Abstract
Pyrimidines are particularly important in dividing tissues as building blocks for nucleic acids, but they are equally important for many biochemical processes, including sucrose and cell wall polysaccharide metabolism. In recent years, the molecular organization of nucleotide biosynthesis in plants has been analyzed. Here, we present a functional analysis of the pyrimidine de novo synthesis pathway. Each step in the pathway was investigated using transgenic plants with reduced expression of the corresponding gene to identify controlling steps and gain insights into the phenotypic and metabolic consequences. Inhibition of expression of 80% based on steady-state mRNA level did not lead to visible phenotypes. Stepwise reduction of protein abundance of Asp transcarbamoylase or dihydro orotase resulted in a corresponding inhibition of growth. This was not accompanied by pleiotropic effects or by changes in the developmental program. A more detailed metabolite analysis revealed slightly different responses in roots and shoots of plants with decreased abundance of proteins involved in pyrimidine de novo synthesis. Whereas in leaves the nucleotide and amino acid levels were changed only in the very strong inhibited plants, the roots show a transient increase of these metabolites in intermediate plants followed by a decrease in the strong inhibited plants. Growth analysis revealed that elongation rates and number of organs per plant were reduced, without large changes in the average cell size. It is concluded that reduced pyrimidine de novo synthesis is compensated for by reduction in growth rates, and the remaining nucleotide pools are sufficient for running basic metabolic processes.
Pyrimidine nucleotides are abundant molecules with essential functions in a multitude of biochemical processes. They are of particular importance in dividing and elongating tissues as building blocks for nucleic acid biosynthesis. In addition, as an energy source or precursors for the synthesis of primary and secondary products, they are participants in various metabolic processes. In particular, the pyrimidine nucleotides are directly involved in plant carbohydrate metabolism providing the energy-rich precursor UDP-Glc for many synthetic reactions, such as Suc and cell wall biosyntheses.
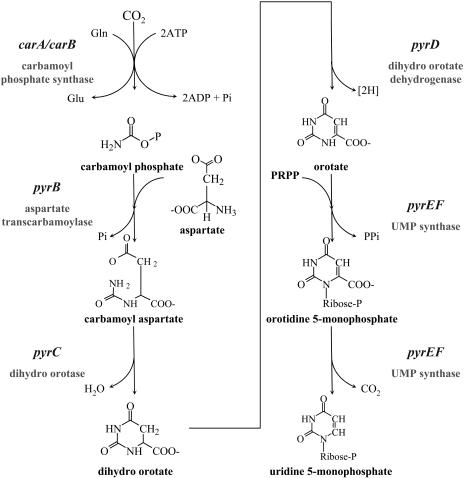
In recent years, the basic processes of plant nucleotide de novo synthesis have been analyzed in some detail (Giermann et al., 2002; Moffatt and Ashihara, 2002; Boldt and Zrenner, 2003; Stasolla et al., 2003; Kafer et al., 2004). Plants utilize the same reactions as those found in other organisms. The so-called orotate pathway of pyrimidine de novo synthesis is defined as the formation of UMP from carbamoyl phosphate, Asp, and phosphoribosyl pyrophosphate. The pathway consists of the six enzymatic reactions as shown in Figure 1. The first reaction is conducted by the Gln hydrolyzing carbamoyl phosphate synthase. This enzyme is not unique to the pyrimidine pathway but is also involved in Arg biosynthesis. It is composed of two different subunits (carbamoyl phosphate synthetase carB large subunit and Gln amidotransferase carA small subunit). The next three steps are catalyzed by three separate enzymes (Asp transcarbamoylase, dihydro orotase, and dihydro orotate dehydrogenase). The fifth and sixth steps are catalyzed by a single, bifunctional enzyme (UMP synthase).
Figure 1.
Pyrimidine de novo synthesis pathway. The orotate pathway of pyrimidine de novo synthesis is the formation of UMP from carbamoyl phosphate, Asp, and phosphoribosyl pyrophosphate. The pathway consists of the six enzymatic reactions catalyzed by carbamoyl phosphate synthase (CPSase), Asp transcarbamoylase (ATCase), dihydro orotase (DHOase), dihydro orotate dehydrogenase (DHODH), and, finally, UMP synthase (UMPSase).
From our molecular studies in tobacco (Nicotiana tabacum) and potato (Solanum tuberosum; Giermann et al., 2002), using exhaustive cDNA cloning and genomic Southern analysis, we showed that there is only one gene present for each single protein subunit. This is supported by analyses of the whole Arabidopsis (Arabidopsis thaliana) genome (Arabidopsis Genome Initiative, 2000) and all publicly available sequences from potato, tobacco, and Lycopersicon esculentum (http://www.ebi.ac.uk; http://www.ncbi.nlm.nih.gov), where in most cases only one gene for each single step per plant haploid genome is found. There are a few exceptions, e.g. the finding of three aspartate transcarbamoylase (ATCase) genes from Pisum sativum (Williamson and Slocum, 1994) and Glycine max or two different cDNAs for uridine monophosphate synthase (UMPSase) from Oryza sativa. From these findings, we conclude that pyrimidine de novo synthesis is an essential process and that complete loss of any one function leads to death of the plant. In order to perform a functional analysis of the whole pyrimidine de novo synthesis pathway, we therefore decided to use antisense and cosuppression strategies to create sets of plants with a progressive stepwise reduction in the expression of each single gene involved in the pathway. These transgenic plants with different levels of expression of a single protein have the advantage over complete knockout mutants with probably lethal phenotypes because analysis of consequences of reduced expression below a certain threshold is possible.
