Abstract
Cell-to-cell tobacco mosaic virus movement protein (TMV MP) mediates viral spread between the host cells through plasmodesmata. Although several host factors have been shown to interact with TMV MP, none of them coresides with TMV MP within plasmodesmata. We used affinity purification to isolate a tobacco protein that binds TMV MP and identified it as calreticulin. The interaction between TMV MP and calreticulin was confirmed in vivo and in vitro, and both proteins were shown to share a similar pattern of subcellular localization to plasmodesmata. Elevation of the intracellular levels of calreticulin severely interfered with plasmodesmal targeting of TMV MP, which, instead, was redirected to the microtubular network. Furthermore, in TMV-infected plant tissues overexpressing calreticulin, the inability of TMV MP to reach plasmodesmata substantially impaired cell-to-cell movement of the virus. Collectively, these observations suggest a functional relationship between calreticulin, TMV MP, and viral cell-to-cell movement.
Following initial infection (usually by mechanical inoculation), many plant viruses, such as tobacco mosaic virus (TMV), spread from cell to cell through plasmodesmata (for review, see Heinlein, 2002; Waigmann et al., 2004), presumably utilizing the existing cellular pathways of plasmodesmal transport. In the case of TMV, this virus encodes a specific cell-to-cell movement protein (MP), which mediates the transport of the viral genomic RNA through plasmodesmata (Deom et al., 1987). To date, MP has been shown to bind TMV RNA (Citovsky et al., 1990, 1992), associate with the cytoskeleton (Heinlein et al., 1995; McLean et al., 1995; Boyko et al., 2000) and endoplasmic reticulum (ER; Heinlein et al., 1998; Reichel and Beachy, 1999), target to plasmodesmata within plant cell walls (Tomenius et al., 1987; Ding et al., 1992a), increase plasmodesmal permeability (Wolf et al., 1989; Waigmann et al., 1994), and undergo negative regulation by phosphorylation (Citovsky et al., 1993; Waigmann et al., 2000; Trutnyeva et al., 2005). To perform many of these functions, TMV MP most likely cooperates with different cellular factors, some of which, such as cell wall pectin methylesterases (PME; Dorokhov et al., 1999; Chen et al., 2000), microtubules (MTs) and actin filaments (Heinlein et al., 1995; McLean et al., 1995; Boyko et al., 2000), and a RIO protein kinase (Yoshioka et al., 2004), are known, while others remain to be discovered. It would be especially interesting to determine whether cellular proteins exist that not only recognize TMV MP but also coreside with it within plasmodesmata.
Using TMV MP as specific ligand in a biochemical purification protocol, we showed that it interacts with plant calreticulin. Calreticulin is a Ca2+-sequestering protein that is highly conserved between different species, including plants (Nelson et al., 1997; Borisjuk et al., 1998, and refs. therein; Michalak et al., 1998). Functionally, calreticulin is involved in Ca2+ storage and signaling, chaperone activity, cell adhesion, and regulation of gene expression (Krause and Michalak, 1997). Although calreticulin was originally detected in the membranes of smooth muscle sarcoplasmic reticulum and nonmuscle ER (Ostwald and MacLennan, 1974), increasing evidence indicates that, in plants, this protein also accumulates within plasmodesmata (Baluska et al., 1999, 2001; Laporte et al., 2003), suggesting its role in cell-to-cell transport. Indeed, our findings suggest that calreticulin can interact directly with TMV MP, and, when overexpressed in transgenic plants, calreticulin interferes with native targeting of TMV MP and significantly delays cell-to-cell movement of the virus.
RESULTS
Identification of Calreticulin Binding to TMV MP
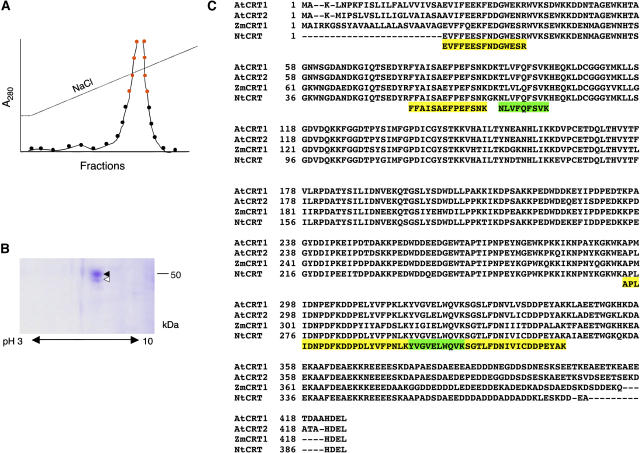
Previously, we utilized affinity chromatography on an immobilized TMV MP column to isolate tobacco (Nicotiana tabacum) cell wall proteins that bind TMV MP (Chen et al., 2000). While in these experiments the major protein species of about 38 kD detected by SDS-PAGE represented PME, other, less abundant bands in the 50- to 60-kD range were also detected (Chen et al., 2000) but remained unidentified. Here, we loaded cell wall extracts on the TMV MP column and, following a wash, eluted the bound protein by applying a continuous gradient of NaCl (Fig. 1A). Proteins contained within the elution peak fractions were subjected to two-dimensional gel electrophoresis, following which the gel region containing protein species with 50- to 60-kD electrophoretic mobility exhibited two adjacent spots (Fig. 1B). Both spots were excised and digested with trypsin, and the resulting peptides were separated by HPLC and analyzed by mass spectrometry for peptide purity and mass. The mass spectrometry profiles of the four most abundant peptides of the top spot (Fig. 1B, black arrowhead; Fig. 1C, yellow highlight) and of the two most abundant peptides of the bottom spot (Fig. 1B, white arrowhead; Fig. 1C, green highlight) contained single-peptide peaks (data not shown), indicating the presence of one peptide species in each fraction. These peptides were then subjected to amino acid sequence analysis. The use of single HPLC peaks in these experiments increased the clarity and confidence of obtained peptide sequences as the observed and predicted masses differed by less than 0.01% (data not shown). All peptide sequences matched the sequences of calreticulin from such diverse plant species as Arabidopsis (Arabidopsis thaliana), maize (Zea mays), and tobacco (Fig. 1C). Potentially, the strongest protein spot of about 50 kD corresponded to the processed form of tobacco calreticulin, whereas the weaker spot with slightly lower electrophoretic mobility was a degradation product or a different isoform of tobacco calreticulin. Thus, calreticulin most likely represents one of the plant proteins that bind TMV MP.
Figure 1.
Purification and identification of TMV MP-interacting protein. A, Purification by affinity chromatography. Protein amounts in the fractions eluted by the indicated continuous NaCl gradient were measured by A280. Fractions pooled for subsequent analysis by two-dimensional gel electrophoresis are indicated by red circles. B, Coomassie blue-stained two-dimensional gel of the pooled fractions indicated in A. Molecular mass expressed in thousands of Daltons is indicated on the right. C, Amino acid sequence of six peptides derived from the purified TMV MP-interacting protein and its alignment with full-length calreticulin sequences from Arabidopsis (AtCRT1, accession no. NM_104513.2, and AtCRT2, accession no. NM_100791.2), maize (ZmCRT1, accession no. Z46772.1), and tobacco (NtCRT, accession no. X85382.1). Alignment was performed by the Clustal algorithm; gaps introduced for alignment are indicated by dashes. Yellow and green boxes indicate peptide sequences derived from the top and bottom protein spots indicated by black and white arrowheads, respectively, in B.
Because calreticulins are conserved between different plant species (Fig. 1C), we elected to perform most of our studies using calreticulin proteins from Arabidopsis to facilitate future studies of TMV MP-calreticulin interactions in this model plant (Meyerowitz and Somerville, 1994; Simon, 1994).
