Abstract
RNA silencing with inverted repeat (IR) constructs has been used to suppress gene expression in various organisms. However, the transitive RNA-silencing effect described in plants may preclude the use of RNA silencing for a gene family. Here, we show that, in rice (Oryza sativa), transitive RNA silencing (spreading of double-stranded RNA along the target mRNA) occurred with the green fluorescent protein transgene but not with the endogenous phytoene desaturase gene. We fused IR copies of unique 3′ untranslated regions derived from the rice OsRac gene family to a strong promoter and stably introduced them into rice. Each of the seven members of the OsRac gene family was specifically suppressed by its respective IR construct. We also examined IR constructs in which multiple 3′ untranslated regions were fused and showed that three members of the OsRac gene family were effectively suppressed by a single construct. Using highly conserved regions of the two members of the OsRac gene family, we also suppressed the expression of all members of the gene family with variable efficiencies. These results suggest that RNA silencing is a useful method for the functional analysis of gene families in rice and other plants.
RNA silencing is a form of gene suppression that occurs at the level of RNA and includes posttranscriptional gene silencing in plants and fungi and RNA interference (RNAi) in Caenorhabditis elegans, Drosophila, and animals (Matzke et al., 2001; Waterhouse et al., 2001; Hannon, 2002; Plasterk, 2002; Baulcombe, 2004). Two types of RNA, double-stranded RNA (dsRNA) and short interfering RNA (siRNA), play key roles in this phenomenon (Fire et al., 1998; Waterhouse et al., 1998; Hamilton and Baulcombe, 1999). Extensive genetic and biochemical studies in various species have yielded a model of RNA silencing in which trigger dsRNA, either introduced into the cell or transcribed from transgenes, is cleaved into small siRNAs of 21 to 26 nucleotides (nt) by an RNase termed Dicer (Bernstein et al., 2001). The siRNAs are incorporated into an RNA-induced silencing complex (RISC) to be associated with the target mRNAs (Hammond et al., 2000), and the activated RISC functions to degrade the target mRNAs and suppress gene expression at various levels (Matzke et al., 2001; Waterhouse et al., 2001; Hannon, 2002; Plasterk, 2002; Meister and Tuschl, 2004).
RNA silencing has been successfully used in plants to suppress specific gene functions (Waterhouse et al., 1998; Chuang and Meyerowitz, 2000; Azevedo et al., 2002; Hayama et al., 2003; Moritoh et al., 2005). One aspect of RNA silencing that has not been well understood is the transitive RNA silencing observed in C. elegans and plants. In C. elegans, a dsRNA trigger can produce new siRNAs 5′ to the region that is initially targeted (Nishikura, 2001; Sijen et al., 2001; Alder et al., 2003). These newly synthesized siRNAs can target other RNAs on the basis of sequence similarity. In plants, it has been shown that siRNA spreads not only in the 3′ to 5′ direction but also in the 5′ to 3′ direction when the green fluorescent protein (gfp) or β-glucuronidase (gus) transgenes are targets of RNA silencing (Braunstein et al., 2002; Klahre et al., 2002; Vaistij et al., 2002; Himber et al., 2003; Van Houdt et al., 2003; Garcia-Perez et al., 2004). However, other reports have shown a lack of such transitive RNA silencing for transgenes. In transgenic tobacco expressing gus-viral satellite DNA, infection with the virus causes the production of siRNA corresponding to the satellite DNA but not to gus located 5′ to the satellite DNA (Wang et al., 2001). A report on viroid-induced RNA silencing in transgenic tobacco (Nicotiana benthamiana) plants also showed that transitive RNA silencing does not occur in these plants (Vogt et al., 2004). This report showed that, when plants stably expressing viroid cDNA fused with the gfp gene at the 3′ end and the free gfp gene was infected by the viroid, no transitive silencing of the gfp RNA was detected. However, when the plants became resistant to the viroid, the siRNA for viroid cDNA was detected. With regard to endogenous genes, there has been only one report on transitive RNA silencing. Vaistij et al. (2002) clearly showed that, in transgenic tobacco, no transitive RNA silencing of the endogenous Rubisco and PDS genes occurred, whereas silencing of the integrated 35S-gfp gene was associated with both 5′ and 3′ spreading of siRNA. A study of β-1,3-glucanase gene silencing in tobacco indicated that protoplasts exhibiting RNA silencing of the β-1,3-glucanase transgene contained siRNA for the endogenous β-1,3-glucanase gene (Sanders et al., 2002). These results were interpreted by the authors to suggest the occurrence of transitive RNA silencing of endogenous β-1,3-glucanase. Therefore, although these studies indicate that transitive RNA silencing occurs in many cases of transgene silencing, there are exceptions. Most importantly, whether transitive RNA silencing occurs in plant endogenous genes remains unclear.
It is assumed that transitive RNA silencing is caused by RNA-dependent RNA polymerase (RdRP; Tang et al., 2003). Studies with mutants of RdRP showed that it is required for RNA silencing in Neurospora (Cogoni and Macino, 1999), Arabidopsis (Arabidopsis thaliana; Dalmay et al., 2000; Mourrain et al., 2000), C. elegans (Smardon et al., 2000; Sijen et al., 2001), and Dictyostelium (Martens et al., 2002). However, RNA silencing induced by inverted repeat (IR) transgenes does not require RdRP (Beclin et al., 2002). One important consequence of transitive RNA silencing is that any RNA that possesses sequence similarity with the original trigger dsRNA may be silenced. Another is that secondary siRNA makes silencing nonautonomous (Himber et al., 2003). These facts may limit the use of RNA silencing in plants because plants often contain gene families with a high sequence similarity and multiple members of a family may be silenced together by a single construct against an individual gene.
In this study, we examined whether transitive RNA silencing machinery is conserved in a monocot, rice (Oryza sativa). Our results showed that transitive RNA silencing of the gfp transgene, but not of endogenous genes, occurs in both 5′ and 3′ orientations in rice. We also examined whether RNA silencing can be used to suppress the expression of individual members of a gene family in rice that have high sequence similarity with one another using a diverged 3′ untranslated region (UTR) as a trigger dsRNA. We also tested whether multiple genes of a gene family can be simultaneously silenced by a single IR construct. We further evaluated whether a single IR construct with high sequence similarity in the conserved region can suppress the expression of other members of the gene family. Our results strongly suggest that the RNA silencing machinery is highly conserved in rice and that dsRNA-mediated RNA silencing is a useful tool for the functional analysis of highly conserved multigene families in rice and plants.
