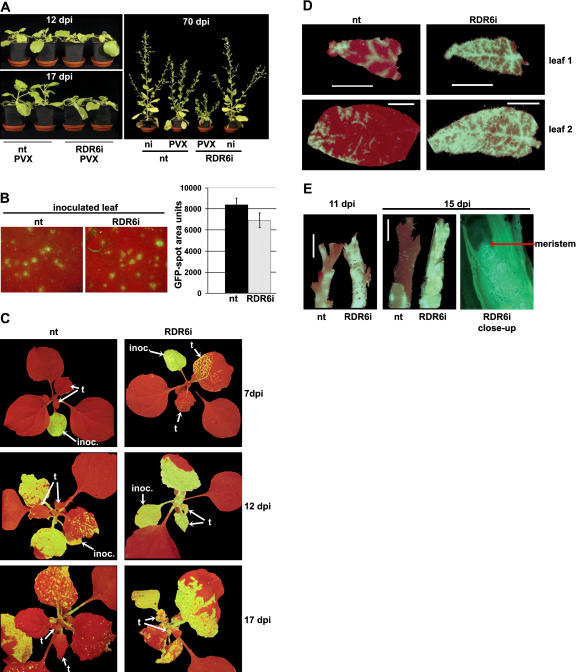
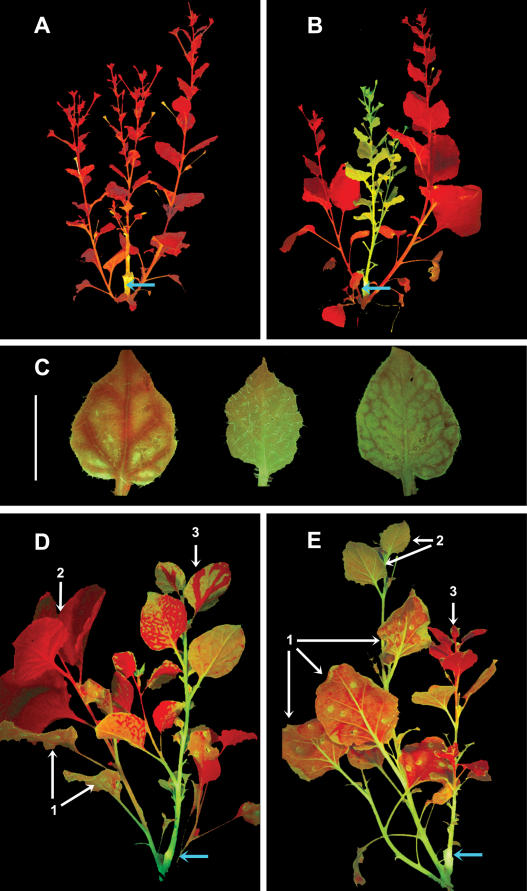
To investigate the role of NbRDR6 in virus defense (Mourrain et al., 2000; Dalmay et al., 2001), we challenged line RDR6i with PVX, PVY, TRV, TMV, TCV, and CMV with and without the Y satellite. The symptoms on RDR6i and nontransformed (nt) plants were the same with TCV, TRV, TMV, or CMV without the Y satellite (TCV and TRV: data not shown; TMV and CMV: Supplemental Fig. 2), but with PVX, PVY, and CMV in combination with the Y satellite symptoms were more severe on RDR6i than on nt plants (Fig. 4A; Supplemental Fig. 2). The symptom difference was particularly striking with PVX; it was first manifested between 12 and 17 d postinoculation (dpi) as heavy stunting and delayed development of new leaves of RDR6i in comparison to nt plants and persisted until the plants died (Fig. 4A). The hypersusceptibility to PVX was not due to saturation of the RNA silencing machinery, because GFP16c and GFP16c/GUSi lines harboring an RNAi construct targeting GUS both exhibited the same PVX symptoms as nt plants (data not shown).
Figure 4.
Comparison of PVX:GFP infection on N. benthamiana nt and RDR6i lines. A, RDR6i and nt plants were inoculated with PVX:GFP and are shown at 12 dpi when there was no difference in appearance of the two plant lines and at 17 dpi when the RDR6i plants were stunted. At 70 dpi, the RDR6i line still exhibited strong symptoms, whereas the nt plants exhibited mild symptoms and were similar to noninoculated plants. B, PVX:GFP infection foci in inoculated leaves of nt and RDR6i plants at 4 dpi imaged under UV light were similar in size and brightness, indicating that replication and cell-to-cell spread of the virus were unaffected by the RDR6i genotype. Spot sizes were measured using the Able Image Analyser software (Mu Labs, Slovenia) and mean values of area units of 55 spots on each plant line at 4 dpi are shown with their ses of the mean. C, PVX:GFP-infected RDR6i and nt plants under UV light. The inoculated leaves indicated by arrows (inoc.). The two topmost leaves (t) showed spread of PVX:GFP into the upper leaves from 7 dpi, whereas in nt plants the uppermost leaves were largely free of GFP fluorescence up to 17 dpi and after. D, Young leaves (leaf 1 and leaf 2 from top) of PVX:GFP-infected nt and RDR6i plants under UV illumination illustrating a difference in PVX:GFP distribution at 12 dpi. White bars = 0.5 cm. E, Stem tips of nt and RDR6i plants infected with PVX:GFP show more extensive viral spread into young tissue in RDR6i than in nt. At 11 dpi, the top 0.5 cm and at 15 dpi the top 1.5 cm of the nt stem appear red, whereas the RDR6i stem exhibits virus-expressed GFP throughout. A longitudinal section of the growing tip of the RDR6i plant (15 dpi, right-hand section) shows GFP fluorescence evenly distributed throughout, including the meristem.