Plasmodesmal Targeting of Calreticulin Requires Its Signal Peptide
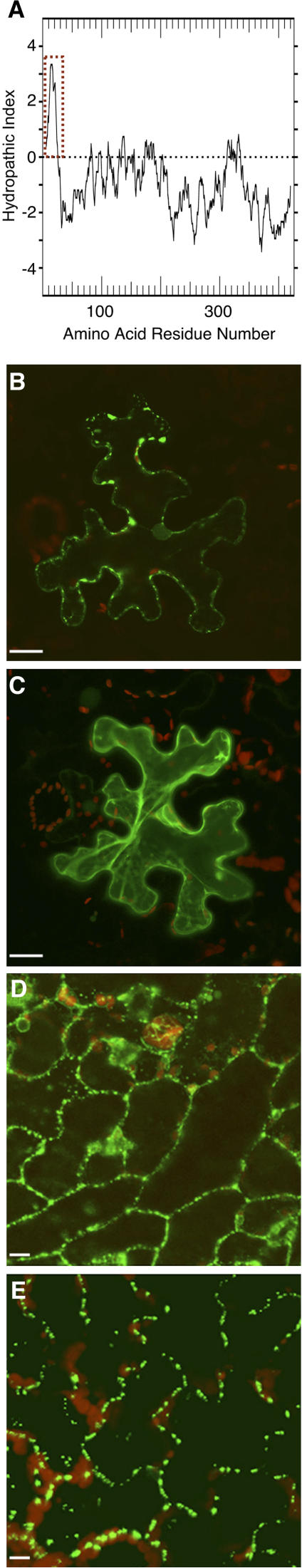
As an ER-based protein, calreticulin carries a hydrophobic N-terminal signal sequence, which is clearly identifiable on a hydropathy profile of Arabidopsis calreticulin 1 (AtCRT1, accession no. NM_104513.2; Fig. 2A). We examined the role of this signal peptide in subcellular targeting of AtCRT1. The full-length protein and its mutant, in which the first 50 N-terminal amino acid residues had been deleted (AtCRT1-del50N), were tagged at their C termini with green fluorescent protein (GFP) and transiently expressed following bombardment of their encoding constructs into Nicotiana benthamiana leaves. Figure 2B shows that the full-length AtCRT1 accumulated within distinct puncta at the cell periphery, a pattern characteristic of plasmodesmal localization (Oparka et al., 1997; Baluska et al., 1999, 2001; Laporte et al., 2003). In contrast, the AtCRT1-del50N mutant lacking its signal peptide remained predominantly cytoplasmic (Fig. 2C), suggesting that the signal peptide and, by implication, ER translocation are necessary for targeting to plasmodesmata.
Figure 2.
Plasmodesmal localization of calreticulin requires signal peptide. A, Hydropathy profile of AtCRT1. Hydrophobic domains are indicated by a positive index and hydrophilic domains by a negative index (Kyte and Doolittle, 1983). Dotted rectangle delineates the signal sequence. B, AtCRT1-GFP transiently expressed in N. benthamiana. C, AtCRT1-del50N-GFP lacking its signal peptide transiently expressed in N. benthamiana. D, AtCRT2-YFP stably expressed in transgenic Arabidopsis from its full-length gene with native regulatory elements. E, TMV MP-GFP stably expressed in transgenic Arabidopsis. YFP, CFP, and GFP signals are indicated in green, and plastid autofluorescence is indicated in red. Images in B and D are single confocal sections; images in C and E are projections of several confocal sections. Bars = 10 μm.
Calreticulin-TMV MP Interaction in Vivo and in Vitro
That calreticulin was biochemically isolated using TMV MP affinity chromatography suggests that these two proteins bind each other. Consistent with this notion, Arabidopsis calreticulin 2 (AtCRT2, accession no. NM_100791.2) and TMV MP were observed within similar subcellular punctate structures, likely the plasmodesmata due to the presence of MP, when tagged with yellow fluorescent protein (YFP; AtCRT2-YFP) and with GFP (TMV MP-GFP) and constitutively expressed in transgenic Arabidopsis plants (Fig. 2, D and E, respectively). To directly demonstrate the calreticulin-TMV MP interaction, we utilized three independent experimental approaches.
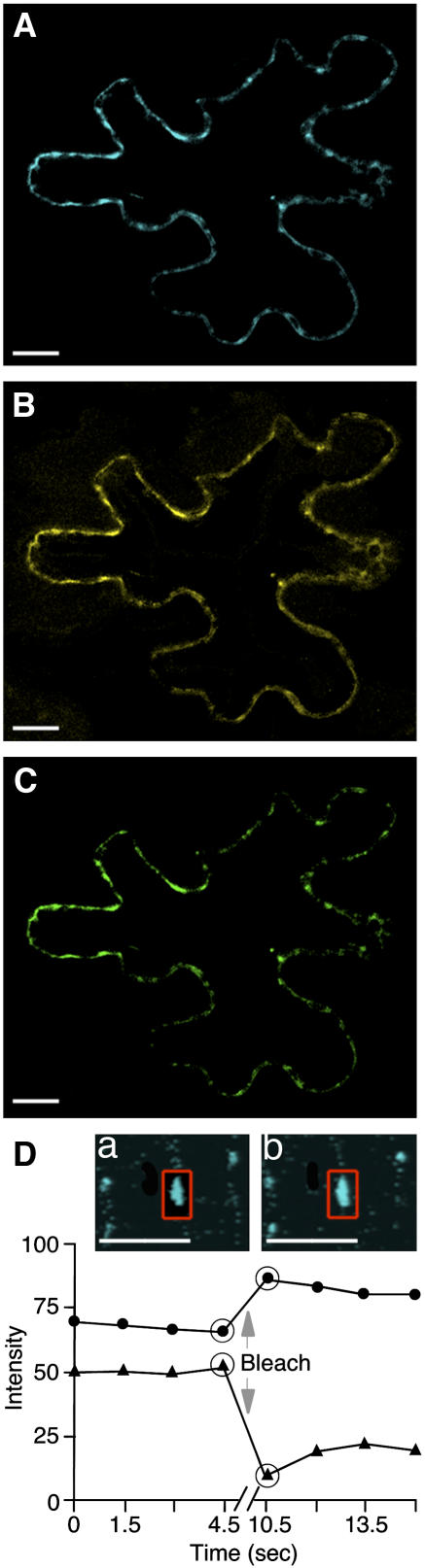
First, we used fluorescence resonance energy transfer (FRET) microscopy, which allows direct detection of protein interactions in planta, within living cells, as well as determination of the subcellular localization of the interacting proteins (Hink et al., 2002; Jares-Erijman and Jovin, 2003). AtCRT2-YFP and TMV MP tagged with a cyan fluorescent protein (CFP) spectral variant of GFP (TMV MP-CFP) were transiently coexpressed in N. benthamiana leaf tissues. FRET between these proteins was detected by acceptor photobleaching in which energy transfer is reduced and donor (i.e. CFP) fluorescence is increased when the acceptor (i.e. YFP) is bleached; such elevated fluorescence is characteristic of FRET because normally bleaching decreases rather than enhances fluorescence (e.g. Kenworthy, 2001; Karpova et al., 2003). Another advantage of acceptor photobleaching is that donor fluorescence cannot be attributed to a bleed-through signal of the acceptor because the latter has been bleached. Figure 3 shows a representative cell coexpressing both tagged proteins; images collected in three separate channels, i.e. the CFP signal (Fig. 3A), the YFP signal (Fig. 3B), and the FRET signal (Fig. 3C), indicate colocalization of and energy transfer between these two proteins at punctate locations at the cell periphery. Quantification of the CFP signal after photobleaching of YFP (Fig. 3D) revealed an increase in the intensity of the donor fluorescence with FRET efficiency (EF) of 19.0% ± 0.4%, which is indicative of FRET (e.g. Kenworthy, 2001; Karpova et al., 2003). Interestingly, this FRET efficiency was higher than that reported for a direct translational fusion between CFP and YFP (7.96% ± 0.38%; Karpova et al., 2003). Based on the FRET efficiency values, we calculated the distance between the interacting molecules of TMV MP and calreticulin to be about 6.4 nm, assuming freely rotating CFP and YFP fluorophores (Gadella et al., 1999).
Figure 3.
FRET microscopy of cells coexpressing TMV MP-CFP and AtCRT2-YFP. A, CFP channel. B, YFP channel. C, FRET channel. D, Acceptor (YFP) photobleaching. a and b, CFP (donor) channel before and after the bleach, respectively. After the bleach, CFP fluorescence increases in the bleach areas indicated by red rectangles. Quantification of donor fluorescent intensity in a representative sample also shows an increase in CFP fluorescence after the bleach. This effect was reproducible over a set of 10 different cells. Circles and triangles indicate the intensity of CFP and YFP signals, respectively; arrow indicates execution of the bleach; and large circles indicate time points immediately before and after the bleach. CFP signal is in blue, YFP signal is in yellow, and FRET signal is in green; plastid autofluorescence was filtered out. All images are single confocal sections. Bars = 10 μm.