RESULTS
Transitive RNA Silencing in Rice
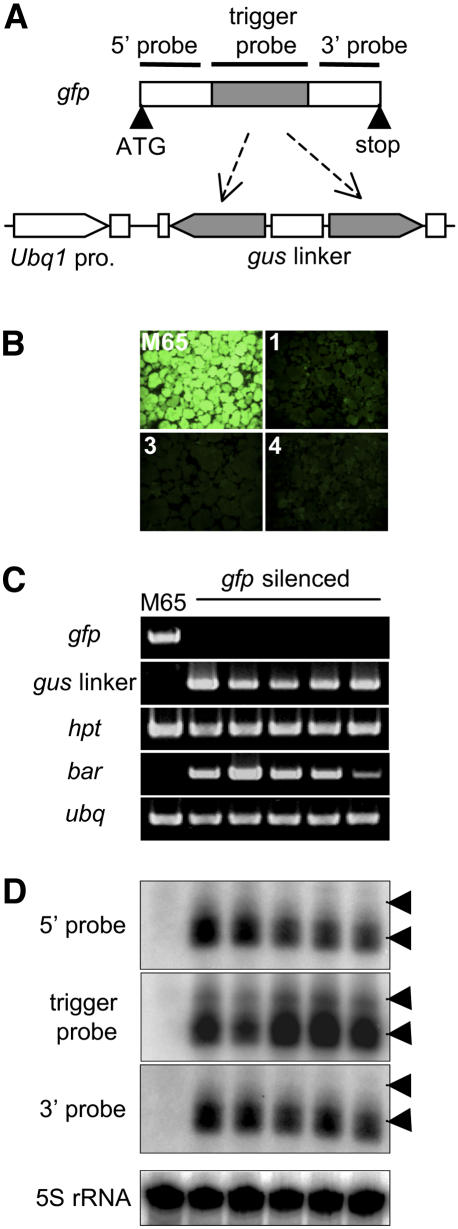
To test whether transitive RNA silencing (spreading of dsRNA along the target mRNA) occurs in rice, we performed an experiment in which the gfp gene was used as the target of RNA silencing. We designed an IR construct that would be transcribed into dsRNA consisting of an internal 314-bp region of the gfp gene (Fig. 1A). As a linker, we inserted a 920-bp fragment of the gus gene to facilitate the quantification of the trigger RNA (Miki and Shimamoto, 2004). We used the maize (Zea mays) ubiquitin (Ubq1) promoter and an intron (Christensen et al., 1992) to produce a high expression of the trigger dsRNA in transgenic rice. We introduced the IR construct into rice calli derived from transgenic rice seeds, M65, which carry the 35S-gfp gene (Fig. 1B). Transgenic rice calli containing both the 35S-gfp and gfp-IR genes were examined for GFP fluorescence and mRNA expression. No fluorescence was detected in three independent transformed transgenic calli (Fig. 1B), and gfp mRNA was clearly suppressed in the five independent transgenic lines examined (Fig. 1C). RNA corresponding to the gus linker region was also detected, indicating that the IR construct was transcribed (Fig. 1C).
Figure 1.
RNA silencing of the 35S-gfp transgene. A, Schematic representation of the gfp gene and IR construct. The central 314 bp (shaded region, nt 201–514; also used as trigger probe) of the 720-bp gfp gene was used as an RNA silencing trigger. The black horizontal lines indicate the regions of the gfp gene used as probes for RNA blotting. The 5′ probe, 196 bp (nt 1–196); the 3′ probe, 200 bp (nt 521–720). Diagram of the gfp IR construct is shown. B, GFP fluorescence in rice calli. GFP fluorescence was observed in the original M65 line and in three independent transgenic lines (1, 3, and 4) carrying the gfp-targeted RNA silencing trigger transgene. C, RT-PCR analysis of gfp mRNA expression in the original M65 line and in five independent transgenic lines carrying the gfp-targeted RNA silencing trigger transgene. gus linker, mRNA derived from the gus linker region of the gfp-IR transgene; hpt, mRNA of the hygromycin resistance gene; bar, the bar gene used for the transformation of the gfp-IR transgene; ubq, mRNA of rice ubiquitin used as a control. D, siRNA analysis of gfp-silenced rice plants. Probes used are shown to the left of each section. Arrowheads indicate the positions of 23- (bottom) and 27-nt (top) DNA oligomers, which are likely to correspond to 21-nt- and 24-nt-size-class siRNA detected in other plants, respectively. The bottom section shows 5S rRNA as a loading control.
siRNA, a molecular marker for dsRNA-based gene silencing, was detected. Two size classes of siRNA have been reported in plants showing RNA silencing (Hamilton et al., 2002; Papp et al., 2003; Miki and Shimamoto, 2004). Therefore, we examined the transformed rice plants for siRNA by purifying their low-molecular-weight RNAs. For siRNA analysis, three probes were used: the region 5′ to the trigger dsRNA, the region used to produce the trigger dsRNA, and the region 3′ to the trigger dsRNA. Interestingly, we detected not only siRNA corresponding to the trigger RNA region but also siRNAs corresponding to regions both 5′ and 3′ to the trigger (Fig. 1D). These secondary siRNAs belong exclusively to the 21-nt siRNA class, while the short and long classes of siRNA (the 21 nt and 24 nt) were detected for the trigger RNA region (Fig. 1D). These results indicate that transitive RNA silencing also occurs for the gfp gene and that spreading of dsRNA along the gfp mRNA occurs in both the 5′ and 3′ orientations in a monocot, rice.
We used DNA oligomers as size markers to detect siRNA signals. Since it has been reported that DNA oligonucleotides migrate approximately 10% faster than RNA markers of equal length (Sambrook et al., 1989; Hamilton et al., 2002), the two classes of siRNA detected in these experiments were considered to be the 21-nt- and 24-nt-size-class siRNAs detected in other plants.
RNA Silencing of an Endogenous Gene, PDS
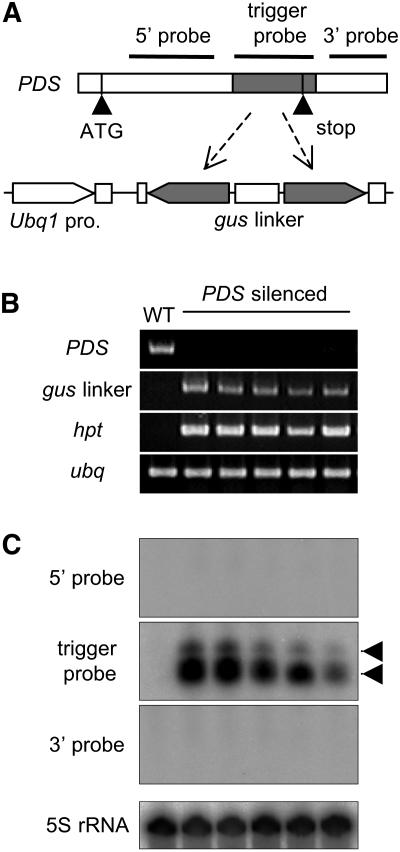
PDS encodes phytoene desaturase and is a single-copy gene in the rice genome. PDS has been used to examine virus-induced gene silencing in both dicots and monocots because of its albino mutant phenotype caused by a lack of carotenoids (Holzberg et al., 2002; Liu et al., 2002). We used the internal 470-bp fragment consisting of the coding and 3′ UTRs to transcribe the trigger dsRNA and tested whether the spreading of siRNA occurred in the 5′ or 3′ direction (Fig. 2A). Rice plants transformed with the PDS IR construct were albino (Miki and Shimamoto, 2004), and PDS mRNA was strongly silenced (Fig. 2B).