We inoculated RDR6i and nt plants with a PVX:GFP vector and monitored GFP fluorescence to find out whether the RDR6i phenotype was associated with altered virus movement. In inoculated leaves the PVX:GFP infection foci showed no significant difference in size between RDR6i and nt plants (Fig. 4B). However, in the systemically infected leaves, there was a difference in GFP fluorescence that was apparent as soon as 7 dpi. In the RDR6i line, 18/22 plants exhibited GFP fluorescence in noninoculated upper leaves, whereas in nt plants, the GFP fluorescence was weak (12/22 plants) or not detectable (10/22; Fig. 4C). By 12 dpi, the newly emerging leaves of RDR6i plants were all fully green fluorescent, while nt leaves emerged with only a few fluorescent areas (Fig. 4, C and D). The differential accumulation of PVX:GFP in upper leaves of RDR6i and nt plants continued until 17 dpi (Fig. 4C). Corresponding to the differential PVX:GFP accumulation in leaves, the stems of RDR6i were uniformly green fluorescent up to and including the apical growing point, whereas nt stems exhibited no, or very low level, fluorescence in the upper 0.5 to 1 cm (Fig. 4E).
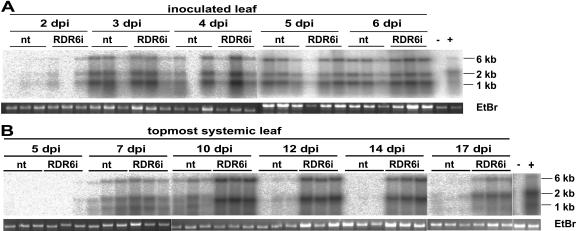
A northern-blot analysis confirmed that the symptom and GFP differences correlated with PVX:GFP RNA accumulation. Thus, in inoculated leaves of nt and RDR6i plants, there was between-plant variation in the PVX:GFP RNA accumulation but, overall, there was no difference between the two lines (Fig. 5A). However, in the upper infected leaves, the PVX:GFP RNA was more abundant in RDR6i. This difference was more pronounced at 10 dpi and later when PVX:GFP RNA levels decreased in the upper leaves of the nt plants but remained high in RDR6i (Fig. 5B).
Figure 5.
Northern-blot analyses of PVX:GFP RNA levels in nt and RDR6i plants. A, PVX:GFP RNA accumulation in inoculated leaves in nt and RDR6i plants in triplicate samples from 2 to 6 dpi. Accumulation of PVX:GFP full-length (6 kb) and major subgenomic RNAs (approximately 1 and 2 kb) were detected with a DNA probe specific for the PVX coat protein sequence. No significant difference in PVX:GFP RNA accumulation apart from plant-to-plant variation was visible. B, PVX:GFP RNA accumulation in the topmost, not yet fully expanded, leaves in triplicate samples from nt and RDR6i plants at the indicated time points from 5 dpi to 17 dpi is showing increasing differences in viral accumulation between the two plant lines throughout the experiment in the newly developing tissue.
These combined GFP and RNA data indicate that the kinetics of viral RNA accumulation and cell-to-cell spread were the same on inoculated leaves of nt plants and RDR6i (Figs. 4B and 5A). It is therefore unlikely that NbRDR6 influences PVX replication or movement between cells. Instead, it seems that NbRDR6 inhibits PVX accumulation in the growing point and newly emerged leaves. Presumably the RDR6i plants are hypersusceptible to PVX because NbRDR6 normally impairs systemic virus movement or is involved in the mechanism that normally excludes PVX and other viruses from the growing point of the infected plant.
Silencing of NbRDR6 Affects Systemic Silencing
An explanation of the NbRDR6-mediated effect on systemic movement of PVX invokes an RNA silencing signal in PVX-infected plants that would move systemically either with or ahead of the virus (Voinnet et al., 2000). This signal would prime an RDR6-mediated silencing mechanism as the virus enters cells that are distant from the site of initial infection. The primed silencing mechanism would inhibit virus accumulation in these cells and impair the spread of the virus.
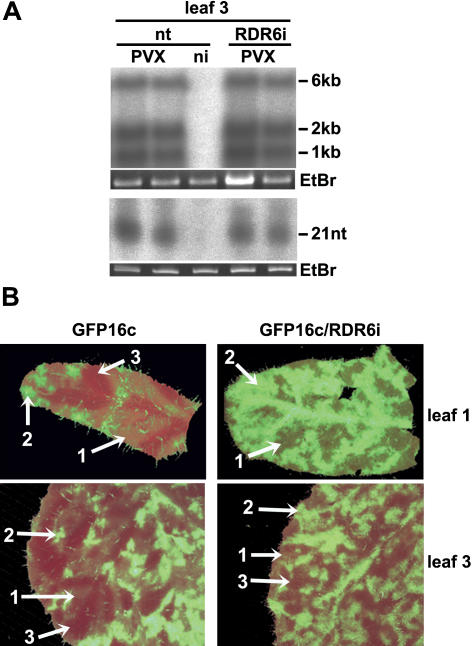
If this hypothesis is correct, RDR6 and the silencing signal would contribute to silencing of viral RNA in cells that are at, but not behind, the systemic infection front. To test this prediction, we monitored PVX-induced silencing in plants either with or without the RDR6i transgene either at or behind the systemic infection front. For analysis of cells behind the infection front, we sampled the third leaf from the top of nt and RDR6i plants at 10 dpi and found no significant difference in PVX:GFP RNA and virus-derived siRNA levels (Fig. 6A). These leaves were further behind the infection front than those used for the analysis in Figure 5 and the absence of a difference in the two lines indicates that RDR6 does not significantly contribute to siRNA production once the infection is established.