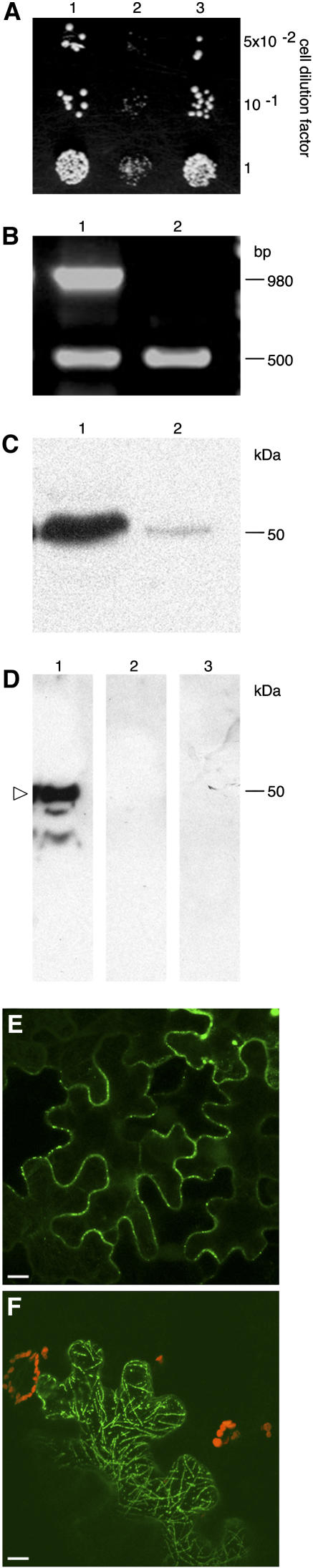
Next, we used the yeast (Saccharomyces cerevisiae) two-hybrid system (Fields and Song, 1989; Hollenberg et al., 1995) to confirm the interaction between TMV MP and calreticulin. Figure 4A shows the interaction between TMV MP and full-length AtCRT1 (lane 1). No interaction was observed between lamin C, known as a nonspecific activator in the two-hybrid system (Bartel et al., 1993), and calreticulin (Fig. 4A, lane 2) or TMV MP (data not shown). An AtCRT1 mutant that lacked its N-terminal signal peptide and thus mimicked the processed form of the protein still bound TMV MP (Fig. 4A, lane 3). Similar results were obtained with MP of another tobamovirus, turnip vein clearing virus (data not shown).
Figure 4.
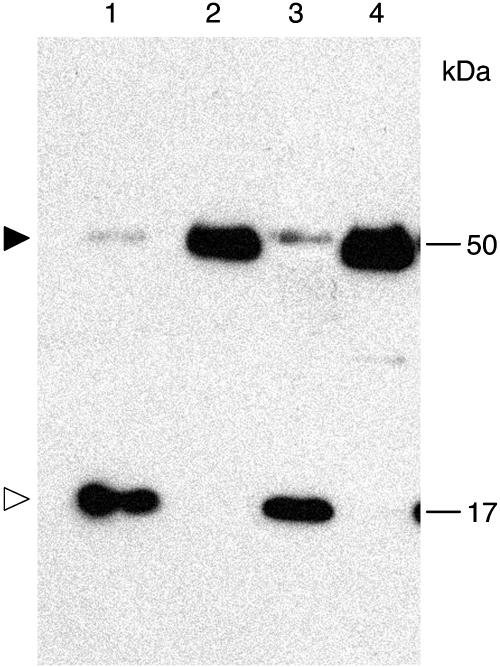
Interaction between calreticulin and TMV MP and effect of calreticulin overexpression on TMV MP subcellular localization. A, Interaction in yeast two-hybrid system. Lane 1, AtCRT1 + TMV MP; lane 2, AtCRT1 + lamin C; lane 3, AtCRT1 lacking signal peptide + TMV MP. B, RT-PCR analysis of ZmCRT1 overexpression in transgenic N. benthamiana. Lane 1, Plants transgenic for ZmCRT1; lane 2, wild-type plants. Top and bottom bands correspond to the ZmCRT1- and actin-specific products. Fragment size in base pairs is indicated on the right. C, Western-blot analysis of ZmCRT1 overexpression in transgenic plants. Lane 1, Plants transgenic for ZmCRT1; lane 2, wild-type plants. Molecular mass expressed in thousands of Daltons is indicated on the right. D, Interaction in renatured-blot overlay assay. Lane 1, Plants transgenic for ZmCRT1 + TMV MP; lane 2, wild-type plants + TMV MP; lane 3, plants transgenic for ZmCRT1 + VirD2. Arrowhead indicates the position of calreticulin. Molecular mass expressed in thousands of Daltons is indicated on the right. E, GFP-TMV MP transiently expressed in wild-type N. benthamiana. F, GFP-TMV MP transiently expressed in transgenic N. benthamiana plants overexpressing ZmCRT1. GFP signal is in green; plastid autofluorescence is in red. Images in E and F are projections of several confocal sections; plastid autofluorescence in E was filtered out. Bars = 10 μm.
Finally, the binding of calreticulin to TMV MP was examined directly using a renatured-blot overlay assay for protein-protein interactions (Dorokhov et al., 1999; Chen et al., 2000). In this approach, calreticulin-containing cell extract is electrophoresed, immobilized on a polyvinylidene difluoride membrane by electroblotting, reacted with purified TMV MP or with an unrelated protein, e.g. VirD2 of Agrobacterium, as a negative control, and binding is detected using anti-TMV MP or anti-VirD2 antibodies. To better approximate the native, processed form of calreticulin, we utilized calreticulin overexpressing transgenic N. benthamiana plants as the source of this protein. From a total of five independently transformed transgenic lines, we present a detailed analysis of one representative line. Figure 4B shows that reverse transcription (RT)-PCR analysis of total RNA obtained from the transgenic plants detected high levels of the calreticulin-specific transcript (lane 1, top band); this product was not detected in wild-type plants (lane 2); this specific detection of the transgenically expressed calreticulin, but not the endogenous calreticulin, was possible because we used maize calreticulin (ZmCRT1, accession no. Z46772.1) as the transgene. Comparable efficiency of RT-PCR in all samples was indicated by similar amounts of actin-specific RT-PCR product in all lanes (Fig. 4, bottom band). Western-blot analysis of the same samples revealed high levels of calreticulin in the overexpressing transgenic plants (indicated by arrowhead in Fig. 4C, lane 1), whereas only low amounts of this protein were detected in the wild-type plants (lane 2). Interestingly, the relative electrophoretic mobility of calreticulin detected in these experiments was approximately 50 kD, corresponding to the processed form of the protein, while its unprocessed form was not detected under our experimental conditions. The overexpressed calreticulin localized to the ER and plasmodesmata in a pattern indistinguishable from that of the endogenous protein (data not shown).
Figure 4D shows that TMV MP specifically interacted with immobilized calreticulin contained within the transgenic cell extracts (lane 1); no interaction was detected using wild-type plant extracts (lane 2), most likely due to low amounts of endogenous calreticulin, which are below the sensitivity of the blot overlay assay. The TMV MP-calreticulin interaction was specific because, when the blot was probed with an unspecific ligand, i.e. purified Agrobacterium VirD2, no binding to calreticulin was observed (Fig. 4D, lane 3). These results strengthened the notion that TMV MP binds calreticulin.
Overexpression of Calreticulin Results in Mislocalization of TMV MP and Impairs TMV Cell-to-Cell Movement
Potentially, high amounts of calreticulin in transgenic plants may interfere with the normal targeting and function of TMV MP. Indeed, TMV MP-GFP transiently expressed in the wild-type N. benthamiana plants accumulated predominantly within the peripheral plasmodesmal puncta (Fig. 4E); furthermore, in these tissues numerous adjacent cells exhibited TMV MP-GFP fluorescence, indicating cell-to-cell movement of the tagged protein. In contrast, transient expression of TMV MP-GFP in transgenic plants overexpressing calreticulin occurred in single cells, showing no intercellular movement, and the tagged protein formed a clear filamentous network, characteristic of the cytoskeleton, together with a vesicular localization, characteristic of the ER, within the expressing cell (Figs. 4F and 5A).
Figure 5.