Figure 2.
RNA silencing of the endogenous PDS gene. A, Schematic representation of the PDS gene and IR construct. The central 470 bp (shaded region, nt 1,260–1,730; also used as trigger probe) of the 2,027-bp PDS cDNA was used as an RNA silencing trigger. The black horizontal lines indicate the regions of the PDS gene used as probes for RNA blotting. The 5′ probe, 475 bp (nt 692–1,166); the 3′ probe, 203 bp (nt 1,768–1,970). A diagram of the PDS IR construct is shown. B, RT-PCR analysis of PDS mRNA expression. Five independent transgenic rice plants were examined by RT-PCR using PDS-specific primers (top). The wild type (WT) is a nontransgenic control. gus linker, mRNA derived from the gus linker region of the PDS-IR transgene; hpt, mRNA of the hygromycin resistance gene; ubq, mRNA of rice ubiquitin used as control. C, Analysis of siRNAs. Probes used are shown to the left of each section. Arrowheads indicate the positions of 23-nt (bottom) and 27-nt (top) DNA oligomers, which are likely to correspond to 21-nt- and 24-nt-size-class siRNA detected in other plants, respectively. The bottom section shows 5S rRNA as a loading control.
For siRNA analysis, three probes were also used: the region 5′ to the trigger dsRNA, the region used to produce the trigger dsRNA, and the region 3′ to the trigger dsRNA. Two size classes of PDS siRNA were detected when the region for the trigger dsRNA was used as a probe (Fig. 2C). However, no signals were detected when either the region 5′ to the trigger dsRNA or the region 3′ to the trigger dsRNA was used as a probe. These results are in sharp contrast to those obtained from the 35S-gfp transgenic rice plants carrying the gfp-IR gene (Fig. 1). These results indicate that no transitive RNA silencing occurs for the endogenous PDS gene and, thus, that spreading of dsRNA does not occur along the PDS mRNA in either the 5′ or the 3′ direction.
Gene-Specific Suppression of the OsRac Gene Family by RNA Silencing
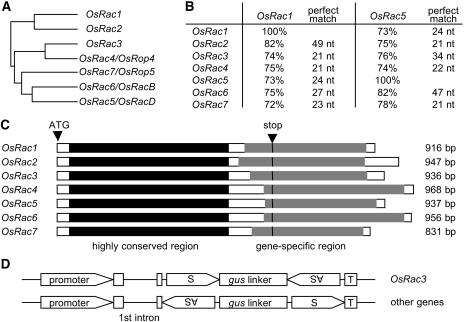
To examine the effectiveness of RNA silencing for the gene-specific suppression of members of a gene family, we chose the OsRac gene family (Fig. 3; Supplemental Fig. 1; Kawasaki et al., 1999; Ono et al., 2001). This family encodes a Rac/Rop-type GTPase, which has multiple functions in various cellular activities, including defense, cell polarity, development, and hormone signaling (Gu et al., 2004). We have identified seven members of the OsRac gene family on the basis of the recently sequenced rice genome and named them OsRac1 through OsRac7 (Fig. 3). Because Christensen et al. (2003) recently named OsRac4 through OsRac7 differently, we show both nomenclatures in Figure 3A. The coding regions of OsRac genes are highly conserved: nucleotide and amino acid sequence identities among the members are 72% to 82% and 78% to 95%, respectively, and there exist contiguous stretches of nucleotides in which the 21 to 49 bases are identical between OsRac1 and the other members of the gene family (Fig. 3B; Supplemental Fig. 2A). However, the 3′ ends of the coding region and the 3′ UTRs are highly divergent; here, the nucleotide sequence identities between members are 44% to 52%, and contiguous stretches of identical nucleotides are only 4 to 8 bases between OsRac1 and the other members of the gene family (Supplemental Fig. 2B).
Figure 3.
Structures of the rice OsRac gene family and constructs for RNA silencing. A, Phylogenetic tree of the OsRac gene family. The dendrogram was generated from the coding region of the nucleotide sequences of OsRac1 to 7 using ClustalW. B, Nucleotide identities between the highly conserved coding sequence of OsRac1 or OsRac5 and those of the other OsRac genes. C, Diagrams of the OsRac genes and regions used to make constructs for RNA silencing. The majority of the coding regions of the OsRac1 to 7 genes are highly conserved, but the 3′ ends of the coding sequences and the 3′ UTRs are highly divergent. The 3′ coding regions and the 3′ UTR gene-specific regions (shaded boxes) were used to generate IR constructs for gene-specific RNA silencing (OsRac1, 309 bp; OsRac2, 328 bp; OsRac3, 316 bp; OsRac4, 332 bp; OsRac5, 246 bp; OsRac6, 367 bp; OsRac7, 221 bp). The conserved regions of OsRac1 and OsRac5 were used to make IR constructs to suppress the other OsRac genes (518 bp, black boxes). The length of cDNA for each of the OsRac genes is shown to the right. D, OsRac gene-targeting RNA silencing IR constructs. These OsRac sequences were placed in the sense (S)-gus linker-antisense (AS) orientation (for the gene-specific OsRac3 construct) or the AS-gus linker-S orientation (for the other constructs). These IR constructs were driven by the maize Ubq1 promoter.
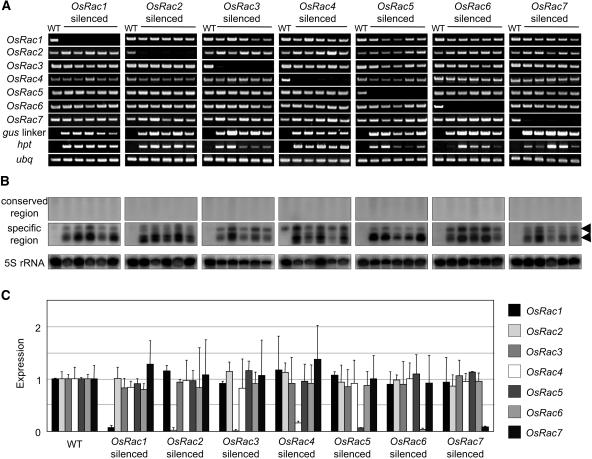
We designed IR constructs transcribing dsRNA consisting of a 220- to 370-bp fragment from the gene-specific region of each OsRac gene (Fig. 3, C and D) and transformed them into rice. Reverse transcription (RT)-PCR analyses of OsRac mRNAs in transgenic rice plants and transgenic suspension cultures showed that each member of the OsRac gene family was suppressed by its corresponding construct (Fig. 4A). Results of western-blot analysis showed that the OsRac1 protein levels in OsRac1-silenced lines were also very low (data not shown). We analyzed a total of 57 independent transgenic plants and 39 suspension cell cultures and observed efficient suppression of OsRac mRNA in all of them.
Figure 4.