Figure 6.
Silencing signal and siRNA production in the presence or absence of NbRDR6 in PVX:GFP-infected plants at 10 dpi. A, Northern-blot analysis of PVX:GFP viral genomic (6 kb) and subgenomic (approximately 1 and 2 kb) RNA and accumulation of PVX:GFP-derived siRNA (21 nt) in fully systemically infected leaves (leaf 3 from the top) shows no difference in viral RNA and siRNA accumulation between nt and RDR6i plants once the infection is fully established. DNA or RNA probes specific for the PVX coat protein sequence were used to detect viral full-length and subgenomic RNAs or virus-derived siRNA, respectively. B, Infection of GFP16c and GFP16c/RDR6i with PVX:GFP for simultaneous monitoring of virus spread, GFP silencing, and transgene-derived GFP expression. Images of first and third leaves (from top) were taken under UV light at 10 dpi. Three levels of GFP fluorescence were discernible: background GFP expression from the GFP transgene (1), bright green areas due to GFP expressed by the replicating virus (2), and dark red areas of GFP silenced tissue (3). In the uppermost leaf 1 of GFP16c, the GFP silencing was evident around the veins, whereas replicating virus produced bright green GFP spots superimposed on the faint fluoresce from the GFP transgene. The same patterns were evident in leaf 3 of GFP16c. However, in GFP16c/RDR6i, the virus-derived GFP was more prevalent and there was only limited evidence of silencing adjacent to the infected areas in the older leaf 3, which was not centered around the veins. A magnified version of the green channel only of this image along with a schematic representation of the areas of different GFP expression levels is available as supplementary data (Supplemental Fig. 3).
However, there was an effect on the silencing signal at the systemic infection front that we observed by comparison of the PVX:GFP-infected lines GFP16c and GFP16c/RDR6i (Fig. 6B; Supplemental Fig. 3). The, young, not yet fully expanded leaves of GFP16c plants (Fig. 6B, leaf 1) exhibited three GFP expression levels indicative of virus- and transgene-derived gene expression. First, there was widespread background GFP fluorescence due to the 35S:GFP transgene (Fig. 6B, arrow 1); second, there were localized spots of intense fluorescence due to strong GFP expression from replicating PVX:GFP (Fig. 6B, arrow 2); and third, in regions around the veins, the GFP fluorescence was lost due to spreading of the silencing signal (Fig. 6B, arrow 3). The GFP16c/RDR6i leaves at the same stage showed more extensive fluorescence due to PVX:GFP than in GFP16c and no evidence for spread of the silencing signal from the veins (Fig. 6B, leaf 1). In older leaves (Fig. 6B, leaf 3), GFP silencing occurred only around infected areas in GFP16c/RDR6i plants but it was more restricted than in the GFP16c plants and did not spread ahead of the virus front alongside the veins. As expected, if a virus-derived silencing signal spreads with or ahead of the virus and prevents virus accumulation, the GFP-silenced tissue in these plants did not subsequently become infected. These results are therefore consistent with a role of RDR6 in either production of the signal or in initiation of silencing in cells that receive the systemic signal.
To investigate the silencing signal in more detail, we carried out grafting assays with RDR6i plants. First, we used an N. benthamiana line that carries the GFP16c transgene and a GFP RNAi construct (GFPi). Plants of this line, designated GFP16c/GFPi, are red under UV light at all stages of growth. When GFP16c scions were grafted onto GFP16c/GFPi stock plants, the GFP silencing spread into the scion after 14 d in 24/24 plants (Fig. 7A). In contrast, none out of 24 GFP16c/RDR6i scions on GFP16c/GFPi rootstocks exhibited GFP silencing after 28 d and, of these, 13/13 plants that were kept for a further 60 d remained fully green fluorescent (Fig. 7B). As expected, GFP-derived siRNA was detectable only in silenced GFP16c scions but not in nonsilenced GFP16c/RDR6i scions (Supplemental Fig. 4). In a replicate experiment, the young leaves in six out of 24 GFP16c/RDR6i scions exhibited a pattern of tightly vein-restricted GFP silencing under UV light. However, this limited silencing did not spread further into the mesophyll at later time-points and eventually it faded (Fig. 7C). This suppression of systemic silencing in the scions was a specific effect of the RDR6i transgene because GFP16c scions with a GUS RNAi transgene were fully competent to receive the GFP silencing signal at the same time as GFP16c scions (data not shown). Our conclusion from these grafting experiments is that NbRDR6 influences the ability to respond to a silencing signal that is translocated out of the GFP16c/GFPi rootstocks.
Figure 7.
Grafting experiments, RDR6i plants as either receptors or producers of a systemic silencing signal. Graft junctions are indicated by blue arrows and all plants are shown under UV illumination. A, A GFP16c plant grafted as scion onto a GFP16c/GFPi stock exhibited complete silencing of GFP within 2 weeks. B, A GFP16c/RDR6i plant grafted as scion onto a GFP16c/GFPi stock remained unsilenced as shown here after 3 weeks and later. C, Young, not yet fully expanded leaves of GFP16c scions (left) showed GFP silencing around the veins as shown here after 1 week that eventually spread so that the whole leaf appeared red under UV light. Leaves of the same stage of GFP16c/RDR6i scions normally did not exhibit any sign of GFP silencing (middle) but in 6/24 plants a pattern of vein-centered silencing was observed that failed to spread into the rest of the leaf (right). White bar = 0.5 cm. D, Transient expression of GFPi in leaves (1) of a GFP16c rootstock induced systemic silencing in the rootstock (2) and a GFP16c scion (3) at 24 d. E, GFP16c/RDR6i rootstocks exhibited local silencing in inoculated leaves, transiently expressing the GFPi construct (1) but no spread of silencing into newly emerging leaves occurred (2). A silencing signal was still produced in these rootstocks, which induced systemic silencing in GFP16c scions after 24 d (3).