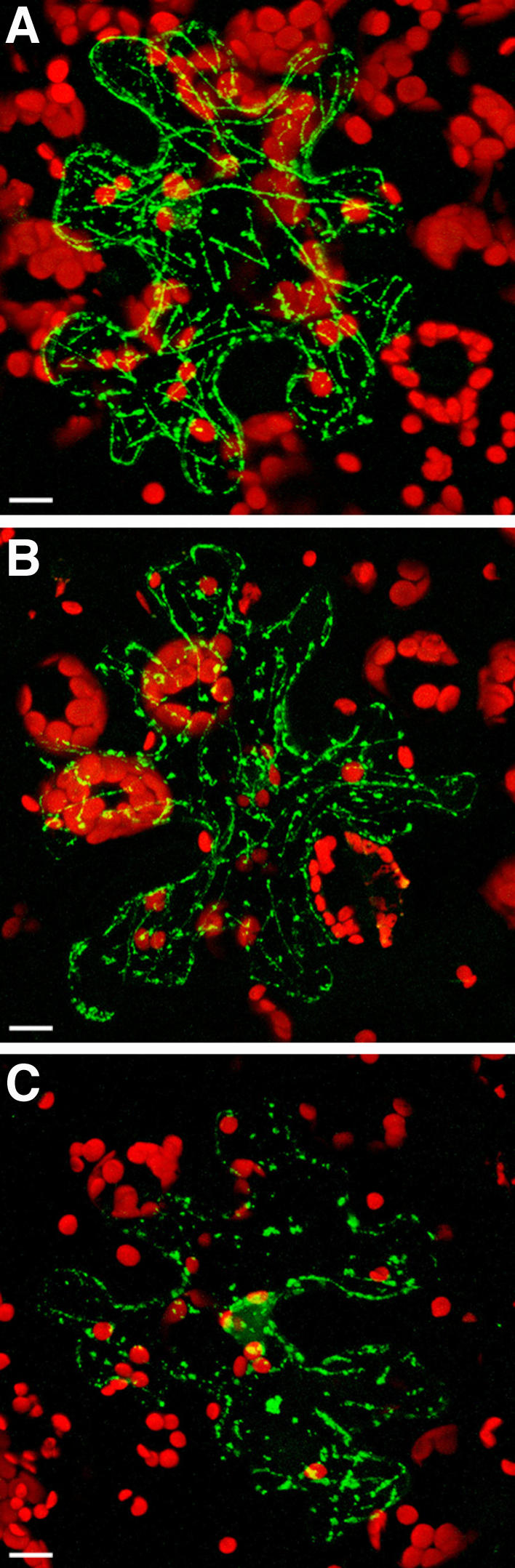
TMV MP associates with MTs in transgenic N. benthamiana plants overexpressing ZmCRT1. A, GFP-TMV MP in mock-treated tissue. B, GFP-TMV MP in tissue treated with 0.2 μM latrunculin A. C, GFP-TMV MP in tissue treated with 5 μm oryzalin. GFP signal is in green; plastid autofluorescence is in red. All images are projections of several confocal sections. Bars = 10 μm.
To distinguish between association of TMV MP-GFP with microfilaments and MTs, we treated the expressing tissues with latrunculin A or oryzalin, drugs that disrupt actin filaments and MTs, respectively (Spector et al., 1983; Morejohn et al., 1987). Figure 5 shows that latrunculin A did not significantly alter the filamentous arrangement of TMV MP-GFP (Fig. 5B), which was comparable to that obtained following mock treatment with water (Fig. 5A), whereas oryzalin disrupted the filaments and left only intracellular bodies (Fig. 5C). Thus, the filamentous arrangement of TMV MP-GFP within plant cells overexpressing calreticulin most likely reflects microtubular association of TMV MP.
Our observations suggest that the presence of large quantities of calreticulin within plant cells most likely redirects TMV MP from plasmodesmata to MTs. How does this effect of overexpressed calreticulin influence viral infection and cell-to-cell movement? To address this question, we used a coat protein (CP) assay in which TMV presence is detected by appearance of the viral CP within the inoculated tissues (Lartey et al., 1997; Ghoshroy et al., 1998; Chen et al., 2000; Waigmann et al., 2000). N. benthamiana leaves were inoculated with TMV, and the appearance of TMV CP due to the cell-to-cell movement of the virus was assayed in leaf areas distal to the inoculation site using western-blot analysis and anti-TMV CP antibodies. The same tissue samples were also analyzed by western blotting for the presence of calreticulin. Figure 6 shows that, only 2 d after inoculation, the wild-type plants accumulated substantial levels of TMV CP (lane 1), which remained fairly constant for another 5 d (lane 3), whereas virtually no TMV CP was detected in transgenic plants overexpressing calreticulin (lanes 2 and 4) even 1 week after inoculation. At later time periods, the inoculated leaves began senescing, preventing reliable detection of TMV CP. Sampling upper, uninoculated leaves demonstrated normal systemic movement of TMV in the wild-type plants, but failed to detect TMV CP in the upper leaves of the calreticulin transgenic plants even 28 d after inoculation (data not shown).
Figure 6.
Delayed cell-to-cell movement of TMV in transgenic N. benthamiana plants overexpressing ZmCRT1. Lane 1, Wild-type plants 2 d postinoculation; lane 2, transgenic plants overexpressing ZmCRT1 2 d postinoculation; lane 3, wild-type plants 7 d postinoculation; lane 4, transgenic plants overexpressing ZmCRT1 7 d postinoculation. Black and white arrowheads indicate positions of calreticulin and TMV CP, respectively. Molecular mass expressed in thousands of Daltons is indicated on the right.
Next, we directly monitored viral cell-to-cell movement using TMV-GFP (Shivprasad et al., 1999). In the wild-type plants, visualization of the inoculated leaves under UV light revealed viral spread to a large number of cells, forming GFP-expressing infection foci of 0.9 ± 0.1 mm 2 d after inoculation. Conversely, the calreticulin overexpressing plants exhibited much smaller infection foci (0.2 ± 0.1 mm), which indicated that, although the virus was able to replicate in the infected cells, its cell-to-cell movement was substantially compromised. Ideally, it would be useful to employ a reciprocal approach and examine the fate of TMV MP and TMV movement in plants that do not express calreticulin, but our experiments using antisense and RNA interference techniques did not yield viable suppressor lines, suggesting that this protein is essential for the survival of the plant.
DISCUSSION
Calreticulin and TMV MP appear to share similar patterns of subcellular localization and transport pathways. Both localize to plasmodesmata (Tomenius et al., 1987; Ding et al., 1992a; Baluska et al., 1999, 2001; Laporte et al., 2003) to which they may be transported via their association with the ER (Ostwald and MacLennan, 1974; Heinlein et al., 1998; Reichel and Beachy, 1999). These common characteristics are consistent with our observations that TMV MP directly recognizes and binds calreticulin from different plant species (i.e. tobacco, Arabidopsis, and maize) in vitro and in vivo. One possibility is that calreticulin, which carries a signal peptide, supplies TMV MP that lacks such a signal sequence (Atkins et al., 1991; Deom et al., 1991) with the ability to enter the ER secretory pathway, which will ultimately deliver both proteins to plasmodesmata. In this regard, it may be significant that calreticulin is not the only ER-associated protein that interacts with TMV MP. Indeed, TMV MP has been shown to interact with PME (Dorokhov et al., 1999; Chen et al., 2000), suggesting that binding to the host cell factors containing signal sequences may represent a general strategy employed by TMV MP to enter the ER secretory pathway; because ER strands span plasmodesmata (Ding et al., 1992b), TMV MP may utilize its association with the ER for transport to plasmodesmata. Obviously, a more active role for calreticulin in viral movement cannot be excluded. For example, TMV MP binding may alter the (as yet unknown) calreticulin activity within plasmodesmata, inducing changes in plasmodesmal permeability.
Interestingly, MP of the grapevine fanleaf virus, a nepovirus that uses a movement mechanism different from that of TMV, failed to coimmunoprecipitate with calreticulin (Laporte et al., 2003); however, these immunoprecipitation studies were performed using microsomal fractions (Laporte et al., 2003), whereas our study focused on the cell wall fractions known to contain plasmodesmata (Ritzenthaler et al., 2000). On the other hand, a potential, albeit indirect, functional link between viral MPs and calreticulin may be inferred from the observations that p8, one of the two MPs of the turnip crinkle virus, interacts with an Arabidopsis protein containing two Arg-Gly-Asp (RGD) cell-attachment sequences (Lin and Heaton, 2001), which are recognized by integrins (Campbell et al., 2000) that, in turn, interact with calreticulin (Dedhar, 1994).
Previous studies indicate that overexpression of one of the interacting proteins can provide valuable insights into the biological effects of such interaction (Reynaud et al., 2000; Bai and Merchant, 2001). So, what happens when the intracellular levels of calreticulin are increased by its overexpression? Our data suggest that an excess of calreticulin severely interferes with plasmodesmal targeting of TMV MP, potentially due to overloading of the TMV MP transport pathway beyond its capacity. Alternatively, accumulation of the overexpressed calreticulin within plasmodesmata may block plasmodesmal sites responsible for accepting TMV MP. In either case, in plant tissues overexpressing calreticulin, the inability of TMV MP to reach its native location results in redirection of this protein to the microtubular network. Supporting this idea, previous studies indicated that TMV MP binding to MTs most likely reflects a movement-unrelated pathway that ultimately leads to TMV MP degradation (Padgett et al., 1996; Reichel and Beachy, 1998; Mas and Beachy, 1999; Gillespie et al., 2002). The effect of calreticulin overexpression on TMV MP localization and TMV spread and the ability of TMV MP to interact with calreticulin provide an indication of the functional relationship between calreticulin, TMV MP, and viral cell-to-cell movement.