Gene-specific RNA silencing of the OsRac gene family in transgenic rice. A, RT-PCR analysis of OsRac1 to 7 mRNA expression. The wild type (WT) is a nontransgenic control. gus linker, mRNA derived from the gus linker region; hpt, mRNA of the hygromycin resistance gene; ubq, mRNA of rice ubiquitin used as control. B, Analysis of siRNAs. Probes used for hybridization are as follows: top, the highly conserved region (Fig. 3C, black boxes); middle, the gene-specific region (Fig. 3C, shaded boxes); bottom, 5S rRNA. Arrowheads indicate the positions of 23-nt (bottom) and 27-nt (top) DNA oligomers, which are likely to correspond to 21-nt- and 24-nt-size-class siRNA detected in other plants, respectively. C, Quantitative analysis on mRNA levels of all the OsRac genes measured by real-time PCR. All of the OsRac1 to 7 values are normalized by the ubq (ubiquitin) control and then normalized to OsRac1 to 7 values in the wild type. Error bars represent the sd calculated from five independent transgenic lines. Bar shadings are identified to the right.
Quantitative analysis of all OsRac mRNA levels by real-time PCR in OsRac-silenced suspension cultures showed that the levels of OsRac mRNAs were less than 10% of those in the wild type (Fig. 4C). No clear morphological abnormalities have been observed in the transgenic rice plants. The orientation of the IR in the vector with respect to the promoter did not affect the efficiency of suppression (Fig. 3, C and D). Taken together, our results show that no detectable transitive RNA silencing occurs in the OsRac gene family.
The presence of OsRac siRNA was evaluated in both the gene-specific region from which each dsRNA was produced and the conserved coding region. Two size classes of siRNA (21 nt and 24 nt) were detected only when the gene-specific regions were probed; siRNA derived from the conserved coding regions was not detected (Fig. 4B), indicating that spreading of dsRNA along the endogenous OsRac mRNA did not occur in the 5′ direction. These results support the above findings that no transitive RNA silencing occurred among the members of the OsRac gene family (Fig. 4A) and are consistent with the results obtained for PDS-silenced plants, in which the siRNAs for the PDS genes were only detected in the region from which the dsRNA was transcribed (Fig. 2C). They further support the observation that transitive RNA silencing of endogenous genes may not occur in rice with high frequency.
Gene-Specific RNA Silencing of Multiple Members of a Gene Family by a Single IR Construct
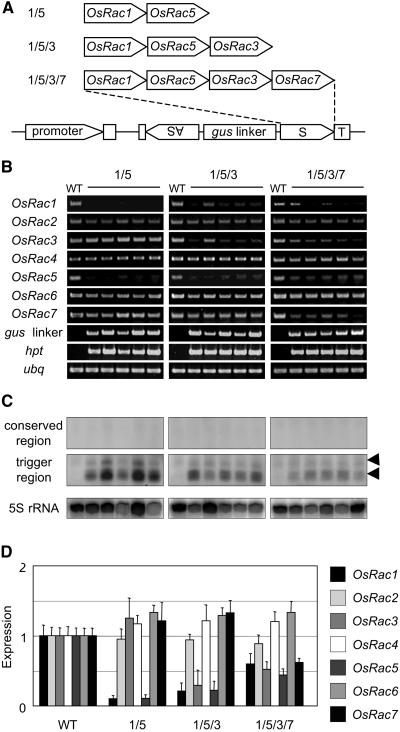
To determine whether a single RNA silencing construct can effectively and specifically suppress the expression of several corresponding genes, we designed IR constructs transcribing dsRNA containing chimeric gene-specific regions of OsRac genes (Fig. 5A; Supplemental Figs. 1 and 2). The OsRac1 and OsRac5 gene-targeting RNA silencing constructs were named 1/5. The other constructs linked the OsRac1, OsRac5, and OsRac3 (1/5/3) or the OsRac1, OsRac5, OsRac3, and OsRac7 (1/5/3/7) gene-specific sequences, respectively (Fig. 5A). RT-PCR analyses of OsRac mRNAs in transgenic rice showed that specific suppression of multiple genes in the OsRac gene family could be achieved by a single chimeric RNA-silencing construct (Fig. 5B). However, the levels of silencing seemed to decline as the number of genes increased (Fig. 5B).
Figure 5.
Gene-specific RNA silencing of multiple genes in the OsRac gene family by a single IR construct. A, Chimeric OsRac sequences used in RNA silencing. Gene-specific regions of OsRac1, 3, 5, and 7 (Fig. 3) were used to generate chimeric RNA silencing constructs. 1/5, OsRac1 and OsRac5 gene-specific regions; 1/5/3, OsRac1, OsRac5, and OsRac3 gene-specific regions; 1/5/3/7, OsRac1, OsRac5, OsRac3, and OsRac7 gene-specific regions. B, RT-PCR analysis of OsRac1 to 7 mRNA expression in five independent transgenic lines. The wild type (WT) is a nontransgenic control. gus linker, mRNA derived from the gus linker region; hpt, mRNA of the hygromycin resistance gene; ubq, mRNA of rice ubiquitin used as a control. C, Analysis of siRNAs. Probes used for hybridization are as follows: top, the highly conserved region of OsRac1 (Fig. 3C, hatched box); middle, regions corresponding to the chimeric RNA silencing trigger sequences in A; bottom, 5S rRNA. Arrowheads indicate the positions of 23-nt (bottom) and 27-nt (top) DNA oligomers, which are likely to correspond to 21-nt- and 24-nt-size-class siRNA detected in other plants, respectively. D, Quantitative analysis of OsRac mRNA expression using real-time PCR. Error bars represent the sd calculated from five independent transgenic lines. Bar shadings are identified to the right.
Results of siRNA analysis showed that two size classes of siRNA were detected when the region for the trigger dsRNA was used as a probe and that no spreading of dsRNA along OsRac mRNAs occurred in the 5′ direction (Fig. 5C). These results were consistent with the results of the single IR constructs (Fig. 4B). Interestingly, the levels of siRNA apparently decreased with increasing the length of the IR region. This observation may explain the lower level of silencing observed with the chimeric 1/5/3/7 construct (Fig. 5, B and D).
For a more accurate quantitative analysis of all OsRac mRNA levels in OsRac-silenced transgenic rice, real-time PCR analysis was performed on five independent transgenic lines for each chimeric RNA-silencing construct. OsRac mRNAs were specifically suppressed by the corresponding multiple OsRac gene constructs (Fig. 5D). However, comparing the levels of OsRac1 and OsRac5 mRNA targeted by the three chimeric constructs, the levels of suppression were more significantly decreased in 1/5/3/7 than in 1/5 (Fig. 5, B and C). In addition to these two-to-four-gene chimeric analyses, we also tested a construct in which all members of the OsRac gene family were targeted by a seven-gene-specific dsRNA. However, no significant target mRNA suppression was detected, and only very weak siRNA signals were found (data not shown). Therefore, these results suggest that a single chimeric IR construct can specifically and efficiently suppress three genes of the OsRac gene family and that the levels of suppression decreased when four genes were fused in a single construct (Fig. 5D).