We tested for the production of a systemic silencing signal in RDR6i by transient expression of the GFPi construct in GFP16c and GFP16c/RDR6i rootstocks. Local GFP silencing was triggered in both plant lines (Supplemental Fig. 1, A and B) and systemic spread of GFP silencing into noninoculated leaves occurred in GFP16c plants, although not in GFP16c/RDR6i (Fig. 7, D and E). This failure of systemic silencing signal in GFP16c/RDR6i was not because the silencing signal was absent. In six out of seven GFP16c plants that were grafted as a scion onto these plants, there was systemic silencing after 24 d (Fig. 7E). NbRDR6 is therefore not required for production of the systemic silencing signal.
We can also rule out that NbRDR6 is required for transport of the silencing signal from a three-way grafting experiment in which 2-cm long RDR6i stems were grafted between a GFP16c/GFPi rootstock and a GFP16c upper scion. In this experiment, the upper scion, but not the middle one, became silenced in seven out of nine plants, beginning at 31 d after grafting of the GFP16 scions (data not shown). Therefore, from these grafting experiments, we conclude that RDR6 affects systemic silencing because it influences a cell's ability to respond to the silencing signal. There is no evidence that this protein affects production or translocation of the signal.
DISCUSSION
In this paper, we describe how NbRDR6, like its Arabidopsis homolog, is implicated in a virus defense mechanism (Mourrain et al., 2000; Dalmay et al., 2001); plants in which NbRDR6 is silenced exhibit hyper-susceptibility to PVX, PVY and CMV in combination with the Y satellite although not to TRV, TMV, TCV, and CMV alone. With PVX, the antiviral role of NbRDR6 is in the growing point and newly emerging leaves of the infected plant rather than at the level of replication or cell to cell movement of the virus in the inoculated leaf (Fig. 4). We discuss below how this observation and our findings that NbRDR6 is implicated in systemic silencing may be informative about the role of RNA silencing in systemic spread and meristem exclusion of plant viruses.
In principle, NbRDR6 might affect susceptibility to PVX because the effector complex of RNA silencing is not formed in RDR6i. However, this effector complex is fully functional in the absence of NbRDR6, as illustrated by virus-induced and RNAi silencing phenotypes of rdr6 in Arabidopsis (Dalmay et al., 2000; Beclin et al., 2002) and as indicated by our observation that dsRNA induced silencing is equally efficient in GFP16c and GFP16c/RDR6i plants (Supplemental Fig. 1C). Abundant dsRNA substrates for DCL are presumably produced independently of NbRDR6 during viral RNA replication or as regions of secondary structure in the positive strand viral RNA (Molnár et al., 2005).
We can also rule out that NbRDR6 is required for systemic signal production (Fig. 7), and a more likely scenario to explain PVX invasion of the meristem in RDR6i plants is based on a proposed role of NbRDR6 in cells that receive the signal. This interpretation is consistent with genetic analysis in Arabidopsis showing that long distance movement of a silencing signal is impaired in rdr6 mutants (Himber et al., 2003). We propose that the PVX hypersusceptibility results because cells at the infection front receive both the viral RNA and a virus-derived silencing signal that could include an siRNA. This signal would be used, perhaps as a primer, for NbRDR6 to produce a dsRNA substrate for Dicer to start producing siRNA as an “immediate early” response to virus infection. Alternatively, the signal could guide the RNA induced silencing complex (RISC) to cleave viral RNA, creating aberrant RNA in the process that lacks either the 5′ cap or the 3′ poly(A) tail and would be recognized by NbRDR6 as a template (Fig. 8). The secondary siRNAs, produced from the NbRDR6-generated templates, would target silencing against the viral RNA and prevent its later accumulation. The potential for an siRNA primed production of dsRNA is illustrated by the ability of an RDR homolog from Neurospora to incorporate siRNA in vitro into long dsRNA (Makeyev and Bamford, 2002). In the RDR6i plants, the incoming silencing signal cannot be processed by NbRDR6 to generate the template for immediate secondary siRNA production. As a result, virus accumulation in the growing points and newly emerging leaves would be enhanced and the virus would spread faster through the plant, as observed (Figs. 4–6).
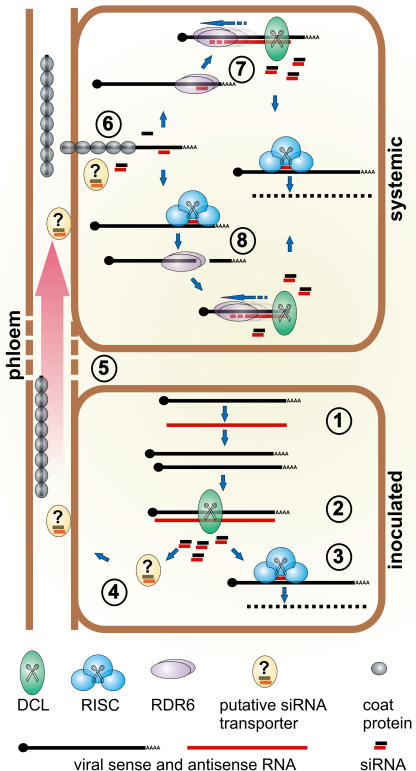
Figure 8.