There are many reports on transgenic plants in which stepwise reduction of one activity has given valuable insight into the in vivo function of specific enzymes in a biochemical pathway (e.g. Stitt and Sonnewald, 1995; Haake et al., 1998). When the impact of changes at different steps within one pathway is compared, substantial differences can also be found for the susceptibility of the pathway to a change in expression at different enzymes (Stitt and Sonnewald, 1995; Strand et al., 2000). In this article, we present the first functional analysis of the whole pyrimidine de novo synthesis pathway, investigating each step in the pathway by using transgenic plants with reduced expression of a single gene. Selected transformants that show a growth reduction are then analyzed to provide insights into the phenotypic and metabolic consequences of reduced pyrimidine de novo synthesis.
RESULTS
Production and Selection of Transgenic Plants with Reduced Expression of Steps Involved in Pyrimidine de Novo Synthesis
The cloning and characterization of all cDNAs from tobacco or potato used in this analysis have been reported by Giermann et al. (2002). All constructs used to transform tobacco or potato plants were made with the binary vector pBinAR (Höfgen and Willmitzer, 1990), a derivative of the pBin19 vector containing the 35S promoter of the cauliflower mosaic virus for constitutive expression of the target gene and the octopine synthase polyadenylation signal. Sense constructs designed for cosuppression and antisense constructs designed for antisense inhibition contained at least 70% of the full-length cDNA sequences. Table I summarizes the cDNA sequences that were used to produce these constructs for plant transformation.
Table I.
Summary of all cDNA sequences used for the constructs to transform plants
All constructs were made with a derivative of the binary vector pBin19, containing the 35S promoter of cauliflower mosaic virus and the octopine synthase polyadenylation signal. CPSaseSU, Small subunit carbamoyl phosphate synthase; CPSaseLU, large subunit carbamoyl phosphate synthase.
| Enzyme | cDNA Accession | Source Organism | Fragment Size | Cloning Sites | Fragment Characteristics | Direction | Construct |
|---|---|---|---|---|---|---|---|
| bp | |||||||
| CPSaseSU | AJ319873 | Tobacco | 1,698 | BamHI/XbaI | Full length | Sense | NtcarAsense |
| CPSaseLU | AJ319872 | Tobacco | 3,315 | SalI/XhoI | 5′end | Antisense | NtcarBanti |
| Tobacco | 3,315 | SalI/XhoI | 5′end | Sense | NtcarBsense | ||
| ATCase | AJ319874 | Potato | 1,380 | BamHI/SalI | Full length | Sense | StpyrBsense |
| Potato | 1,380 | BamHI/Asp-718 | Full length | Antisense | StpyrBanti | ||
| DHOase | AX093580 | Potato | 1,328 | XbaI/SalI | Full length | Sense | StpyrCsense |
| Potato | 984 | BamHI/Asp-718 | 5′end | Antisense | StpyrCanti | ||
| DHODH | AX093582 | Tobacco | 1,371 | BamHI/XbaI | 3′end | Sense | NtpyrDsense |
| Tobacco | 1,371 | BamHI/Asp-718 | 3′end | Antisense | NtpyrDanti | ||
| UMPSase | U22260 | Tobacco | 1,388 | SalI/BamHI | 3′end | Antisense | NtpyrEFanti |
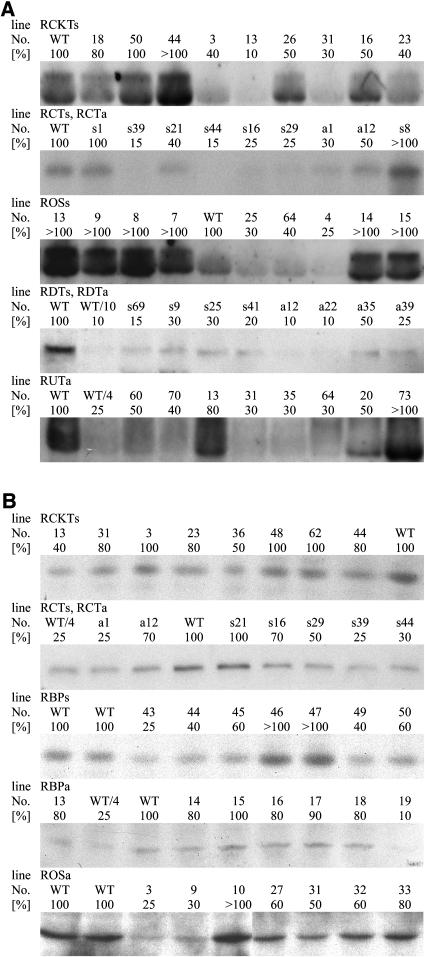
Either N. tabacum L. cv SNN or S. tuberosum L. cv Desirée and cv Solara or both species were used as target organisms. For each targeted enzymatic step, >60 transformed plants were regenerated and analyzed. A complete summary of the expression levels found in all 538 transformants is provided in Table II. The expression levels are given in percentage of wild type in rounded numbers, and the proportion of plants showing a particular reduction is given. Plants were initially analyzed by measuring mRNA and/or protein abundance. Representative results from each analysis of differences in mRNA expression, using northern hybridizations, and protein abundance, using western analysis, are presented in Figure 2.
Table II.