RNA Silencing by the Highly Conserved Coding Region of OsRac1 or OsRac5
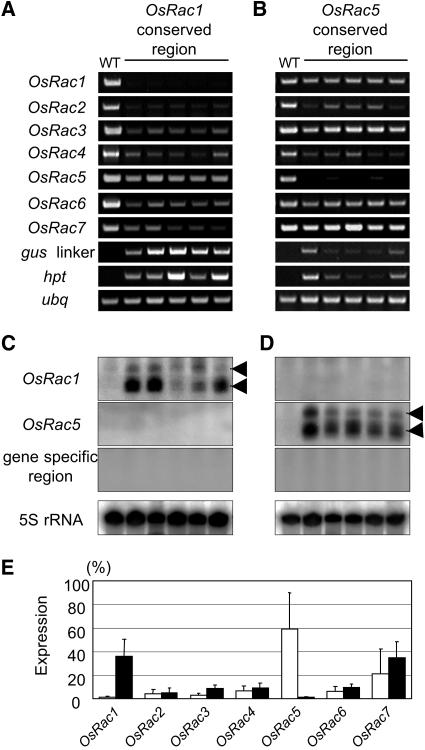
To determine whether a single dsRNA species derived from the highly conserved coding region of one OsRac gene can effectively suppress the expression of the entire gene family, we used the highly conserved coding regions of OsRac1 or OsRac5 as trigger dsRNAs (Fig. 3, B and C; Supplemental Figs. 1 and 2). This region was 518 bp; the relative nucleotide identities of the corresponding region between OsRac1 and the other six members of the gene family are 72% to 82%, and those between OsRac5 and the other members are 73% to 82% (Fig. 3B; Supplemental Fig. 2A). The mRNA levels of OsRac1 and OsRac5 were highly suppressed in transgenic lines expressing the conserved regions of OsRac1 and OsRac5 dsRNA, respectively, while the mRNA levels of the other members of the OsRac gene family were reduced to variable degrees (Fig. 6, A, B, and E).
Figure 6.
RNA silencing of the OsRac gene family by the highly conserved sequences of OsRac1 or OsRac5. A and B, RT-PCR analysis. Wild type (WT) is a nontransgenic control. Five independent transgenic rice plants expressing the highly conserved region of OsRac1 (A) or OsRac5 (B) were analyzed. C and D, RNA gel-blot analysis to detect siRNAs. Probes used are the conserved regions of OsRac1 (top) or OsRac5 (middle) and the gene-specific region of OsRac1 (bottom; Fig. 3C). Arrowheads indicate the positions of 23-nt (bottom) and 27-nt (top) DNA oligomers. The bottom section shows 5S rRNA as a loading control. E, Expression levels of all the OsRac genes, measured by real-time PCR analysis. Relative expression of each OsRac gene in transgenic rice is shown with the expression of their corresponding wild-type genes taken as 100%. White bars, Transgenic plants expressing dsRNA of the OsRac1 conserved region; black bars, transgenic plants expressing dsRNA of the OsRac5 conserved region. Error bars represent the sd calculated from 11 (OsRac1) or 10 (OsRac5) independent transgenic lines.
siRNA analysis showed that no spreading of dsRNA along OsRac1 or OsRac5 mRNA occurred in the 3′ direction (Fig. 6, C and D). In transgenic plants expressing OsRac1 coding region dsRNA, no siRNA for the OsRac5 coding region was detected (Fig. 6C), although the nucleotide identity between the conserved region of OsRac1 and OsRac5 was 73% (Fig. 3B) and some reduction of mRNA levels was detected (Fig. 6E). Similarly, no siRNA for the OsRac1 coding region was detected in the transgenic plants expressing dsRNA from the OsRac5 coding region (Fig. 6D). These results suggest that suppression of the other members of the OsRac gene family was caused by the OsRac1 or OsRac5 coding region siRNA and not by transitive RNA silencing. Transgenic rice plants were generally weaker and shorter than nontransgenic control plants, suggesting that the OsRac gene family may have important functions in growth and development and that their functions are redundant (C. Letian, T. Togashi, D. Miki, T. Kawasaki, and K. Shimamoto, unpublished data).
To accurately quantify the expression levels of all the OsRac genes in transgenic suspension lines, real-time PCR was performed with nine independent transgenic lines for the OsRac1 coding region dsRNA and 10 lines for the OsRac5 coding region dsRNA (Fig. 6E). This analysis indicated that levels of OsRac1 and OsRac5 mRNA in the transgenic lines were less than 1% of those in the wild type. In the OsRac1 coding region dsRNA plants, mRNA levels were reduced to less than 10% for OsRac2, OsRac3, OsRac4, and OsRac6, while the levels of OsRac5 and OsRac7 mRNAs were relatively high. In OsRac5 coding region dsRNA plants, the mRNAs for OsRac2, OsRac3, OsRac4, and OsRac6 were low, while those for OsRac1 and OsRac7 were higher. These results suggest that the suppression levels generally correlate with the overall sequence similarity between the trigger region and the targets (Fig. 3B; Supplemental Fig. 2A). However, other factors must also play a role. Although OsRac6 mRNA was effectively suppressed by the OsRac1 coding region dsRNA, the sequence similarity between the two was similar to that between OsRac5 and OsRac7 (Figs. 3B and 6E).
Together, these results demonstrate that it is possible to suppress the expression of an entire gene family by using a conserved region to transcribe the dsRNA; however, the levels of suppression achieved may depend partly on the degree of sequence identity.
DISCUSSION
Lack of Transitive RNA Silencing of Endogenous Genes in Rice
In the experiments described here, we could not observe any evidence for transitive RNA silencing of endogenous genes in a monocot, rice. On the other hand, transitive RNA silencing of the gfp transgene in rice was clearly demonstrated. These findings are consistent with our observations that no spreading of siRNA along endogenous target RNA occurred in either the 5′ or the 3′ direction. To confirm the lack of transitivity in the RNA silencing of OsRac genes, an RNase protection assay, which is more sensitive than northern hybridization, was performed, and the results are shown in Supplemental Figure 3. The results indicate that no significant spreading of siRNA signals in either the 5′ or the 3′ direction was detected. Our results support the conclusion of Vaistij et al. (2002) that transitive RNA silencing of the gfp transgene, but not of the small subunit of Rubisco or PDS endogenous genes, occurs in tobacco. We have also been unable to detect any evidence of transitive RNA silencing in rice when the endogenous OsGEN-L gene is suppressed by RNA silencing (Moritoh et al., 2005). These data suggest that siRNA spreading activity is absent or very weak when endogenous genes are targeted in rice.
In C. elegans, endogenous genes are clearly subjected to transitive RNAi (Alder et al., 2003). The transitive RNAi in C. elegans has directionality. It was only observed in the 5′ direction and acted over a short distance. By contrast, transitive RNA silencing of the gfp and gus transgenes in plants often occurs in both the 5′ and 3′ directions (Braunstein et al., 2002; Klahre et al., 2002; Vaistij et al., 2002; Van Houdt et al., 2003). These results suggest that there are differences in the mechanisms of transitive RNA silencing (RNAi) among different organisms. Indeed, unambiguous cases of transitive RNA silencing of plant endogenous genes have not been described. In the case of β-1,3-glucanase gene silencing in tobacco protoplasts, the level of RNA silencing of the β-1,3-glucanase endogenous gene was much lower than that of the β-1,3-glucanase transgene, although siRNA for the endogenous β-1,3-glucanase gene was detected (Sanders et al., 2002). These results suggest that RNAs derived from endogenous genes may be less efficiently used for RNA silencing.