Model for RDR6 function in antiviral defense against PVX. In the inoculated cell (lower box) PVX replicates, forming dsRNA with genomic or subgenomic and antisense RNA in the process (1). DCL recognizes the dsRNA as a substrate and produces virus-derived siRNA (2), which guides RISC to the viral RNA (3). siRNA, probably bound to a transporter protein (4), are translocated through the phloem stream into upper parts of the plant alongside the coated viral RNA (5). Viral RNA and virus-derived siRNA are unloaded simultaneously into cells in the upper part of the plant and the virus-derived siRNA can anneal to the unpacked viral RNA immediately (6). Annealed siRNA could either be used as a primer by RDR6 to produce the dsRNA substrate for DCL (7) or they could guide RISC to cleave the viral RNA, leaving an aberrant (noncapped or nonpolyadenylated respectively) RNA that is recognized by RDR6 for unprimed synthesis of the dsRNA substrate for DCL (8). Both pathways lead to immediate production of siRNA to target viral RNA for RISC-mediated destruction. If no signal or no RDR6 is present in the receiving tissue, this immediate response is not possible and production of viral siRNA relies on viral replication, giving the virus a head start before the silencing machinery. Once the infection is fully established, the viral replicase produces abundant substrate for DCL and the RDR6 pathway does not contribute significantly to virus-derived siRNA production anymore.
This model (Fig. 8) requires that PVX produces a systemic silencing signal and that this signal spreads through the plant either with or ahead of the virus front. Consistent with this prediction, PVX:GFP produces a signal that travels slightly ahead of the virus front (Fig. 6). The proposal that a signal is produced from PVX:GFP differs from a suggestion that the p25 silencing suppressor protein of PVX prevents long range silencing in grafting assays (Voinnet et al., 2000). According to this earlier hypothesis, p25 would prevent all signal production in PVX-infected cells. However, the grafting assay tested for a silencing signal that would travel many centimeters away from the infected cells, whereas our present hypothesis requires only that the signal is at or slightly ahead of the virus infection front. The signal we observed in PVX:GFP-infected GFP16c plants (Fig. 6B) could have been generated inside the vasculature in such amounts that p25 is not sufficient to stop it from moving short distances and spreading into the mesophyll.
A silencing signal-based mechanism of virus defense could operate anywhere at the infection front. However, it might be expected that this process would be particularly important in the growing point. This part of the plant is a strong photosynthetic sink and, consequently, a preferred transport destination of both viruses and silencing signals. In effect, this silencing signal hypothesis proposes that the well-established phenomenon of meristem exclusion (Matthews, 1991) is a variation of the silencing-related “recovery” process in which the upper leaves of a virus-infected plant are symptom-free and immune to secondary infection (Covey et al., 1997; Ratcliff et al., 1997); classical meristem exclusion would be recovery that is restricted to the growing point of the infected plant, whereas recovery would be meristem exclusion that operated not only in the meristem but also in the uppermost leaves of the plant.
Further supporting evidence that silencing is involved in meristem exclusion is from the previous findings that RNA viruses acquire the ability to invade meristems if they are inoculated to plants expressing viral suppressors of silencing (Foster et al., 2002; A. Martin-Hernandez and D.C. Baulcombe, unpublished data). A difference between viruses that exhibit the restricted meristem exclusion or the more extensive recovery process might be related to the amount of silencing signal produced by the virus and the nature of the viral silencing suppressor protein(s), which might interfere with signaling. Consistent with this idea, the outcome of a virus infection—normal meristem exclusion or recovery—can be influenced by transgenic expression of a virus-derived gene (Lindbo et al., 1993). Presumably, in the lines showing recovery, the virus-specific silencing signal is produced by both the virus and the transgene, whereas in the lines that exhibit the normal response to virus infection, there is very little signal from the transgene.
It is not clear at present why RDR6i plants are not hypersusceptible to all tested viruses. Only PVX, PVY, and CMV in combination with its Y satellite exhibited enhanced virulence on RDR6i plants (Fig. 4; Supplemental Fig. 2). Perhaps the signal from the cells infected with TMV, TRV, and other viruses is produced at too low levels or too late to have an effect on virus spread. Alternatively, the RNA of the other viruses may have sequence elements or structures that prevent its being used as a template by NbRDR6. There are five other RDR proteins in Arabidopsis and it may be that one or more of them use other viral RNAs. Consistent with this idea, the ortholog of RDR1 has been shown to contribute to defense against TMV and PVX in N. tabacum (Xie et al., 2001) and the lack of a functional ortholog of this enzyme is at least partly responsible for the hypersusceptibility of N. benthamiana plants to TMVs (Yang et al., 2004). Transforming N. benthamiana with a functional copy of RDR1 increased resistance against TMVs but not against CMV or PVX (Yang et al., 2004), illustrating the differential role of RDR proteins in defense against viruses belonging to different groups.
For a full understanding of RNA silencing in antiviral defense, it will be necessary to generate plants that are mutant or silenced for the remaining RDR homologs and also for other genes including AGO and DCL homologs that are required for RNA silencing. Our analysis described here shows how the effects of these proteins on virus susceptibility will need to be assessed in the whole plant rather than simply at the level of the cell.
MATERIALS AND METHODS
Transgenic Plants and DNA Constructs
Nicotiana benthamiana line GFP16c has been described as line 16c previously (Ratcliff et al., 2001). Line GFP16c/RDR6i was obtained by transformation of GFP16c with a construct, containing a 245-bp cDNA fragment of the sequence of N. benthamiana RDR6 in an inverted repeat configuration. This fragment was amplified from N. benthamiana total RNA by RT-PCR using primers 5′-agactagtggcgcgccTGACGTGGCTTTTGATG-3′ and 5′-tcggatccatttaaaTCTTGAATAAAGCATTGGCC-3′, and cloned into the AscI/SwaI and XbaI/XmaI sites of RNAi vector pFGC5941 (GenBank accession no. AY310901).