Summary of the expression differences found in all transformed plants
Plants were grown for 4 weeks in 1.5-L pots of soil in the greenhouse. Samples were taken from the fourth expanded leaf from the top of the plant after 6 h of illumination and quickly frozen in liquid N2. All values are single determinations per plant. The expression levels are given in percentage of wild type in rounded numbers, and the proportion of plants showing a particular reduction is given. n.d., Not determined.
| Enzyme | Construct | Transformed Species | Line Name | Total Number | >50% | RNA 20%–50% | <20% | >50% | Protein 20%–50% | <20% |
|---|---|---|---|---|---|---|---|---|---|---|
| CPSaseSU | NtcarAsense | Tobacco | RCKTs | 64 | 84% | 15% | 1% | 92% | 8% | 0% |
| CPSaseLU | NtcarBsense | Tobacco | RCTs | 77 | 86% | 12% | 2% | 93% | 7% | 0% |
| NtcarBanti | Tobacco | RCTa | 14 | 93% | 7% | 0% | 93% | 7% | 0% | |
| ATCase | StpyrBsense | Tobacco | RBPs | 61 | n.d. | n.d. | n.d. | 87% | 10% | 0% |
| StpyrBanti | Potato | RBPa | 63 | n.d. | n.d. | n.d. | 91% | 6% | 3% | |
| DHOase | StpyrCsense | Tobacco | ROSs | 53 | 94% | 6% | 0% | n.d. | n.d. | n.d. |
| StpyrCanti | Potato | ROSa | 44 | n.d. | n.d. | n.d. | 90% | 7% | 3% | |
| DHODH | NtpyrDsense | Tobacco | RDTs | 46 | 76% | 20% | 4% | n.d. | n.d. | n.d. |
| NtpyrDanti | Tobacco | RDTa | 50 | 64% | 30% | 6% | n.d. | n.d. | n.d. | |
| UMPSase | NtpyrEFanti | Tobacco | RUTa | 66 | 77% | 23% | 0% | n.d. | n.d. | n.d. |
Figure 2.
Representative results of expression differences in various transformed plants. For assignment of the screening of each transformation series summarized in Table II, representative parts of the blots are shown with untransformed controls and various transformants. A, Steady-state mRNA determination using northern analysis. If not indicated, same amounts of RNA were loaded on the gel, blotted, and hybridized with the respective cDNAs. B, Protein determination using SDS-PAGE and immunoblotting with specific antibodies. If not indicated, same amounts of protein were loaded, blotted, and detected with the respective antibodies. Plants were grown for 4 weeks in 1.5-L pots of soil in the greenhouse. Samples were taken from the fourth expanded leaf from the top of the plant after 6 h of illumination and quickly frozen in liquid N2. All values are single determinations per plant. For assignment of the lines, please refer to Table II.
For screening purposes, we prefer the analysis on protein level if appropriate antibodies are available because it is faster and closer to the expected difference in protein abundance and activity. Only in case of both subunits of carbamoyl phosphate synthase (CPSase) did we initially screen mRNA and protein level. We found reduced steady-state mRNA levels of 50% to 80% in 7% to 15% of the transformants and a reduction of >80% in 0% to 2% of the transformants. In case of the RCKTs plants, where a full-length construct was used, the values for reductions on steady-state mRNA level caused by cosuppression might be underestimated because we have not analyzed the appearance of small interfering RNAs to visualize that posttranscriptional gene silencing has already started although high levels of steady-state mRNAs are still present (Fig. 2; line RCKTs 44 has >100% mRNA and 80% protein compared to line RCKTs 31 with 30% mRNA and 80% protein). Such a scenario has been reported for other transgene expression studies (Schubert et al., 2004). Furthermore, as the analysis of the RCTs and RCTa lines revealed no clear correlation between the steady-state mRNA and the protein amounts in lines with reduced expression (Fig. 2; line RCTs s39 has 15% mRNA and 25% protein compared to line RCTa a1 with 30% mRNA and 25% protein), we opted for screening on protein abundance. Only in cases where this was not possible because no suitable antibody was available did we use the analysis on steady-state mRNA.
Sense (cosuppression) and antisense constructs led to a similar inhibition. A reduction of 50% to 80% of expression at the mRNA level was found in 16% of the 187 species-specific sense transformants (RCKTs, RCTs, and RDTs) and in 20% of the 237 antisense transformants (RCTa, RDTa, and RUTa). A reduction of >80% of expression at the mRNA level was found in 2% of the sense and the antisense transformants. At the protein level, a reduction of 50% to 80% was found in 7% of the sense and the antisense transformants. A reduction of >80% at the protein level was only seen in 2% of the plants when antisense transformations of potato with potato cDNAs were performed. These values match results from comparable studies with enzymes in pathways (Stitt and Sonnewald, 1995; Zrenner et al., 1995, 1996; Haake et al., 1998).
In the cases where full-length constructs were used in sense orientation (RCKTs, RBPs, and ROSs) a small number of overexpressors could be found. In 8% of the species-specific RCKTs transformants, a steady-state mRNA level of >100% was found, but there was no increase at the protein level (Table II, Fig. 2). In the line ROSs, where the potato dihydro orotase (DHOase) is expressed in tobacco, an increase of steady-state mRNA was found in 60% of the plants (Table II, Fig. 2). However, it was not possible to detect DHOase-specific proteins in the tobacco transformants (data not shown). In the RBPs transformants, where the potato ATCase is expressed in tobacco, 3% of the plants showed an increase in ATCase-specific protein (Table II, Fig. 2). All these overexpression lines are not further analyzed within this study.