One possible reason for the apparent lack of transitive RNA silencing of plant endogenous genes may be the low concentration of the RNA substrates (templates) for RdRP. However, since the Rubisco small subunit is not affected by the spreading of siRNA despite being the most abundant mRNA in plants (Vaistij et al., 2002), the mRNA level might not be a factor affecting transitive RNA silencing. Another explanation is that the gfp gene is exceptional in that its RNA directs new dsRNA very efficiently in both the 5′ and 3′ directions or that mRNA originating from endogenous genes is not efficiently used for the production of siRNA, as suggested by Vaistij et al. (2002). This could be caused by differences in the structure of mRNA with proteins and/or other reasons. Indeed, it was previously shown that a 200-nt fragment derived from the 3′ end of gus mRNA was not sensitive to virus-induced gene silencing (Braunstein et al., 2002). Alternatively, endogenous mRNA may be uniquely marked so that it is not efficiently subjected to dsRNA synthesis by RdRP.
Two Classes of siRNAs in Rice
We show here that two distinct classes of siRNA can be detected in a monocot, rice, in which dsRNA-mediated RNA silencing occurs. Two siRNA-generating Dicer-like (DCL) activities have been identified in wheat-germ extract using dsRNA as a substrate (Tang et al., 2003). Therefore, they might represent two DCLs involved in siRNA generation in cereals. The two classes of siRNAs are assumed to possess different roles and are processed by different DCLs (Xie et al., 2004). We identified five DCL genes in the rice genome (D. Miki and K. Shimamoto, unpublished data), whereas Arabidopsis contains four genes (Schauer et al., 2002; Xie et al., 2004). It will be of interest to know the function of an additional DCL gene in rice. In contrast to plants, C. elegans and humans have only one Dicer gene, and it has both miRNA and siRNA functions (Grishok et al., 2001; Ketting et al., 2001; Knight and Bass, 2001; Provost et al., 2002; Zhang et al., 2002).
We showed that the siRNA derived from primary dsRNAs has two sizes, short and long (Figs. 1D, 2C, 4B, 5C, and 6C), whereas gfp-derived 5′ and 3′ secondary siRNAs exclusively have a short size (Fig. 1D). These results are consistent with the previous findings that secondary siRNAs produced in Arabidopsis and tobacco are exclusively of the 21-nt size (Himber et al., 2003; Garcia-Perez et al., 2004). Five RdRP genes are identified in the rice genome (D. Miki and K. Shimamoto, unpublished data), and they may be involved in the production of a different siRNA.
A long siRNA signal was detected in the nontransgenic control (wild type) when it was probed with the OsRac4 gene-specific region (Fig. 4B). No other siRNA signal was detected in the wild type. We searched for genomic sequences that could possibly transcribe dsRNA that has homology with the OsRac4 gene-specific region in the rice genomic sequence database Rice BLAST (http://RiceBLAST.dna.affrc.go.jp/), but we were not able to identify such sequences. Instead, a hairpin structure in which 47 of 57 nt could potentially form a double-stranded structure could be predicted in the 3′ UTR of OsRac4 by the RNA secondary structure prediction program (http://www.genebee.msu.su/services/rna2_reduced.html; data not shown). Therefore, it is possible that the predicted hairpin structure found in the 3′ UTR region of OsRac4 is a precursor of the long siRNA, although a transposon-derived dsRNA is another possibility.
Gene-Specific Suppression of Multiple Members in a Gene Family by a Single IR Construct
We could demonstrate that a single IR construct having gene-specific regions of multiple OsRac genes can simultaneously suppress the expression of multiple genes (Fig. 5). These results show that double and triple gene knock down may be possible by a single RNA silencing construct. In our study, three genes were the maximum that could be efficiently suppressed by a single construct. The suppression efficiency was reduced when the number of fused genes was increased. The levels of silencing appeared to be proportional to the levels of siRNA detected, suggesting that the generation of siRNA was a limiting factor for efficient RNA silencing in this method. It might be more difficult for a longer RNA trigger to fold into a stable hairpin RNA structure, which is a precursor for siRNA. Alternatively, long precursor RNA may be simply less stable than shorter hairpin RNA. By further studying various parameters for IR constructs, such as the length of the trigger sequence and choice of promoters, this method could be improved and become useful for studying gene function in rice and other plants.
Simultaneous Suppression of Multiple Members of a Gene Family by RNA Silencing with Highly Conserved Sequences
We found that a highly conserved region of OsRac1 or OsRac5, used for the IR construct, could suppress the mRNA expression of all seven OsRac genes to variable degrees (Fig. 6). By choosing a highly conserved sequence, it is thus possible to suppress multiple genes in a gene family with a single gene construct. The suppression efficiency was generally correlated with the level of homology between trigger and target sequences. However, other factors may also play a role. For example, although OsRac2 to 4 mRNAs were highly suppressed by OsRac5 coding region dsRNA, the sequence similarity of OsRac2 to 4 was lower than that of OsRac7 with respect to OsRac5 (Figs. 3B and 6E). In other words, suppression efficiency does not exclusively depend on sequence homology between trigger and target sequences. One factor influencing the silencing efficiency could be the concentration of the target mRNA. The mRNA expression levels of OsRac2, 3, and 4 are relatively high, while that of OsRac7 is low among the seven OsRac genes (data not shown). Therefore, it is possible that genes with higher levels of mRNA expression may be better silenced than those with lower levels of expression. This could be due to easy access of mRNA with higher amounts to siRNA and RISC; however, this idea should be tested in other genes with variable expression levels. These results suggest one possible strategy to suppress an entire gene family using a limited number of IR constructs by using highly conserved sequences in each clade of a gene family and to construct a silencing vector for each clade. This strategy could also be used to suppress the expression of a set of genes that share certain conserved sequences but do not belong to the same gene family.
RNA Silencing as a Useful Method for Functional Genomics in Rice
The rice genome was recently sequenced, and, since a large collection of full-length cDNAs is available (Feng et al., 2002; Sasaki et al., 2002; Kikuchi et al., 2003; Rice Chromosome 10 Sequencing Consortium, 2003; Yu et al., 2005), many efforts to systematically identify the functions of rice genes have been and are being undertaken (Hirochika, 2001; Shimamoto and Kyozuka, 2002). However, the number of available knockout mutants is far from being saturated at present, and it will be practically impossible to obtain tagged lines for all members of a gene family in the near future. Therefore, the RNA silencing approaches described here, using 3′ UTRs for single and multiple gene-specific suppression or using highly conserved gene sequences, can be used as alternative means for identifying gene function in rice. We recently developed efficient Gateway vectors for RNA silencing of rice genes (Miki and Shimamoto, 2004). RNA silencing can be used to complement existing tagged lines to suppress all of the members of a gene family.