The same fragment was also cloned blunt-ended into the SmaI site of a TRV-based gene silencing vector (Jones et al., 1999) to generate TRV:RDR6. N. benthamiana line RDR6i was obtained by back-crossing line GFP16c/RDR6i to nt N. benthamiana. Construct GFPi carries a 400-bp fragment of the GFP sequence, amplified by primers 5′-AGTAAAGGAGAAGAACTTTTCACT-3′ and 5′-TTCCGTCCTCCTTGAAATCGA-3′ in forward and reverse orientation in RNAi vector pFGC5941. Line GFP16c was transformed with GFPi to obtain line GFP16c/GFPi. The GUS RNAi construct GUSi was made by cloning a 287-bp GUS-fragment, amplified using primers 5′-GGACTAGTGGCGCGCCGGATACGTTAGCCGGGCT-3′ and 5′-ACGGATCCCATTTAAATGTTTGCCTCCCTGCTGCGG-3′ into pFGC5941. Line GFP16c was transformed with this construct to obtain line GFP16c/GUSi. The activity of the RNAi construct in this line was verified in a northern-blot assay of GUS-derived siRNA formation. All transgenic plants were used in homozygous state.
To obtain construct GF:invTerm, the octopine synthase terminator from construct GFPi was amplified using primers 5′-ATCCGTCACTACGTGTAGTCCCTAGAGTCCTGTC-3′ and 5′-TGCATCCACGTAGTGCAGTCACGACGTTGTAAAAC-3′, cloned into the DraIII site of construct 35S:GF (Lu et al., 2003), screening for inverse orientation with respect to the GF-cassette. For infection of plants by transient Agrobacterium-mediated expression of PVX constructs under the control of the cauliflower mosaic virus 35S promoter, constructs pGR107 (GenBank accession no. AY297842; Lu et al., 2003) and pPVX-GFP were used. Construct pGR107 contains the full-length PVX genome, while pPVX-GFP (Baulcombe et al., 1995) carries the mGFP5 modified jellyfish GFP (Haseloff et al., 1997) under the control of a duplicate coat protein promoter.
For transient expression in N. benthamiana, pGreen- and pBin-based constructs were mobilized into Agrobacterium strain GV3101 (containing the pSoup helper plasmid) or C58C1 (pCH32 helper plasmid), respectively.
Plant Growth Conditions
All plants were grown in a glasshouse with 16-h supplemental lighting (HQI halide lights) at a constant temperature of 22/20°C (day/night).
Inoculation of Plants with Recombinant Viruses and Transient Expression Vectors
Inoculation of N. benthamiana with Agrobacterium carrying constructs for transient expression of transgenes or viruses was done as described previously (English et al., 1997).
For the assessment of NbRDR6 function in N. benthamiana, 2- to 3-week-old plants of line 16c were systemically silenced for GFP by agro-inoculating GFPi. Six weeks postinoculation, when the plants were completely silenced and appeared red under UV light, lower leaves were agro-inoculated with TRV:RDR6 and upper, noninoculated leaves were monitored for breakdown of GFP silencing under UV light, which usually appeared 4 to 6 weeks postinoculation with the VIGS vector in all five to 10 inoculated plants.
Grafting
For grafting of N. benthamiana, 3- to 4-week-old plants were used. Scion stems were cut to a wedge shape that was then inserted into a vertical slit cut into the stem of the rootstock about 2 cm above soil level. The grafting junction was wrapped with Parafilm and plants were kept humid under a plastic cover for 1 week or until the grafts had taken.
GFP Imaging
GFP expression in plants was photographed under UV light using a Nikon D1X digital camera with a Kodak Wratten filter number 8 and a B-100AP longwave-UV lamp (Ultra-Violet Products, Upland, CA) or using a Leica MZ-FLIII dissecting microscope with GFP-filter and a Leica DC200 digital camera (Leica, Solms, Germany).
Nucleic Acid Extraction and Gel-Blot Analysis
Total nucleic acid was extracted from plant tissue by either using TRI Reagent (Sigma, St. Louis) or, for viral RNA analyses, following the procedure of White and Kaper (White and Kaper, 1989). Briefly, leaf tissue (100–150 mg) was homogenized in 600 μL extraction buffer (100 mm Gly-NaOH, pH 9.5, 10 mm EDTA, 100 mm NaCl, 2% [w/v] SDS) and added to 600 μL buffer-saturated phenol. The aqueous phase was subsequently extracted with equal volumes of phenol/chloroform and chloroform before precipitating in Na-acetate/ethanol.
For the detection of siRNAs, equal amounts of total RNA (approximately 10 μg) were separated on 8% polyacrylamide gels in 1× Tris-borate/EDTA with 50% (w/v) urea and transferred onto Zeta-Probe GT membranes (Bio-Rad, Hercules, CA) by overnight capillary transfer in 20× SSC. In vitro transcribed 32P-labeled RNA probes of cloned fragments of GFP, GUS, RDR6, or the PVX-coat protein sequence were used to detect corresponding siRNA over night at 42°C using PerfectHyb hybridization buffer (Sigma) following a 2-h prehybridization in the same buffer.
Viral RNA was detected by separating 5 μg total nucleic acid on formaldehyde agarose gels and transferring to Zeta-Probe GT membranes (Bio-Rad) by overnight capillary transfer according to standard laboratory protocols (Sambrook and Russel, 2001). For the detection of PVX RNA, DNA fragments of the cloned PVX coat protein sequence were amplified by PCR and labeled with 32P by standard laboratory methods (Sambrook and Russel, 2001). Hybridization was carried out in PerfectHyb buffer at 68°C overnight.