As already mentioned, there is only one gene per enzymatic function involved in pyrimidine de novo synthesis present in Solanaceous species. Therefore, a reduction in expression is expected to cause some differences in plant growth and metabolism. All plants that showed <20% of expression at the steady-state mRNA level or at the protein abundance level compared to wild-type plants were analyzed further.
Tobacco Plants with Reduced Expression of CPSase, Dihydro Orotate Dehydrogenase, and UMPSase Do Not Show Phenotypical Differences
Primary transformant 13 of line RCKTs showed a reduction of 60% in the protein amount of the small subunit of CPSase (Fig. 2B), but no reduction in the large subunit of CPSase (data not shown) was observed. It is known from studies in Escherichia coli that the large subunit of CPSase is less stable when the small subunit is missing (Guillou et al., 1989; Cervera et al., 1993), but the coregulation of protein expression and stability of both subunits of CPSase of plants is not a subject of this study. Primary transformants RCTs 39 and RCTs 44 showed reductions of 75% and 70% in the protein amount of the large subunit of CPSase (Fig. 2B), respectively, but no reduction in the small subunit of CPSase (data not shown). These lines with reduction in CPSase-specific steady-state mRNA accumulation and CPSase protein did not show any obvious phenotypic differences when compared to wild-type plants or transformants without reduction in expression (data not shown). Analysis of the offspring (12 to 16 plants) of these lines confirmed that there were no significant differences in height, leaf number, or the above ground fresh weight when grown under standardized conditions in growth chambers on sand culture for 25 d (data not shown). Thus, a reduction of >80% in mRNA expression level and 60% and 75% on protein level of the small or large subunit of CPSase does not lead to visual phenotypic changes.
Lines RDTs 69 and 41 and lines RDTa 12 and 9 showed the largest reduction (>85%) in dihydro orotate dehydrogenase (DHODH)-specific steady-state mRNA accumulation. They did not show any obvious phenotypic differences when compared to wild-type plants or transformants without reduction in expression (data not shown). This result was confirmed by analyzing plant height, leaf number, and shoot fresh weight in the offspring of these lines.
Among all antisense transformants using the cDNA of UMP synthase, no plants with >70% of reduction in steady-state mRNA accumulation could be found. Line RUTa 31 showed the largest reduction in UMPSase-specific steady-state mRNA accumulation (>70%) and lines RUTa 24 and 25 showed intermediate reduction (60%). These lines did not show any obvious phenotypic differences when compared to wild-type plants or transformants without reduction in expression (data not shown). This result was confirmed by analyzing plant height, leaf number, and shoot fresh weight in the offspring of these lines.
Potato Plants with Reduced Expression of Either ATCase or DHOase, the Second and Third Steps of Pyrimidine de Novo Synthesis, Show Decreased Growth
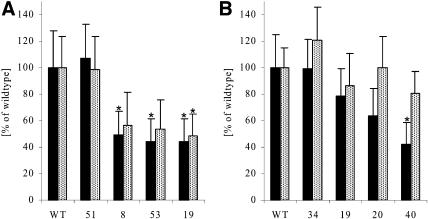
As already pointed out earlier, only in the case of the antisense transformations of potato using potato cDNAs was there a reduction of >80% in protein level observed (Table II). These potato plants transformed with antisense constructs of either ATCase or DHOase were analyzed further, including lines with a reduction of >80% in protein level and plants from the same transformation series with a less severe reduction in protein amount. As explained in the beginning, there is no clear correlation of steady-state mRNA data and remaining protein amounts, dependent on specific parameters like mRNA stability, translation efficiency, or protein stability. In order to obtain a reliable picture of what amount of the specific enzyme is left in the transgenic plants, we included the results on protein level, as these are the more significant data. A progressive reduction in the ATCase (Fig. 3A) or DHOase (Fig. 3B) protein abundance in source leaves was linked to a corresponding reduction in plant size (Fig. 3, C and D). Decreases in plant height were already apparent when 50% to 60% of the ATCase or DHOase protein was missing. Plants were usually propagated and maintained in tissue culture on Murashige and Skoog medium containing 2% Suc for 3 weeks and were than transferred to sand culture and watered daily with nutrient solution. After about 3 weeks on sand culture, when growth differences were already established, no other pleiotropic effects were visible (Fig. 3, C and D); no chlorosis or necrosis could be detected, and chlorophyll content and anthocyanin content on a leaves fresh weight basis were unchanged (data not shown; anthocyanin only increased in ROSa 40). When DHOase protein amount was <10% of that in wild-type plants, growth was severely inhibited after transfer to soil or sand, and the transformants eventually died without flowering or tuberization.
Figure 3.
Phenotypical analysis of plants with reduced ATCase (RBPa) or DHOase (ROSa) protein using the antisense strategy. A, Protein determination using SDS-PAGE and immunoblotting with ATCase-specific antibodies. The arrow indicates the ATCase protein at approximately 39 kD. One representative Western analysis is shown. Plants were grown for 3 weeks in 1.5-L pots of sand in the growth chamber. Samples were taken from the fourth leaf from the top of the plant after 6 h of illumination and quickly frozen in liquid N2. WT, Wild type. B, Protein determination using SDS-PAGE and immunoblotting with DHOase-specific antibodies. The arrow indicates the DHOase protein at approximately 36 kD. One representative Western analysis is shown. Plants were grown for 3 weeks in 0.5-L pots of sand in the growth chamber. Samples were taken from the fourth leaf from the top of the plant after 6 h of illumination and quickly frozen in liquid N2. For better assignment of the lines, values are also given as means and sd of four replicates in percentage of the wild type amount. sd is below 25% of the mean, and all lines show significantly reduced protein amount (P < 0.05) when compared to the wild type with the exception of line ROSa 34. C, Representative phenotype of plants transformed with a construct for antisense reduction of ATCase (RBPa). D, Representative phenotype of plants transformed with a construct for antisense reduction of DHOase (ROSa).