MATERIALS AND METHODS
Accession Numbers
The accession numbers for the sequences described in this study are as follows: PDS, AF049356; ubq, D12629; 5S rRNA, D26370; OsRac1, BAA84492; OsRac2, BAA84493; OsRac3, BAA84494; OsRac4/OsRop4, AK061102; OsRac5/OsRacD, AK067504; OsRac6/OsRacB, AK100842; and OsRac7/OsRop5, AK058414.
Constructs and Plant Materials
In all of the RNA-silencing-triggered IR constructs made, the IR regions were amplified using the specific primers shown in Supplemental Table I and then subcloned into the pENTR/D-TOPO cloning vector (Invitrogen, Carlsbad, CA) to yield entry vectors. RNA silencing constructs carrying fragments of endogenous genes were made using the pANDA vector (Miki and Shimamoto, 2004), and the construct for silencing the 35S-gfp transgene (Hashizume et al., 1999) was made using the pANDA-β vector (Miki and Shimamoto, 2004; http://bsw3.naist.jp/simamoto/pANDA/real/pANDA_top.htm). The final RNA silencing vectors were produced by an LR clonase reaction between an entry vector and the pANDA or pANDA-β vector. Transgenic rice (Oryza sativa) plants were generated by Agrobacterium-mediated transformation of rice calli (cv Kinmaze), performed according to a published protocol (Hiei et al., 1994). Plants were regenerated from transformed calli by selecting for hygromycin resistance. Regenerated transgenic rice plants were grown in a greenhouse at 28°C.
Detection of GFP Fluorescence
GFP fluorescence of transgenic rice calli was observed with a fluorescence stereomicroscope (MZ FL III; Leica Microsystems, Wetzlar, Germany) under blue light (425/60 nm).
RT-PCR
Total RNA was extracted using the guanidinium method (Chomczynski and Sacchi, 1987). For the synthesis of first-strand cDNA, 1 μg of total RNA was reverse transcribed using an oligo poly-T primer and SuperScript II (Invitrogen) in 20 μL total volume. One microliter of the synthesized first-strand cDNA was used for PCR analysis with different sets of gene-specific primers. The primers, number of cycles, and annealing temperatures used for RT-PCR are shown in Supplemental Table I.
Gel-Blot Analysis siRNA
siRNA detection was performed as described previously (Miki and Shimamoto, 2004), following a published protocol (Hamilton and Baulcombe, 1999). To purify small-sized RNA, a protocol for the isolation of low-molecular-weight RNA (QIAGEN, Valencia, CA) was used with the QIAGEN RNA/DNA kit (QIAGEN). Probes used for the detection of siRNA were DNA fragments amplified using specific primers (Supplemental Table I) and labeled with 32P-dCTP. DNA oligomers of 23 and 27 nt were used as molecular size markers. Hybridization signals were detected using a BAS-2500 Bioimaging analyzer (Fuji, Tokyo).
Real-Time PCR
For real-time RT-PCR analysis, the SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) and ABI PRISM 7700 sequence detector (Applied Biosystems) were used according to the manufacturer's instructions. The PCR conditions were 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. For every experiment, 1 μL of first-strand cDNA solution was used for PCR amplification with primers specific for each gene (Supplemental Table I).
Acknowledgments
We thank Junko Kyozuka of the University of Tokyo for pGUS27 and p2K-1+, and members of the Plant Molecular Genetics Lab in Nara Institute of Science and Technology (NAIST) for their suggestions and participation in discussions.
This work was supported by the Research for the Future Program of the Japan Society for the Promotion of Science (grant no. JSPS–RFTF 00L01604) and by the Ministry of Agriculture, Forestry, and Fisheries of Japan (Rice Genome Project).
The online version of this article contains Web-only data.
References
- Alder MN, Dames S, Gaudet J, Mango SE (2003) Gene silencing in Caenorhabditis elegans by transitive RNA interference. RNA 9: 25–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo C, Sadanandom A, Kitagawa K, Freialdenhoven A, Shirasu K, Schulze-Lefert P (2002) The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295: 2073–2076 [DOI] [PubMed] [Google Scholar]
- Baulcombe D (2004) RNA silencing in plants. Nature 431: 356–363 [DOI] [PubMed] [Google Scholar]
- Beclin C, Boutet S, Waterhouse P, Vaucheret H (2002) A branched pathway for transgene-induced RNA silencing in plants. Curr Biol 12: 684–688 [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366 [DOI] [PubMed] [Google Scholar]
- Braunstein TH, Moury B, Johannessen M, Albrechtsen M (2002) Specific degradation of 3′ regions of GUS mRNA in posttranscriptionally silenced tobacco lines may be related to 5′-3′ spreading of silencing. RNA 8: 1034–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162: 156–159 [DOI] [PubMed] [Google Scholar]
- Christensen AH, Sharrock RA, Quail PH (1992) Maize polyubiquitin genes: structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol Biol 18: 675–689 [DOI] [PubMed] [Google Scholar]
- Christensen TM, Vejlupkova Z, Sharma YK, Arthur KM, Spatafora JW, Albright CA, Meeley RB, Duvick JP, Quatrano RS, Fowler JE (2003) Conserved subgroups and developmental regulation in the monocot rop gene family. Plant Physiol 6: 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang CF, Meyerowitz EM (2000) Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc Natl Acad Sci USA 97: 4985–4990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoni C, Macino G (1999) Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature 399: 166–169 [DOI] [PubMed] [Google Scholar]
- Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe DC (2000) An RNA-dependent RNA polymerase gene in Arabidopsis is required for post-transcriptional gene silencing mediated by a transgene but not by a virus. Cell 101: 543–553 [DOI] [PubMed] [Google Scholar]
- Feng Q, Zhang Y, Hao P, Wang S, Fu G, Huang Y, Li Y, Zhu J, Liu Y, Hu X, et al (2002) Sequence and analysis of rice chromosome 4. Nature 420: 316–320 [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811 [DOI] [PubMed] [Google Scholar]
- Garcia-Perez RD, Houdt HV, Depicker A (2004) Spreading of post-transcriptional gene silencing along the target gene promotes systemic silencing. Plant J 38: 594–602 [DOI] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC (2001) Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106: 23–34 [DOI] [PubMed] [Google Scholar]
- Gu Y, Wang Z, Yang Z (2004) ROP/RAC GTPase: an old new master regulator for plant signaling. Curr Opin Plant Biol 7: 527–536 [DOI] [PubMed] [Google Scholar]
- Hamilton A, Voinnet O, Chappell L, Baulcombe D (2002) Two classes of short interfering RNA in RNA silencing. EMBO J 21: 4671–4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC (1999) A species of small antisense RNA in post-transcriptional gene silencing in plants. Science 286: 950–952 [DOI] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ (2000) An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404: 293–296 [DOI] [PubMed] [Google Scholar]
- Hannon GJ (2002) RNA interference. Nature 418: 244–251 [DOI] [PubMed] [Google Scholar]
- Hashizume F, Tsuchiya T, Ugaki M, Niwa Y, Tachibana N, Kowyama Y (1999) Efficient Agrobacterium-mediated transformation and the usefulness of a synthetic GFP reporter gene in leading varieties of japonica rice. Plant Biotechnol 16: 397–401 [Google Scholar]
- Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K (2003) Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422: 719–722 [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Himber C, Dunoyer P, Moissiard G, Ritzenthaler C, Voinnet O (2003) Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO J 22: 4523–4533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirochika H (2001) Contribution of the Tos17 retrotransposon to rice functional genomics. Curr Opin Plant Biol 4: 118–122 [DOI] [PubMed] [Google Scholar]
- Holzberg S, Brosio P, Gross C, Pogue GP (2002) Barley stripe mosaic virus-induced gene silencing in a monocot plant. Plant J 30: 315–327 [DOI] [PubMed] [Google Scholar]
- Kawasaki T, Henmi K, Ono E, Hatakeyama S, Iwano M, Satoh H, Shimamoto K (1999) The small GTP-binding protein Rac is a regulator of cell death in plants. Proc Natl Acad Sci USA 96: 10922–10926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH (2001) Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev 15: 2654–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi S, Satoh K, Nagata T, Kawagashira N, Doi K, Kishimoto N, Yazaki J, Ishikawa M, Yamada H, Ooka H, et al (2003) Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 301: 376–379 [DOI] [PubMed] [Google Scholar]
- Klahre U, Crete P, Leuenberger SA, Iglesias VA, Meins F Jr (2002) High-molecular-weight RNAs and small interfering RNAs induce systemic post-transcriptional gene silencing in plants. Proc Natl Acad Sci USA 99: 11981–11986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SW, Bass BL (2001) A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 293: 2269–2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP (2002) Virus-induced gene silencing in tomato. Plant J 31: 777–786 [DOI] [PubMed] [Google Scholar]
- Martens H, Novotny J, Oberstrass J, Steck TL, Postlethwait P, Nellen W (2002) RNAi in Dictyostelium: the role of RNA-directed RNA polymerases and double-stranded RNase. Mol Biol Cell 13: 445–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke M, Matzke AJ, Kooter JM (2001) RNA: guiding gene silencing. Science 293: 1080–1083 [DOI] [PubMed] [Google Scholar]
- Meister G, Tuschl T (2004) Mechanisms of gene silencing by double-stranded RNA. Nature 431: 343–349 [DOI] [PubMed] [Google Scholar]
- Miki D, Shimamoto K (2004) Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol 45: 445–450 [DOI] [PubMed] [Google Scholar]
- Moritoh S, Miki D, Akiyama M, Kawahara M, Izawa T, Maki H, Shimamoto K (2005) RNAi-mediated silencing of OsGEN-L (OsGEN-like), a new member of the RAD2/XPG nuclease family, causes male sterility by defect of microspore development in rice. Plant Cell Physiol 46: 699–715 [DOI] [PubMed] [Google Scholar]
- Mourrain P, Beclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Jouette D, Lacombe AM, Nikic S, Picault N, et al (2000) Arabidopsis SGS2 and SGS3 genes are required for post-transcriptional gene silencing and natural virus resistance. Cell 101: 533–542 [DOI] [PubMed] [Google Scholar]
- Nishikura K (2001) A short primer on RNAi: RNA-directed RNA polymerase acts as a key catalyst. Cell 107: 415–418 [DOI] [PubMed] [Google Scholar]
- Ono E, Wong HL, Kawasaki T, Hasegawa M, Kodama O, Shimamoto K (2001) Essential role of the small GTPase Rac in disease resistance of rice. Proc Natl Acad Sci USA 98: 759–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp I, Mette MF, Aufsatz W, Daxinger L, Schauer SE, Ray A, Van Der Winden J, Matzke M, Matzke AJ (2003) Evidence for nuclear processing of plant micro RNA and short interfering RNA precursors. Plant Physiol 132: 1382–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasterk RH (2002) RNA silencing: the genome's immune system. Science 296: 1263–1265 [DOI] [PubMed] [Google Scholar]
- Provost P, Dishart D, Doucet J, Frendewey D, Samuelsson B, Radmark O (2002) Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J 21: 5864–5874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice Chromosome 10 Sequencing Consortium (2003) In-depth view of structure, activity, and evolution of rice chromosome 10. Science 300: 1566–1569 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Sanders M, Maddelein W, Depicker A, Van Montagu M, Cornelissen M, Jacobs J (2002) An active role for endogenous beta-1,3-glucanase genes in transgene-mediated co-suppression in tobacco. EMBO J 21: 5824–5832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Matsumoto T, Yamamoto K, Sakata K, Baba T, Katayose Y, Wu J, Niimura Y, Cheng Z, Nagamura Y, et al (2002) The genome sequence and structure of rice chromosome 1. Nature 420: 312–316 [DOI] [PubMed] [Google Scholar]
- Schauer SE, Jacobsen SE, Meinke DW, Ray A (2002) DICER-LIKE 1: blind men and elephants in Arabidopsis development. Trends Plant Sci 7: 487–491 [DOI] [PubMed] [Google Scholar]
- Shimamoto K, Kyozuka J (2002) Rice as a model for comparative genomics of plants. Annu Rev Plant Biol 53: 399–419 [DOI] [PubMed] [Google Scholar]
- Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH, Fire A (2001) On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107: 465–476 [DOI] [PubMed] [Google Scholar]
- Smardon A, Spoerke JM, Stacey SC, Klein ME, Mackin N, Maine EM (2000) EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr Biol 10: 169–178 [DOI] [PubMed] [Google Scholar]
- Tang G, Reinhart BJ, Bartel DP, Zamore PD (2003) A biochemical framework for RNA silencing in plants. Genes Dev 17: 49–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaistij FE, Jones L, Baulcombe DC (2002) Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell 14: 857–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Houdt H, Bleys A, Depicker A (2003) RNA target sequences promote spreading of RNA silencing. Plant Physiol 131: 245–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt U, Pelissier T, Putz A, Razvi F, Fischer R, Wassenegger M (2004) Viroid-induced RNA silencing of GFP-viroid fusion transgenes does not induce extensive spreading of methylation or transitive silencing. Plant J 38: 107–118 [DOI] [PubMed] [Google Scholar]
- Wang MB, Wesley SV, Finnegan EJ, Smith NA, Waterhouse PM (2001) Replicating satellite RNA induces sequence-specific DNA methylation and truncated transcripts in plants. RNA 7: 16–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse PM, Graham MW, Wang MB (1998) Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc Natl Acad Sci USA 95: 13959–13964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse PM, Wang MB, Finnegan EJ (2001) Role of short RNAs in gene silencing. Trends Plant Sci 6: 297–301 [DOI] [PubMed] [Google Scholar]
- Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC (2004) Genetic and functional diversification of small RNA pathways in plants. PLoS Biol 2: e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Wang J, Lin W, Li S, Li H, Zhou J, Ni P, Dong W, Hu S, Zeng C, et al (2005) The genomes of Oryza sativa: a history of duplications. PLoS Biol 3: e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W (2002) Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J 21: 5875–5885 [DOI] [PMC free article] [PubMed] [Google Scholar]