Real-Time RT-PCR
Transcript levels of RDR genes were analyzed by quantitative real-time RT-PCR using the Chromo4 detector in combination with a PTC-200 Thermal Cycler (MJ Research/Bio-Rad). Total RNA was extracted from four pools of five plants at six-leaf-stage for each plant line using TRI Reagent (Sigma) according to manufacturer's instructions. Synthesis of cDNA from 5 μg of total RNA was performed with random hexanucleotides (Roche, Mannheim, Germany) and Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA) following manufacturer's instructions. Reactions without reverse transcriptase or without template were included as controls. For the quantitative real time PCR, cDNA corresponding to 500 ng of total RNA was used in 50-μL reactions using DyNAmo HS SYBR Green qPCR kit (Finnzymes, Espoo, Finland) according to manufacturer's instructions. Primers were designed to amplify similar sized regions of GAPDH, serving as an internal standard, and the N. benthamiana orthologs of RDR6, RDR1 (GenBank accession no. AY574374; Yang et al., 2004), and RDR2 (GenBank accession no. AY722009). Primer sequences were: GAPDH, 5′-GGAGGAGGGAACAACAAGAGG-3′ and 5′-AGATGCCGTCAGTGCCGA-3′ (amplicon length, 238 bp); RDR6, 5′-CTCAGCTTGGGGACCTCA-3′ and 5′-CAGCCTCCAGAATCCTCAC-3′ (amplicon length, 261 bp); RDR1, 5′-GCATTGAACACGCCTTGGA-3′ and 5′-GCAGAACCCGATTGGATACG-3′ (amplicon length, 225 bp); RDR2, 5′-GGTGTAGAGAAGAGAGTTA-3′ and 5′-GTTAGAATGAGTTGGTGC-3′ (amplicon length, 260 bp). The reverse primer for RDR6 anneals outside the region of RDR6, which was used for the RDR6i construct while the forward primers anneals inside that region. Amplicons were also analyzed on agarose gels to ensure formation of single PCR products. Efficiencies of the real time PCR reactions for all four primer combinations were analyzed using the LineRegPCR software (Ramakers et al., 2003), average efficiencies ranged from 1.89 to 1.95 for all primer combinations used.
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number DQ093875.
Supplementary Material
Acknowledgments
The authors thank Louise Chappell for CMV Y satellite inoculations and images, Attila Molnar and Natalya Elina for critical reading of the manuscript, and Sharyn Carter for excellent technical assistance.
This work was supported by the European Commission Research Directorate (contract no. QLG2–CT–2002–01673). The Sainsbury Laboratory is supported by the Gatsby Charitable Foundation.
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.063537.
References
- Astier-Manifacier S, Cornuet P (1974) RNA-dependent RNA polymerase in Chinese cabbage. Biochim Biophys Acta 232: 484–493 [DOI] [PubMed] [Google Scholar]
- Baulcombe D (2004) RNA silencing in plants. Nature 431: 356–363 [DOI] [PubMed] [Google Scholar]
- Baulcombe DC, Chapman SN, Santa Cruz S (1995) Jellyfish green fluorescent protein as a reporter for virus infections. Plant J 7: 1045–1053 [DOI] [PubMed] [Google Scholar]
- Beclin C, Boutet S, Waterhouse P, Vaucheret H (2002) A branched pathway for transgene-induced RNA silencing in plants. Curr Biol 12: 684–688 [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366 [DOI] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM (2005) Principles of microRNA target recognition. PLoS Biol 3: e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell MA, Hannon GJ (2004) RNase III enzymes and the initiation of gene silencing. Nat Struct Mol Biol 11: 214–218 [DOI] [PubMed] [Google Scholar]
- Chan SW-L, Zilberman D, Xie Z, Johansen LK, Carrington JC, Jacobsen SE (2004) RNA silencing genes control de novo DNA methylation. Science 303: 1336. [DOI] [PubMed] [Google Scholar]
- Covey SN, Al-Kaff NS, Langara A, Turner DS (1997) Plants combat infection by gene silencing. Nature 385: 781–782 [Google Scholar]
- Dalmay T, Hamilton AJ, Rudd S, Angell S, Baulcombe DC (2000) An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101: 543–553 [DOI] [PubMed] [Google Scholar]
- Dalmay TD, Horsefield R, Braunstein TH, Baulcombe DC (2001) SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis. EMBO J 20: 2069–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- English JJ, Davenport GF, Elmayan T, Vaucheret H, Baulcombe DC (1997) Requirement of sense transcription for homology-dependent virus resistance and trans-inactivation. Plant J 12: 597–603 [Google Scholar]
- Fagard M, Vaucheret H (2000) Systemic silencing signal(s). Plant Mol Biol 43: 285–293 [DOI] [PubMed] [Google Scholar]
- Foster TM, Lough TJ, Emerson SJ, Lee RH, Bowman JL, Forster RLS, Lucas WJ (2002) A surveillance system regulates selective entry of RNA into the shoot apex. Plant Cell 14: 1497–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden TA, Schauer SE, Lang JD, Pien S, Mushegian AR, Grossniklaus U, Meinke DW, Ray A (2002) SHORT INTEGUMENTS1/SUSPENSOR1/CARPEL FACTORY, a Dicer homolog, is a maternal effect gene required for embryo development in Arabidopsis. Plant Physiol 130: 808–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseloff J, Siemering KR, Prasher DC, Hodge S (1997) Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc Natl Acad Sci USA 94: 2122–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr AJ, Jensen MB, Dalmay T, Baulcombe D (2005) RNA polymerase IV directs silencing of endogenous DNA. Science 308: 118–120 [DOI] [PubMed] [Google Scholar]