Detailed Growth Analysis of Potato Plants with Reduced Expression of Either ATCase or DHOase
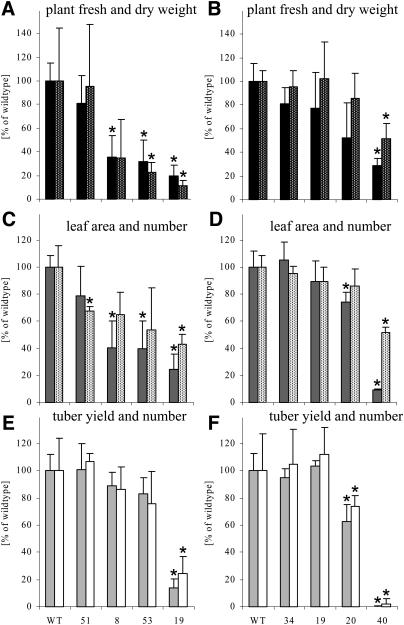
More information about the inhibition of growth in lines with reduced expression of either ATCase or DHOase is provided in Figure 4. With reduced protein abundance of ATCase (or DHOase) the total fresh weight and also the plant dry weight was reduced by the 23rd day (or 19th day for DHOase) after the plants were transferred to sand and watered daily with nutrient solution (Fig. 4, A and B). Under the applied conditions, root growth on a fresh weight basis was even more reduced than shoot growth (data not shown). At the indicated time of harvest, the total leaf area per plant decreased with abundance of ATCase or DHOase protein, and the leaf number was also reduced, although to a lesser extent (Fig. 4, C and D).
Figure 4.
Growth analysis of plants with reduced expression of either ATCase (RBPa) or DHOase (ROSa). A, Fresh weight (black bars) and dry weight (black bars with white dots) of RBPa plants. B, Fresh weight (black bars) and dry weight (black bars with white dots) of ROSa plants. C, Leaf area (gray bars) and leaf number (white bars with black dots) of RBPa plants. D, Leaf area (gray bars) and leaf number (white bars with black dots) of ROSa plants. E, Tuber yield (light gray bars) and tuber number (white bars) of RBPa plants. WT, Wild type. F, Tuber yield (light gray bars) and tuber number (white bars) of ROSa plants. Plants were grown for 23 d (RBPa) or 19 d (ROSa) in 1.5-L pots of sand in the growth chamber. For fresh and dry weight determinations and for leaf area and number measurements, plants were harvested after 6 h of illumination (RBPa) or at the end of the day (ROSa) and quickly frozen in liquid N2. For tuber yield, plants were grown for at least 16 weeks until complete senescence. For better comparison, all values are given as means and sd of 4 to 10 replicates in percentage of the wild-type amount. For assignment of the lines, please refer to Table II and Figure 3. Significant differences (P < 0.05) are marked with an asterisk.
Depending on the growth conditions, there were no obvious differences in the development of plants with or without reduction in the amount of ATCase or DHOase protein. The plants started to tuberize at approximately the same time, but with decreasing ATCase or DHOase protein the tuber yield per plant was reduced at the end of the growing period. Although line RBPa 51, with 40% of ATCase protein remaining, showed no reduction in tuber yield or tuber number per plant, lines RBPa 8, 53, and 19 showed a decrease in yield of approximately 15%, 20%, and 85% and a concomitant reduction in tuber number per plant of approximately 15%, 25%, and 75%, respectively, when compared to wild-type plants grown for 15 weeks in 1.5-L pots in sand culture (Fig. 4E). Comparable results were found for the ROSa transformants (Fig. 4F) and also for RBPa and ROSa transformants grown in bigger pots in soil in the greenhouse during summer or winter (data not shown).
Metabolite Analysis Reveals Different Responses in Roots and Shoots of Plants with Decreased Abundance of Proteins Involved in Pyrimidine de Novo Synthesis
We investigated whether reduced growth rates of plants with decreased abundance of enzymes involved in pyrimidine de novo synthesis were accompanied by changes in free nucleotide levels. The nucleotide content and nucleotide ratios of leaves and roots were measured in RBPa plants after 6 h of illumination, 23 d after transfer to sand culture. Table III shows a summary of these results. Although there were no striking changes, a reduction in the nucleotide content of leaves from plants showing strongest reduction of ATCase protein was observed, but without any change in the purine-to-pyrimidine ratio. In roots, there was a slight increase in nucleotide levels in the intermediate lines and a decrease in the strongest inhibited line. This change occurs for both adenine and uridine nucleotides. Comparable results were found in leaves and roots for ROSa transformants (data not shown; roots of intermediate lines were analyzed). In general, there were no specific effects measurable on pyrimidine nucleotide levels, despite the plants having reduced amounts of either ATCase or DHOase protein, which are involved in pyrimidine de novo synthesis. In the few examples where a slight change of pyrimidines was observed, the purine levels were changed in the same way.
Table III.