MATERIALS AND METHODS
TMV MP Affinity Purification of Calreticulin
TMV MP (100 μg)—produced in Escherichia coli, purified to near homogeneity, and verified by western-blot analysis as described (Citovsky et al., 1990, 1992, 1993)—was immobilized on Affi-Gel 10 (Bio-Rad Laboratories, Hercules, CA), according to the manufacturer's instructions, and packed into a 1-mL column equilibrated in binding buffer (140 mm NaCl, 10 mm Tris-HCl, pH 7.4, 2 mm EDTA, 0.1% Tween 20, 2 mm dithiothreitol [DTT]). Cell wall extract purified from 500 g of leaf tissue of wild-type tobacco plants (Nicotiana tabacum cv Turk), as described (Chen et al., 2000), was concentrated to 5 mL by ultrafiltration, mixed with 1 volume of double-strength denaturation buffer (7 m guanidine hydrochloride [Valeant Pharmaceuticals International, Costa Mesa, CA], 2 mm EDTA, 50 mm DTT, 50 mm Tris-HCl, pH 8.3), and incubated for 2 h at 4°C. After incubation, the protein extract was renatured by overnight dialysis against 2 L of the binding buffer. The resulting sample was loaded on the TMV MP column at a flow rate of 0.5 mL/min. The column was then washed with 50 mL binding buffer and eluted with a continuous gradient of NaCl of 0.1 to 2.5 m in the binding buffer. The eluted fractions corresponding to the protein peak were pooled, dialyzed against buffer H (0.1 m HEPES, pH 7.4, 5 mm DTT, 10 mm EDTA, 1 mm phenylmethane sulfonyl fluoride, 1 mm leupeptin, 1 mm pepstatin), and concentrated to 0.25 mL by ultrafiltration.
Two-Dimensional Gel Electrophoresis and Peptide Sequence Analysis
The concentrated fractions from the TMV MP affinity column were electrophoresed on a 12.5% SDS polyacrylamide gel (Laemmli, 1970); the gel region corresponding to protein electrophoretic mobility of 50 to 60 kD was excised and sent for further custom processing to the Campus Chemical Instrument Center (CCIC) Mass Spectrometry and Proteomics Facility of The Ohio State University (www.ccic.ohio-state.edu/MS). At this facility, the samples were separated by two-dimensional gel electrophoresis, and the most prominent protein spots were excised and digested in gel with sequencing grade trypsin from Promega (Madison, WI) using the Montage in-gel digestion kit from Millipore (Bedford, MA), following the manufacturer's recommended protocols. Briefly, bands were trimmed as close as possible to minimize background polyacrylamide material. The gels were washed in 50% methanol/5% acetic acid for several hours. The gel bands were dried with acetonitrile and reconstituted with DTT solution to reduce the Cys. Iodoacetamide was added to alkylate the Cys and the gel was washed again with cycles of acetonitrile and ammonium bicarbonate. Trypsin was added and digested at room temperature overnight. The peptides were extracted from the polyacrylamide with 50% acetonitrile and 5% formic acid several times and pooled together. The extracted pools were concentrated in a speed vac to approximately 25 μL.
Capillary liquid chromatography (LC)-nanospray tandem mass spectrometry (MS/MS) was performed on a Micromass hybrid quadrupole time-of-flight Q-TOF II (Micromass, Wythenshawe, UK) mass spectrometer equipped with an orthogonal nanospray source from New Objective (Woburn, MA) operated in positive ion mode. The LC system was an UltiMat Plus with a FAMOS autosampler and Switchos column switcher (LC-Packings/Dionex, Sunnyvale, CA). Solvent A was water containing 50 mm acetic acid and solvent B was acetonitrile. Each sample (2.5 μL) was first injected into the trapping column, and then washed with 50 mm acetic acid. The injector port was switched to inject, and the peptides were eluted off the trap onto the column. A 5-cm × 75-μm ID ProteoPep C18 column packed directly in the nanospray tip was used for chromatographic separations. Peptides were eluted directly off the column into the Q-TOF system using a gradient of 2% to 80% B over 30 min, with a flow rate of 40 μL/min with a precolumn split to about 500 nL/min. Total run time was 55 min. The nanospray capillary voltage was set at 3.0 kV and the cone voltage at 55 V. The source temperature was maintained at 100°C. Mass spectra were recorded using MassLynx 4.0 with automatic switching functions. Mass spectra were acquired from mass 400 to 2,000 D/s with a resolution of 8,000 (full-width half-maximum). When the desired peak was detected at a minimum of 15 ion counts, the mass spectrometer automatically switched to acquire the collision-induced dissociation MS/MS spectrum of the individual peptide. Collision energy was set dependent on charge state recognition properties. Sequence information from the MS/MS data was processed using Mascot Distiller. Database searches were performed using Mascot from Matrix Science and PEAKS from Bioinformatics Solutions.
Sequence information from the MS/MS data was processed using Mascot Distiller to form a peak list (.mgf file). Data processing was performed following the guidelines in Molecular & Cellular Proteomics. Briefly, data were minimally processed, a three-point smooth was applied, and the centroid was calculated from the top 80% of the peak height. The charge state of each ion selected for MS/MS was calculated; however, the peaks were not deisotoped. Assigned peaks have a minimum of five counts (S/N of 3) and must show the corresponding C13 ion to be considered valid. The mass accuracy of the precursor ions was set to 1.2 D to accommodate accidental selection of the C13 ion and the fragment mass accuracy was set to 0.3 D. Considered modifications (variable) were Met oxidation and carbamidomethyl Cys.
Expression of Fluorescently Tagged Calreticulin and TMV MP
For transient expression, the coding sequences of TMV MP (derived from pETP30; Citovsky et al., 1990) and the sGFP(S65T) codon-optimized variant of GFP (Chiu et al., 1996; Niwa et al., 1999) were cloned as PCR-amplified SalI-EcoRI and EcoRI-PstI fragments, respectively, into the SalI-PstI sites of a plant expression vector, pCd, under the control of the 35S promoter (Tzfira et al., 2001); this construct generated a translational fusion of TMV MP to the N terminus of GFP (TMV MP-GFP). For a translational fusion of TMV MP to the N terminus of CFP (TMV MP-CFP), the coding sequence of TMV MP was first cloned as a SalI-EcoRI PCR-amplified fragment into the XhoI-EcoRI sites of pSAT1-MCS (Tzfira et al., 2005), and, into the SalI-BamHI sites of the resulting construct, the coding sequence of CFP (derived from pECFP-N1; CLONTECH Laboratories, Mountain View, CA) was then cloned as a SalI-BglII PCR-amplified fragment. The coding sequences of the Arabidopsis (Arabidopsis thaliana) calreticulin CRT1 (AtCRT1, accession no. NM_104513.2) and its mutant lacking the first 50 amino acid residues (AtCRT1-del50N) were PCR amplified and cloned into the BglII-SalI sites of pSAT6-EGFP-N1 (Tzfira et al., 2005); these constructs produced translational fusions of AtCRT1 and AtCRT1-del50N to the N terminus of GFP (AtCRT1-GFP and AtCRT1-del50N-GFP). All PCR reactions were performed as described (Kang et al., 1995), using a high-fidelity Pfu DNA polymerase (Stratagene, La Jolla, CA) and verified by DNA sequencing. The resulting constructs were bombarded into Nicotiana benthamiana leaves at a pressure of 150 psi using a portable Helios gene gun system (model PDS-1000/He; Bio-Rad Laboratories) as described (Tzfira et al., 2001), followed by incubation for 24 to 28 h at 25°C to allow expression of the transfected DNA. For latrunculin A (Sigma, St. Louis) and oryzalin (Sigma) treatments, 0.2 and 5 μm water solutions, respectively, of these drugs were syringe infiltrated into the adaxial surfaces of the bombarded leaves 24 h after bombardment, and their effect on the TMV MP subcellular localization was examined 2 h later; mock treatment by infiltration with water was used as negative control. All fluorescence microscopy, except for FRET assays, was performed using a Zeiss LSM 5 Pascal confocal laser-scanning microscope (Carl Zeiss, Thornwood, NY). Experiments were repeated at least three times.
For expression in transgenic plants, the full-length AtCRT2 gene (AGI code At1g09210) with its native regulatory elements was internally (30 bp upstream of the stop codon) tagged with the citrine variant of YFP (Griesbeck et al., 2001) using the fluorescent tagging of full-length proteins technique (Tian et al., 2004) with P1, P2, P3, and P4 primers: 5′-GCTCGATCCACCTAGGCTTTTGGGAGCTCTGTTCTGATCCT-3′, 5′-CACAGCTCCACCTCCACCTCCAGGCCGGCCGTCTTTCTCAGAGGTTTCCTCGCTAT-3′, 5′-TGCTGGTGCTGCTGCGGCCGCTGGGGCCGCCACCGCTCATGTATGCTA-3′, and 5′-CGTAGCGAGACCACAGGATACAAGAAGTGGGAGGGAAAATA-3′, respectively. Our construct included the entire intergenic region (1,235 bp) upstream of the AtCRT2 translation initiation codon and 974 bp downstream of the stop codon. All PCR conditions were as described (Tian et al., 2004). The resulting AtCRT2-YFP gene was verified by DNA sequencing and transferred into pBIN-GW by Gateway recombination as described (Tian et al., 2004). In addition, we transferred the TMV MP-GFP expression cassette from the pCd vector into the Asp-718-XbaI sites of pBIN19 (accession no. U09365.1). These binary constructs were introduced into Agrobacterium tumefaciens GV3101, used to transform Arabidopsis (Col-0 strain) plants by floral dipping (Kim et al., 2003), and kanamycin-resistant T1 transformants were analyzed by confocal microscopy for calreticulin and TMV MP expression and subcellular localization.