- Himber C, Dunoyer P, Moissiard G, Ritzenthaler C, Voinnet O (2003) Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO J 22: 4523–4533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L, Hamilton AJ, Voinnet O, Thomas CL, Maule AJ, Baulcombe DC (1999) RNA-DNA interactions and DNA methylation in post-transcriptional gene silencing. Plant Cell 11: 2291–2302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM (2005) Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433: 769–773 [DOI] [PubMed] [Google Scholar]
- Lindbo JA, Silva-Rosales L, Proebsting WM, Dougherty WG (1993) Induction of a highly specific antiviral state in transgenic plants: implications for regulation of gene expression and virus resistance. Plant Cell 5: 1749–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2TΔΔC Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lu R, Malcuit I, Moffett P, Ruiz MT, Peart J, Wu A-J, Rathjen JP, Bendahmane A, Day L, Baulcombe D (2003) High throughput virus-induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO J 22: 5690–5699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev EV, Bamford DH (2002) Cellular RNA-dependent RNA polymerase involved in posttranscriptional gene silencing has two distinct activity modes. Mol Cell 10: 1417–1427 [DOI] [PubMed] [Google Scholar]
- Matthews REF (1991) Plant Virology, Ed 3. Academic Press, San Diego
- Mlotshwa S, Voinnet O, Mette MF, Matzke M, Vaucheret H, Ding SW, Pruss G, Vance VB (2002) RNA silencing and the mobile silencing signal. Plant Cell (Suppl) 14: S289–S301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnár A, Csorba T, Lakatos L, Várallyay É, Lacomme C, Burgyán J (2005) Plant virus derived siRNAs predominantly originate from highly structured single-stranded viral RNAs. J Virol 79: 7812–7818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motamedi MR, Verdel A, Colmenares SU, Gerber SA, Gygi SP, Moazed D (2004) Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell 119: 789–802 [DOI] [PubMed] [Google Scholar]
- Mourrain P, Beclin C, Elmayan T, Feuerbach F, Godon C, Morel J-B, Jouette D, Lacombe AM, Nikic S, Picault N, et al (2000) Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101: 533–542 [DOI] [PubMed] [Google Scholar]
- Palauqui JC, Elmayan T, Pollien JM, Vaucheret H (1997) Systemic acquired silencing: transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J 16: 4738–4745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS (2004) SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev 18: 2368–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Lekanne Deprez RH, Moorman AFM (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339: 62–66 [DOI] [PubMed] [Google Scholar]
- Ratcliff F, Harrison BD, Baulcombe DC (1997) A similarity between viral defense and gene silencing in plants. Science 276: 1558–1560 [DOI] [PubMed] [Google Scholar]
- Ratcliff F, Martin-Hernandez AM, Baulcombe DC (2001) Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J 25: 237–245 [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbour-joining method: a new method for reconstruction of phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russel DW (2001) Molecular Cloning: A Laboratory Manual, Ed 3. Cold Spring Harbor Laboratory Press, New York
- Schiebel W, Haas B, Marinkovic S, Klanner A, Sanger HL (1993. a) RNA-directed RNA polymerase from tomato leaves. I. Purification and physical properties. J Biol Chem 268: 11851–11857 [PubMed] [Google Scholar]
- Schiebel W, Haas B, Marinkovic S, Klanner A, Sanger HL (1993. b) RNA-directed RNA polymerase from tomato leaves. II. Catalytic in vitro properties. J Biol Chem 268: 11858–11867 [PubMed] [Google Scholar]
- Schiebel W, Pelissier T, Reidel L, Thalmeir S, Schiebel R, Kempe D, Lottspeich F, Sanger HL, Wassenegger M (1998) Isolation of an RNA-directed RNA polymerase-specific cDNA clone from tomato. Plant Cell 10: 2087–2102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Haley B, Zamore PD (2002) Evidence that siRNAs function as guides, not primers, in the Drosophila and human RNAi pathways. Mol Cell 10: 537–548 [DOI] [PubMed] [Google Scholar]
- Vaucheret H, Vazquez F, Crete P, Bartel DP (2004) The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev 18: 1187–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert J-L, Bartel DP, Crete P (2004) Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell 16: 69–79 [DOI] [PubMed] [Google Scholar]
- Voinnet O, Baulcombe DC (1997) Systemic signalling in gene silencing. Nature 389: 553. [DOI] [PubMed] [Google Scholar]
- Voinnet O, Lederer C, Baulcombe DC (2000) A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103: 157–167 [DOI] [PubMed] [Google Scholar]
- Voinnet O, Vain P, Angell S, Baulcombe DC (1998) Systemic spread of sequence-specific transgene RNA degradation is initiated by localised introduction of ectopic promoterless DNA. Cell 95: 177–187 [DOI] [PubMed] [Google Scholar]
- White JL, Kaper JM (1989) A simple method for detection of viral satellite RNAs in small tissue samples. J Virol Methods 23: 83–94 [DOI] [PubMed] [Google Scholar]
- Xie Z, Fan B, Chen CH, Chen Z (2001) An important role of an inducible RNA-dependent RNA polymerase in plant antiviral defense. Proc Natl Acad Sci USA 98: 6516–6521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC (2004) Genetic and functional diversification of small RNA pathways in plants. PLoS Biol 2: 642–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SJ, Carter SA, Cole AB, Cheng NH, Nelson RS (2004) A natural variant of a host RNA-dependent RNA polymerase is associated with increased susceptibility to viruses by Nicotiana benthamiana. Proc Natl Acad Sci USA 101: 6297–6302 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.