Nucleotide levels and ratios in leaves and roots of plants with reduced expression of ATCase (RBPa)
Plants were grown for 23 d in 1.5-L pots of sand in the growth chamber. Plants were harvested after 6 h of illumination (RBPa) and quickly frozen in liquid N2. P, All purine nucleotides; U, all pyrimidine nucleotides. Values are given as means and sd of four replicates; significant differences (P < 0.05) are marked with an asterisk. WT, Wild type; gfw, grams fresh weight.
| Leaves
|
WT
|
51
|
8
|
53
|
19
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | ||||
| nmol·gfw−1 | |||||||||||||
| UDPG | 101.000 | 6.400 | 99.000 | 9.640 | 113.000 | 5.580 | 69.000 | 30.700 | 69.000 | 22.400 | * | ||
| ADP | 32.300 | 7.040 | 34.100 | 4.540 | 37.600 | 8.500 | 23.700 | 11.100 | 24.200 | 0.490 | * | ||
| UDP | 3.000 | 0.960 | 2.600 | 0.550 | 2.700 | 0.630 | 2.710 | 1.100 | 1.600 | 0.280 | * | ||
| GDP | 4.600 | 1.490 | 3.400 | 0.870 | 4.600 | 1.260 | 3.300 | 0.940 | 3.400 | 0.800 | |||
| UTP | 22.700 | 2.190 | 25.700 | 3.520 | 20.600 | 3.370 | 15.800 | 6.740 | 12.700 | 1.390 | * | ||
| ATP | 94.900 | 13.200 | 121.000 | 23.600 | 108.000 | 5.980 | 59.800 | 26.100 | 72.000 | 1.280 | |||
| GTP | 30.200 | 3.530 | 38.900 | 9.010 | 37.900 | 5.440 | 20.200 | 8.770 | * | 22.400 | 2.350 | * | |
| Total | 290.000 | 14.400 | 325.000 | 50.400 | 325.000 | 21.600 | 194.000 | 85.300 | 206.000 | 24.500 | * | ||
| P | 162.000 | 9.870 | 198.000 | 37.400 | 189.000 | 16.200 | 107.000 | 46.900 | 122.000 | 1.330 | * | ||
| U | 127.000 | 7.740 | 127.000 | 13.000 | 136.000 | 6.520 | 87.700 | 38.400 | 83.900 | 23.300 | |||
| ATP/ADP | 3.000 | 0.830 | 3.500 | 0.240 | 3.000 | 0.640 | 2.500 | 0.110 | 2.900 | 0.100 | |||
| UTP/UDP | 8.400 | 4.250 | 9.900 | 2.090 | 8.000 | 3.290 | 5.700 | 0.200 | 8.000 | 1.800 | |||
| UTP/UDPG | 0.224 | 0.024 | 0.259 | 0.017 | 0.183 | 0.031 | 0.231 | 0.020 | 0.195 | 0.054 | |||
| UDP/UDPG | 0.030 | 0.008 | 0.026 | 0.005 | 0.024 | 0.005 | 0.039 | 0.004 | 0.025 | 0.008 | |||
| UTP/ATP | 0.244 | 0.050 | 0.214 | 0.019 | 0.188 | 0.022 | 0.266 | 0.019 | 0.177 | 0.020 | |||
| UDP/ADP | 0.095 | 0.023 | 0.078 | 0.018 | 0.074 | 0.017 | 0.117 | 0.011 | 0.067 | 0.011 | |||
| U/P | 0.395 | 0.026 | 0.361 | 0.043 | 0.370 | 0.026 | 0.309 | 0.103 | 0.377 | 0.087 | |||
| Roots | |||||||||||||
| ADPG | 2.000 | 1.120 | 1.200 | 0.220 | 0.300 | 0.090 | * | 0.080 | 0.050 | * | 0.060 | 0.030 | * |
| UDPG | 103.000 | 22.400 | 155.000 | 30.700 | 194.000 | 29.600 | * | 111.000 | 21.400 | 63.000 | 9.990 | * | |
| ADP | 17.900 | 1.710 | 25.000 | 4.340 | 35.400 | 8.180 | * | 20.000 | 2.260 | 13.700 | 3.030 | ||
| UDP | 7.900 | 0.950 | 11.700 | 1.830 | 14.900 | 2.040 | * | 10.900 | 3.370 | 6.200 | 1.010 | ||
| GDP | 3.100 | 0.880 | 4.300 | 0.790 | 5.100 | 0.730 | * | 1.870 | 0.730 | 1.600 | 1.120 | ||
| UTP | 59.300 | 9.290 | 75.400 | 5.580 | 91.200 | 6.600 | * | 68.800 | 7.410 | 44.700 | 5.070 | ||
| ATP | 75.800 | 11.600 | 105.000 | 12.000 | 130.000 | 16.200 | * | 91.400 | 9.880 | 61.000 | 8.910 | ||
| GTP | 25.000 | 6.720 | 35.800 | 3.720 | 44.500 | 6.070 | * | 34.100 | 4.400 | 20.100 | 5.350 | ||
| Total | 292.000 | 37.700 | 413.000 | 55.000 | 515.000 | 62.700 | * | 338.000 | 49.000 | 211.000 | 33.300 | ||
| P | 122.000 | 17.400 | 170.000 | 20.200 | 215.000 | 26.800 | * | 147.000 | 17.100 | 96.600 | 17.700 | ||
| U | 170.000 | 24.300 | 242.000 | 35.400 | 300.000 | 36.600 | * | 191.000 | 31.900 | 114.000 | 15.600 | ||
| ATP/ADP | 4.200 | 0.600 | 4.200 | 0.240 | 3.700 | 0.630 | 4.500 | 0.120 | 4.400 | 0.330 | |||
| UTP/UDP | 7.500 | 0.920 | 6.500 | 0.600 | 6.100 | 0.600 | 6.500 | 1.240 | 7.200 | 0.950 | |||
| UTP/UDPG | 0.598 | 0.146 | 0.496 | 0.085 | 0.475 | 0.054 | 0.626 | 0.061 | 0.704 | 0.030 | |||