Detection of Protein Interaction by FRET Microscopy
Constructs (1:1 m ratio) expressing AtCRT2-YFP from its full-length gene with native regulatory elements and TMV MP-CFP were bombarded into N. benthamiana leaves, and, after 16 to 24 h of transient expression, FRET measurements using the acceptor photobleaching method (Kenworthy, 2001) were performed essentially as described (Karpova et al., 2003). Cells were examined under a Zeiss LSM 510 Meta laser-scanning confocal microscope with a high numerical aperture (1.2–1.3) water immersion objective (63×). The function of multitrack in the Zeiss AIM software was employed to detect the fluorescence. Track 1 (CFP channel) was configured as CFP (donor) detection only excited by a 458-nm argon laser line at 20% laser power and collected emission fluorescence at 460 to 500 nm. Track 2 (YFP channel) was configured as YFP (acceptor) detection only excited by a 514-nm argon laser line at 10% laser power and collected emission fluorescence at 525 to 600 nm. Track 3 (FRET channel) was configured as FRET detection excited by a 458-nm argon laser line at 20% laser power and collected emission fluorescence at 525 to 600 nm. This multitrack configuration, which minimizes the cross-talk between CFP and YFP, was set based on the results of control experiments for CFP-tagged TMV MP and YFP-tagged calreticulin bombarded alone (Karpova et al., 2003). Cells were bleached in the YFP channel by scanning a region of interest (ROI) 100 times using a 514-nm argon laser line at 100% laser power. The time of bleach ranged from 5 to 15 s, depending on the number and localization of bleach ROIs. Before and after the bleach, CFP images were collected to assess changes in the donor fluorescence. Time lapse recording combining with the bleaching was performed to collect four CFP/YFP image pairs before and after the bleach to ensure that bleaching due to imaging was minimal (Karpova et al., 2003). The intensity of fluorescence over time was automatically measured by using the function of ROI mean of the microscope. Each measurement was conducted on a set of 10 different cells. The percentage of FRET efficiency (EF) was calculated as EF = (In+1 − In) × 100/In+1, where In and In+1 are the CFP intensities at the time points between which the bleach had occurred (Karpova et al., 2003). Because the bleached areas contained an entire punctum with extremely low fluorescence of the immediate surroundings, we did not subtract the background signal to minimize data manipulation. The distance (R) between donor and acceptor was calculated from the EF values and an average Förster radius (R0) of approximately 5.0 nm for the CFP-YFP pair (Tsien, 1998; Gadella et al., 1999) using the equation R = R0 × (1/EF − 1)1/6 (Gadella et al., 1999). Note that the EF and R values, while indicative of the degree of FRET and the distance separating two interacting molecules, respectively, also depend on the microscope optics, the local relative concentrations of the donor and acceptor, and the insertion sites of the CFP and YFP tags within the protein molecules (Gadella et al., 1999).
Detection of Protein Interaction by Yeast Two-Hybrid Assay
TMV MP fusion with LexA in pBTM116 (TRP1+; Hollenberg et al., 1995) has been described (Chen et al., 2000). AtCRT2 and its mutant lacking the sequence coding for the first 50 amino acids were PCR amplified and cloned into the EcoRI-SalI sites of pGAD424 (LEU3+; CLONTECH). Plasmids expressing the potential interactors were introduced into yeast (Saccharomyces cerevisiae) strain L40 (Hollenberg et al., 1995) and grown for 2 d at 30°C on His-, Leu-, and Trp-deficient medium. For negative control, we utilized a fragment of the human lamin C gene (Bartel et al., 1993) in pBTM116.
Calreticulin-Overexpressing Transgenic Plants
The coding sequence of maize calreticulin CRT1 (ZmCRT1, accession no. Z46772.1) was PCR amplified, cloned into the SalI-EcoRI sites of pCd, transferred as a BamHI-XbaI fragment into the BamHI-XbaI sites of pBIN19, and the resulting binary plasmid introduced into A. tumefaciens C1C58, which was then used to transform N. benthamiana plants as described (Horsch et al., 1985). The resulting transgenic plants were selected on a kanamycin-containing medium and maintained for 1 month under sterile conditions on a Murashige and Skoog basal medium (Murashige and Skoog, 1962) with no exogenous growth regulators. Plants were then transferred to soil in a greenhouse, allowed to set seed, and the transgenic progeny were selected by germinating the seeds on Murashige and Skoog agar in the presence of kanamycin and then maintained on soil.
RT-PCR
Total RNA was extracted with TRIzol reagent (Invitrogen, Carlsbad, CA) from 200 mg of leaf tissue derived from the wild-type or calreticulin-overexpressing N. benthamiana plants, treated with RQ1 RNase-free DNase (Promega), reverse-transcribed with Moloney murine leukemia virus reverse transcriptase (New England Biolabs, Beverly, MA), and the resulting cDNAs were PCR amplified and detected by ethidium bromide staining of agarose gels. ZmCRT1-specific forward, 5′-CATCTAGAGCTCGTCGTGCTGCTT-3′, and reverse, 5′-ATGGCGATCCGCAAGGGGTCTTCG-3′, primers generated a 1,300-bp product from the ZmCRT1 transcript. As a control, in the same RT-PCR reactions, we amplified tobacco actin (GenBank accession no. X63603; Thangavelu et al., 1993) using forward and reverse primers, 5′-TCACTGAAGCACCTCTTAACC-3′ and 5′-CAGCTTCCATTCCAATCATTG-3′, respectively, which generated a 500-bp product.
Western-Blot Analysis
Leaf samples (50 mg fresh weight) from healthy or TMV-infected N. benthamiana plants were ground in 200 μL of extraction buffer (75 mm Tris/HCl, pH 6.8, 9 m urea, 4.3% SDS, and 7.5% β-mercaptoethanol) and boiled for 10 min. The extracts were then centrifuged at 10,000g for 10 min and 10 μL of the supernatant was analyzed by SDS-PAGE on 12.5% gels (Laemmli, 1970). Following electrophoresis, the presence of calreticulin or TMV CP was detected by western-blot analysis using antibodies against maize calreticulin (a generous gift from Dr. Wendy F. Boss, North Carolina State University, Raleigh, NC) or anti-TMV CP antibodies, respectively, and the ECL kit (Amersham, Piscataway, NJ).
Detection of Protein Interaction by Renatured-Blot Overlay Assay
Leaf extracts (50 μg protein) were resolved on a 12.5% SDS polyacrylamide gel and electroblotted (135 mA for 1 h at room temperature) onto a 7.5 cm × 15 cm polyvinylidene difluoride Immobilon P membrane in transfer buffer (50 mm Tris-base, 192 mm Gly, 20% methanol, 0.01% SDS). The membrane was then washed for 15 min with gentle shaking in buffer 30 mm Tris-HCl, pH 7.4, 0.05% Tween 20. The washed membrane was denatured for 2 h at room temperature in 50 mL of denaturation buffer, and, after a 5-min wash in Tris-buffered saline (TBS; 140 mm NaCl, 30 mm Tris-HCl, pH 7.4), the blotted proteins were renatured by an overnight incubation at 4°C in 250 mL of renaturation buffer (140 mm NaCl, 10 mm Tris-HCl, pH 7.4, 2 mm EDTA, 1% bovine serum albumin, 0.1% Tween 20, 2 mm DTT) with gentle shaking (10 rpm). Renaturation in the presence of 1% bovine serum albumin also blocked potential nonspecific protein adsorption sites on the membrane. For TMV MP binding, the renatured blot was incubated for 90 min at room temperature with 10 mL of purified 10-μg/mL TMV MP in the renaturation buffer. Following incubation, the membrane was washed four times (5 min at room temperature) with TBS plus 0.1% Tween 20 and incubated for 1 h at room temperature with anti-TMV MP antibody for 1 h. After four washes with TBS plus Tween, TMV MP binding was detected using the ECL kit.