| UDP/UDPG | 0.078 | 0.012 | 0.076 | 0.007 | 0.076 | 0.001 | 0.097 | 0.013 | 0.098 | 0.014 | |||
| UTP/ATP | 0.785 | 0.086 | 0.720 | 0.046 | 0.704 | 0.040 | 0.753 | 0.009 | 0.735 | 0.026 | |||
| UDP/ADP | 0.443 | 0.044 | 0.467 | 0.020 | 0.427 | 0.052 | 0.537 | 0.109 | 0.458 | 0.077 | |||
| U/P | 0.858 | 0.068 | 0.944 | 0.053 | 1.030 | 0.029 | 0.878 | 0.063 | 0.792 | 0.026 | |||
We further investigated whether reduced growth rates of plants with decreased abundance of enzymes involved in pyrimidine de novo synthesis were accompanied by changes in carbohydrate levels. The Glc, Fru, Suc, and starch content was measured in leaves and roots of ROSa plants at the beginning and end of the day, in plants 19 d after transfer to sand culture. The results from the end of the day are shown in Figure 5, A and C. There was an increase in both soluble sugars and starch in the leaves, whereas in the roots, the content of all carbohydrates was reduced at every time point analyzed (Fig. 5; data not shown). Essentially the same results were obtained for RBPa plants harvested after 6 h of illumination, 23 d after transfer to sand culture (Fig. 5, B and D).
Figure 5.
Carbohydrate content of plants with reduced expression of either ATCase (RBPa) or DHOase (ROSa). A, Soluble sugars (Glc plus Fru plus Suc; black bars) and starch (black bars with white dots) of leaves of RBPa plants. B, Soluble sugars (Glc plus Fru plus Suc; black bars) and starch (black bars with white dots) of leaves of ROSa plants. C, Soluble sugars (Glc plus Fru plus Suc; black bars) and starch (black bars with white dots) of roots of RBPa plants. WT, Wild type. D, Soluble sugars (Glc plus Fru plus Suc; black bars) and starch (black bars with white dots) of roots of ROSa plants. Plants were grown for 23 d (RBPa) or 19 d (ROSa) in 1.5-L pots of sand in the growth chamber. Samples were taken from the fourth leaf from the top of the plant after 6 h of illumination (RBPa) or at the end of the day (ROSa) and from the whole roots at the same time point as indicated, briefly washed with water and dried, and quickly frozen in liquid N2. For better comparison, all values are given as means and sd of four replicates in percentage of the wild-type amount. For assignment of the lines, please refer to Table II and Figure 3. Significant differences (P < 0.05) are marked with an asterisk.
In order to analyze if reduced growth rates are caused by altered nitrogen and amino acid availability, the metabolite analysis was extended to include the steady-state amino acid content of leaves and roots of RBPa plants harvested after 6 h of illumination, 23 d after transfer to sand culture. The results are summarized in Table IV. No large overall change in amino acid content of leaves was apparent when compared to the untransformed control plants, and only a reduction in the amino acid content of leaves from plants showing strongest reduction of ATCase protein was observed. Although there were no striking changes in amino acid levels from roots, a slight increase in intermediate lines and a decrease in roots from plants showing strongest reduction of ATCase protein was observed. Comparable results were found in leaves and roots for ROSa transformants (data not shown; only roots of intermediate lines were analyzed).
Table IV.
Amino acid levels in leaves and roots of plants with reduced expression of ATCase (RBPa)
Plants were grown for 23 d in 1.5-L pots of sand in the growth chamber. Plants were harvested after 6 h of illumination (RBPa). Values are given as means and sd of four replicates; significant differences (P < 0.05) are marked with an asterisk. WT, Wild type; gfw, grams fresh weight.