Infection of Plants and Detection of TMV Cell-To-Cell Movement
Infectious cDNA clones of the wild-type TMV (pTMV304; kindly provided by Dr. William O. Dawson, University of Florida, Lake Alfred, FL) or of GFP-expressing TMV (Shivprasad et al., 1999; pTMV30BGFP; a generous gift from Dr. Dennis Lewandowski, University of Florida, Lake Alfred, FL) were linearized with KpnI. Infectious transcripts were produced from the SP6 (pTMV304) or T7 (pTMV30BGFP) promoter and capped with 7 m-diguanosine triphosphate using a RiboMax large-scale RNA production system kit, according to the directions of the manufacturer (Promega). Lower, fully expanded leaves of wild-type and calreticulin transgenic N. benthamiana plants were mechanically inoculated with 10 μL of the resulting 100-μg/mL RNA solution supplemented with 3 mg/mL of celite. Three plants of each tested line were inoculated in each experiment.
To assay for cell-to-cell spread of the virus within the inoculated leaf, the inoculation was performed on one side of the leaf blade. In plants inoculated with in vitro-produced transcripts of the wild-type TMV, 50-mg tissue samples were removed from the leaf areas distal to the inoculation site and at the same distance from it in all sampled plants. The presence of TMV CP in these samples was detected by western-blot analysis. In plants inoculated with in vitro-produced transcripts of TMV-GFP, the leaves were directly observed and photographed under a Leica MZ FLIII epifluorescence stereoscope (Leica Microsystems, Bannockburn, IL).
Acknowledgments
We thank Drs. Bill Dawson, Dennis Lewandowski, and Wendy Boss for their generous gifts of pTMV304, pTMV30BGFP, and anti-maize calreticulin antibodies, respectively.
This work was supported by grants from the National Institutes of Health, the National Science Foundation, the U.S. Department of Agriculture, and the U.S.-Israel Binational Science Foundation to V.C., and from the U.S.-Israel Binational Agricultural Research and Development Fund to V.C. and Y.G.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.064386.
References
- Atkins D, Hull R, Wells B, Roberts K, Moore P, Beachy RN (1991) The tobacco mosaic virus 30 K movement protein in transgenic tobacco plants is localized to plasmodesmata. J Gen Virol 72: 209–211 [DOI] [PubMed] [Google Scholar]
- Bai L, Merchant JL (2001) ZBP-89 promotes growth arrest through stabilization of p53. Mol Cell Biol 21: 4670–4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baluska F, Cvrckova F, Kendrick-Jones J, Volkmann D (2001) Sink plasmodesmata as gateways for phloem unloading. Myosin VIII and calreticulin as molecular determinants of sink strength? Plant Physiol 126: 39–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baluska F, Samaj J, Napier R, Volkmann D (1999) Maize calreticulin localizes preferentially to plasmodesmata in root apex. Plant J 19: 481–488 [DOI] [PubMed] [Google Scholar]
- Bartel P, Chien C, Sternglanz R, Fields S (1993) Elimination of false positives that arise in using the two-hybrid system. Biotechniques 14: 920–924 [PubMed] [Google Scholar]
- Borisjuk N, Sitailo L, Adler K, Malysheva L, Tewes A, Borisjuk L, Manteuffel R (1998) Calreticulin expression in plant cells: developmental regulation, tissue specificity and intracellular distribution. Planta 206: 504–514 [DOI] [PubMed] [Google Scholar]
- Boyko V, Ferralli J, Ashby J, Schellenbaum P, Heinlein M (2000) Function of microtubules in intercellular transport of plant virus RNA. Nat Cell Biol 2: 826–832 [DOI] [PubMed] [Google Scholar]
- Campbell KD, Reed WA, White KL (2000) Ability of integrins to mediate fertilization, intracellular calcium release, and parthenogenetic development in bovine oocytes. Biol Reprod 62: 1702–1709 [DOI] [PubMed] [Google Scholar]
- Chen MH, Sheng J, Hind G, Handa A, Citovsky V (2000) Interaction between the tobacco mosaic virus movement protein and host cell pectin methylesterases is required for viral cell-to-cell movement. EMBO J 19: 913–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu W, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J (1996) Engineered GFP as a vital reporter in plants. Curr Biol 6: 325–330 [DOI] [PubMed] [Google Scholar]
- Citovsky V, Knorr D, Schuster G, Zambryski PC (1990) The P30 movement protein of tobacco mosaic virus is a single-strand nucleic acid binding protein. Cell 60: 637–647 [DOI] [PubMed] [Google Scholar]
- Citovsky V, McLean BG, Zupan J, Zambryski PC (1993) Phosphorylation of tobacco mosaic virus cell-to-cell movement protein by a developmentally regulated plant cell wall-associated protein kinase. Genes Dev 7: 904–910 [DOI] [PubMed] [Google Scholar]
- Citovsky V, Wong ML, Shaw A, Prasad BVV, Zambryski PC (1992) Visualization and characterization of tobacco mosaic virus movement protein binding to single-stranded nucleic acids. Plant Cell 4: 397–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedhar S (1994) Novel functions for calreticulin: interaction with integrins and modulation of gene expression? Trends Biochem Sci 19: 269–271 [DOI] [PubMed] [Google Scholar]
- Deom CM, Shaw MJ, Beachy RN (1987) The 30-kilodalton gene product of tobacco mosaic virus potentiates virus movement. Science 237: 389–394 [DOI] [PubMed] [Google Scholar]
- Deom CM, Wolf S, Holt CA, Lucas WJ, Beachy RN (1991) Altered function of the tobacco mosaic virus movement protein in a hypersensitive host. Virology 180: 251–256 [DOI] [PubMed] [Google Scholar]
- Ding B, Haudenshield JS, Hull RJ, Wolf S, Beachy RN, Lucas WJ (1992. a) Secondary plasmodesmata are specific sites of localization of the tobacco mosaic virus movement protein in transgenic tobacco plants. Plant Cell 4: 915–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B, Turgeon R, Parthasarathy MV (1992. b) Substructure of freeze-substituted plasmodesmata. Protoplasma 169: 28–41 [Google Scholar]
- Dorokhov YL, Makinen K, Frolova OY, Merits A, Saarinen J, Kalkkinen N, Atabekov JG, Saarma M (1999) A novel function for a ubiquitous plant enzyme pectin methylesterase: the host-cell receptor for the tobacco mosaic virus movement protein. FEBS Lett 461: 223–228 [DOI] [PubMed] [Google Scholar]
- Fields S, Song O (1989) A novel genetic system to detect protein-protein interactions. Nature 340: 245–246 [DOI] [PubMed] [Google Scholar]
- Gadella TW Jr, van der Krogt GN, Bisseling T (1999) GFP-based FRET microscopy in living plant cells. Trends Plant Sci 4: 287–291 [DOI] [PubMed] [Google Scholar]
- Ghoshroy S, Freedman K, Lartey R, Citovsky V (1998) Inhibition of plant viral systemic infection by non-toxic concentrations of cadmium. Plant J 13: 591–602 [DOI] [PubMed] [Google Scholar]
- Gillespie T, Boevink P, Haupt S, Roberts AG, Toth R, Valentine T, Chapman S, Oparka KJ (2002) Movement protein reveals that microtubules are dispensable for the cell-to-cell movement of Tobacco mosaic virus. Plant Cell 14: 1207–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griesbeck O, Baird GS, Campbell RE, Zacharias DA, Tsien RY (2001) Reducing the environmental sensitivity of yellow fluorescent protein. Mechanism and applications. J Biol Chem 276: 29188–29194 [DOI] [PubMed] [Google Scholar]
- Heinlein M (2002) The spread of Tobacco mosaic virus infection: insights into the cellular mechanism of RNA transport. Cell Mol Life Sci 59: 58–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinlein M, Epel BL, Padgett HS, Beachy RN (1995) Interaction of tobamovirus movement proteins with the plant cytoskeleton. Science 270: 1983–1985 [DOI] [PubMed] [Google Scholar]
- Heinlein M, Padgett HS, Gens JS, Pickard BG, Casper SJ, Epel BL, Beachy RN (1998) Changing patterns of localization of the tobacco mosaic virus movement protein and replicase to the endoplasmic reticulum and microtubules during infection. Plant Cell 10: 1107–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hink MA, Bisselin T, Visser AJ (2002) Imaging protein-protein interactions in living cells. Plant Mol Biol 50: 871–883 [DOI] [PubMed] [Google Scholar]
- Hollenberg SM, Sternglanz R, Cheng PF, Weintraub H (1995) Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol Cell Biol 15: 3813–3822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsch RB, Fry JE, Hoffman NL, Eichholtz D, Rogers SG, Fraley RT (1985) A simple and general method for transferring genes into plants. Science 227: 1229–1231 [DOI] [PubMed] [Google Scholar]
- Jares-Erijman EA, Jovin TM (2003) FRET imaging. Nat Biotechnol 21: 1387–1395 [DOI] [PubMed] [Google Scholar]
- Kang J, Kuhn JE, Schafer P, Immelmann A, Henco K (1995) Quantification of DNA and RNA by PCR. In MJ McPherson, BD Hames, GR Taylor, eds, PCR 2, A Practical Approach. IRL Press, Oxford
- Karpova TS, Baumann CT, He L, Wu X, Grammer A, Lipsky P, Hager GL, McNally JG (2003) Fluorescence resonance energy transfer from cyan to yellow fluorescent protein detected by acceptor photobleaching using confocal microscopy and a single laser. J Microsc 209: 56–70 [DOI] [PubMed] [Google Scholar]
- Kenworthy AK (2001) Imaging protein-protein interactions using fluorescence resonance energy transfer microscopy. Methods 24: 289–296 [DOI] [PubMed] [Google Scholar]
- Kim JY, Yuan Z, Jackson D (2003) Developmental regulation and significance of KNOX protein trafficking in Arabidopsis. Development 130: 4351–4362 [DOI] [PubMed] [Google Scholar]
- Krause KH, Michalak M (1997) Calreticulin. Cell 88: 439–443 [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF (1983) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157: 105–132 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 277: 680–685 [DOI] [PubMed] [Google Scholar]
- Laporte C, Vetter G, Loudes AM, Robinson DG, Hillmer S, Stussi-Garaud C, Ritzenthaler C (2003) Involvement of the secretory pathway and the cytoskeleton in intracellular targeting and tubule assembly of Grapevine fanleaf virus movement protein in tobacco BY-2 cells. Plant Cell 15: 2058–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartey R, Ghoshroy S, Ho J, Citovsky V (1997) Movement and subcellular localization of a tobamovirus in Arabidopsis. Plant J 12: 537–545 [DOI] [PubMed] [Google Scholar]
- Lin B, Heaton LA (2001) An Arabidopsis thaliana protein interacts with a movement protein of Turnip crinkle virus in yeast cells and in vitro. J Gen Virol 82: 1245–1251 [DOI] [PubMed] [Google Scholar]
- Mas P, Beachy RN (1999) Replication of tobacco mosaic virus on endoplasmic reticulum and role of the cytoskeleton and virus movement protein in intracellular distribution of viral RNA. J Cell Biol 147: 945–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean BG, Zupan J, Zambryski PC (1995) Tobacco mosaic virus movement protein associates with the cytoskeleton in tobacco cells. Plant Cell 7: 2101–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerowitz EM, Somerville CR , eds (1994) Arabidopsis, Vol Monograph 27. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Michalak M, Mariani P, Opas M (1998) Calreticulin, a multifunctional Ca2+ binding chaperone of the endoplasmic reticulum. Biochem Cell Biol 76: 779–785 [DOI] [PubMed] [Google Scholar]
- Morejohn LC, Bureau TE, Molè-Bajer J, Bajer AS, Fosket DE (1987) Oryzalin, a dinitroaniline herbicide, binds to plant tubulin and inhibits microtubule polymerization in vitro. Planta 172: 252–264 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nelson DE, Glaunsinger B, Bohnert HJ (1997) Abundant accumulation of the calcium-binding molecular chaperone calreticulin in specific floral tissues of Arabidopsis thaliana. Plant Physiol 114: 29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa Y, Hirano T, Yoshimoto K, Shimizu M, Kobayashi H (1999) Non-invasive quantitative detection and applications of non-toxic, S65T-type green fluorescent protein in living plants. Plant J 18: 455–463 [DOI] [PubMed] [Google Scholar]
- Oparka KJ, Prior DAM, Santa-Cruz S, Padgett HS, Beachy RN (1997) Gating of epidermal plasmodesmata is restricted to the leading edge of expanding infection sites of tobacco mosaic virus (TMV). Plant J 12: 781–789 [DOI] [PubMed] [Google Scholar]
- Ostwald TJ, MacLennan DH (1974) Isolation of a high affinity calcium-binding protein from sarcoplasmic reticulum. J Biol Chem 249: 974–979 [PubMed] [Google Scholar]
- Padgett HS, Epel BL, Kahn TW, Heinlein M, Watanabe Y, Beachy RN (1996) Distribution of tobamovirus movement protein in infected cells and implications for cell-to-cell spread of infection. Plant J 10: 1079–1088 [DOI] [PubMed] [Google Scholar]
- Reichel C, Beachy RN (1998) Tobacco mosaic virus infection induces severe morphological changes of the endoplasmic reticulum. Proc Natl Acad Sci USA 95: 11169–11174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichel C, Beachy RN (1999) The role of the ER and cytoskeleton in plant viral trafficking. Trends Plant Sci 4: 458–462 [DOI] [PubMed] [Google Scholar]
- Reynaud EG, Leibovitch MP, Tintignac LA, Pelpel K, Guillier M, Leibovitch SA (2000) Stabilization of MyoD by direct binding to p57Kip2. J Biol Chem 275: 18767–18776 [DOI] [PubMed] [Google Scholar]
- Ritzenthaler C, Findlay K, Roberts K, Maule AJ (2000) Rapid detection of plasmodesmata in purified cell walls. Protoplasma 211: 165–171 [Google Scholar]
- Shivprasad S, Pogue GP, Lewandowski DJ, Hidalgo J, Donson J, Grill LK, Dawson WO (1999) Heterologous sequences greatly affect foreign gene expression in tobacco mosaic virus-based vectors. Virology 255: 312–323 [DOI] [PubMed] [Google Scholar]
- Simon AE (1994) Interactions between Arabidopsis thaliana and viruses. In EM Meyerowitz, CR Somerville, eds, Arabidopsis, Vol Monograph 27. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 685–704
- Spector I, Shochet NR, Kashman Y, Groweiss A (1983) Latrunculins: novel marine toxins that disrupt microfilament organization in cultured cells. Science 219: 493–495 [DOI] [PubMed] [Google Scholar]
- Thangavelu M, Belostotsky D, Bevan MW, Flavell RB, Rogers HJ, Lonsdale DM (1993) Partial characterization of the Nicotiana tabacum actin gene family: evidence for pollen-specific expression of one of the gene family members. Mol Gen Genet 240: 290–295 [DOI] [PubMed] [Google Scholar]
- Tian GW, Mohanty A, Chary SN, Li S, Paap B, Drakakaki G, Kopec CD, Li J, Ehrhardt D, Jackson D, et al (2004) High-throughput fluorescent tagging of full-length Arabidopsis gene products in planta. Plant Physiol 135: 25–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomenius K, Clapham D, Meshi T (1987) Localization by immunogold cytochemistry of the virus coded 30 K protein in plasmodesmata of leaves infected with tobacco mosaic virus. Virology 160: 363–371 [DOI] [PubMed] [Google Scholar]
- Trutnyeva K, Bachmaier R, Waigmann E (2005) Mimicking carboxyterminal phosphorylation differentially affects subcellular distribution and cell-to-cell movement of Tobacco mosaic virus movement protein. Virology 332: 563–577 [DOI] [PubMed] [Google Scholar]
- Tsien RY (1998) The green fluorescent protein. Annu Rev Biochem 67: 509–544 [DOI] [PubMed] [Google Scholar]
- Tzfira T, Tian GW, Lacroix B, Vyas S, Li J, Leitner-Dagan Y, Krichevsky A, Taylor T, Vainstein A, Citovsky V (2005) pSAT vectors: a modular series of plasmids for fluorescent protein tagging and expression of multiple genes in plants. Plant Mol Biol 57: 503–516 [DOI] [PubMed] [Google Scholar]
- Tzfira T, Vaidya M, Citovsky V (2001) VIP1, an Arabidopsis protein that interacts with Agrobacterium VirE2, is involved in VirE2 nuclear import and Agrobacterium infectivity. EMBO J 20: 3596–3607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waigmann E, Chen MH, Bachmaier R, Ghoshroy S, Citovsky V (2000) Phosphorylation of tobacco mosaic virus cell-to-cell movement protein regulates viral movement in a host-specific fashion. EMBO J 19: 4875–4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waigmann E, Lucas W, Citovsky V, Zambryski PC (1994) Direct functional assay for tobacco mosaic virus cell-to-cell movement protein and identification of a domain involved in increasing plasmodesmal permeability. Proc Natl Acad Sci USA 91: 1433–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waigmann E, Ueki S, Trutnyeva K, Citovsky V (2004) The ins and outs of non-destructive cell-to-cell and systemic movement of plant viruses. Crit Rev Plant Sci 23: 195–250 [Google Scholar]
- Wolf S, Deom CM, Beachy RN, Lucas WJ (1989) Movement protein of tobacco mosaic virus modifies plasmodesmatal size exclusion limit. Science 246: 377–379 [DOI] [PubMed] [Google Scholar]
- Yoshioka K, Matsushita Y, Kasahara M, Konagaya KI, Nyunoya H (2004) Interaction of Tomato mosaic virus movement protein with tobacco RIO kinase. Mol Cells 17: 223–229 [PubMed] [Google Scholar]