| Leaves
|
WT
|
51
|
8
|
53
|
19
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | |||||
| nmol·gfw−1 | ||||||||||||||
| Asp | 650.0 | 233.0 | 714.0 | 250.0 | 773.0 | 112.0 | 714.0 | 160.0 | 474.0 | 143.0 | ||||
| Glu | 2320.0 | 456.0 | 2684.0 | 952.0 | 1854.0 | 301.0 | 2199.0 | 214.0 | 1272.0 | 211.0 | * | |||
| Asn | 194.0 | 48.7 | 178.0 | 96.6 | 96.0 | 25.0 | * | 159.0 | 67.2 | 183.0 | 148.0 | |||
| Ser | 328.0 | 67.2 | 332.0 | 178.0 | 180.0 | 34.5 | * | 177.0 | 45.8 | * | 164.0 | 41.8 | * | |
| Gln | 397.0 | 144.0 | 468.0 | 253.0 | 524.0 | 111.0 | 360.0 | 83.0 | 338.0 | 130.0 | ||||
| Gly | 33.1 | 10.6 | 27.5 | 9.3 | 21.6 | 1.8 | 18.9 | 7.1 | 40.2 | 10.8 | ||||
| Thr | 91.8 | 26.8 | 109.7 | 55.3 | 107.1 | 14.9 | 100.6 | 15.1 | 71.1 | 11.4 | ||||
| His | 28.6 | 22.4 | 21.1 | 16.2 | 12.0 | 7.1 | 12.6 | 4.5 | 10.4 | 4.3 | ||||
| Ala | 273.0 | 99.1 | 303.0 | 117.0 | 340.0 | 49.5 | 289.0 | 41.2 | 205.0 | 71.9 | ||||
| Arg | 50.3 | 21.2 | 36.4 | 13.9 | 19.4 | 2.6 | * | 31.4 | 13.0 | 35.3 | 16.9 | |||
| Tyr | 19.1 | 7.2 | 8.1 | 1.9 | * | 6.2 | 1.2 | * | 13.7 | 12.8 | 6.4 | 1.3 | * | |
| Val | 40.4 | 9.6 | 37.4 | 12.0 | 28.5 | 4.2 | 47.9 | 21.4 | 26.6 | 3.8 | ||||
| Met | 7.2 | 2.6 | 5.8 | 2.4 | 3.6 | 1.7 | 5.8 | 2.0 | 4.8 | 2.5 | ||||
| Trp | 118.0 | 17.8 | 98.0 | 25.4 | 77.0 | 13.7 | 82.0 | 12.3 | 43.0 | 15.9 | ||||
| Phe | 52.8 | 15.8 | 49.7 | 16.4 | 58.5 | 11.2 | 59.7 | 25.7 | 37.9 | 2.3 | ||||
| Ile | 31.4 | 8.8 | 26.6 | 7.8 | 18.5 | 4.7 | 31.6 | 11.7 | 25.2 | 1.7 | ||||
| Leu | 19.5 | 7.8 | 15.7 | 5.6 | 5.2 | 0.7 | * | 24.3 | 15.5 | 8.1 | 4.9 | |||
| Lys | 26.0 | 9.0 | 26.4 | 9.5 | 19.8 | 4.5 | 20.3 | 5.7 | 16.2 | 6.0 | ||||
| Total | 4753.0 | 828.0 | 5203.0 | 2009.0 | 4235.0 | 689.0 | 4422.0 | 495.0 | 3013.0 | 480.0 | ||||
| Roots | ||||||||||||||
| Asp | 542.0 | 63.6 | 590.0 | 149.0 | 441.0 | 60.9 | 485.0 | 50.9 | 205.0 | 14.6 | * | |||
| Glu | 1715.0 | 355.0 | 2283.0 | 484.0 | 2196.0 | 516.0 | 1274.0 | 371.0 | 723.0 | 38.6 | * | |||
| Asn | 635.0 | 337.0 | 681.0 | 311.0 | 279.0 | 42.2 | 312.0 | 201.0 | 86.0 | 12.3 | * | |||
| Ser | 186.0 | 51.9 | 228.0 | 47.4.0 | 201.0 | 59.3 | 134.0 | 27.3 | 67.0 | 8.4 | * | |||
| Gln | 1184.0 | 380.0 | 1933.0 | 717.0 | 1345.0 | 480.0 | 746.0 | 523.0 | 300.0 | 35.8 | * | |||
| Gly | 59.0 | 16.3 | 87.4 | 16.5 | 104.3 | 55.3 | 36.7 | 21.5 | 21.5 | 5.8 | * | |||
| Thr | 96.3 | 18.3 | 121.2 | 20.2 | 117.3 | 33.1 | 78.9 | 27.7 | 24.2 | 1.6 | * | |||
| His | 160.0 | 61.2 | 206.3 | 65.9 | 121.7 | 29.6 | 76.2 | 35.5 | 20.8 | 10.9 | * | |||
| Ala | 237.0 | 97.5 | 390.0 | 56.6 | 545.0 | 279.0 | 248.0 | 101.0 | 119.0 | 44.7 | * | |||
| Arg | 66.4 | 22.5 | 73.9 | 9.8 | 37.8 | 2.9 | 48.3 | 17.2 | 14.4 | 1.5 | * | |||
| Tyr | 20.0 | 7.6 | 22.7 | 3.8 | 16.2 | 5.2 | 12.7 | 3.3 | 4.4 | 0.5 | * | |||
| Val | 93.8 | 24.6 | 114.0 | 23.6 | 86.3 | 15.1 | 77.2 | 31.7 | 25.3 | 1.8 | * | |||
| Met | 37.7 | 5.5 | 38.3 | 1.9 | 31.7 | 4.0 | 25.0 | 7.0 | 18.9 | 3.5 | * | |||
| Trp | 61.5 | 23.4 | 68.8 | 5.9 | 48.8 | 14.2 | 33.5 | 11.8 | 16.2 | 11.8 | * | |||
| Phe | 56.9 | 15.1 | 63.8 | 7.3 | 59.0 | 14.5 | 34.7 | 8.8 | 18.3 | 9.4 | * | |||
| Ile | 73.6 | 39.7 | 77.6 | 10.0 | 44.6 | 6.8 | 47.9 | 13.0 | 17.2 | 8.0 | * | |||
| Leu | 72.9 | 39.4 | 64.9 | 13.4 | 35.2 | 6.1 | 54.1 | 14.9 | 12.7 | 2.6 | * | |||
| Lys | 51.3 | 12.7 | 57.1 | 8.1 | 40.2 | 9.5 | 44.6 | 16.3 | 10.4 | 2.3 | * | |||
| Total | 5565.0 | 1269.0 | 7446.0 | 2016.0 | 6188.0 | 1838.0 | 4013.0 | 1566.0 | 1780.0 | 132.0 | ||||
The Elongation Rate and Number of Roots per Plant Is Reduced in Plants with Decreased Protein Abundance of Enzymes in the Pathway of Pyrimidine de Novo Synthesis
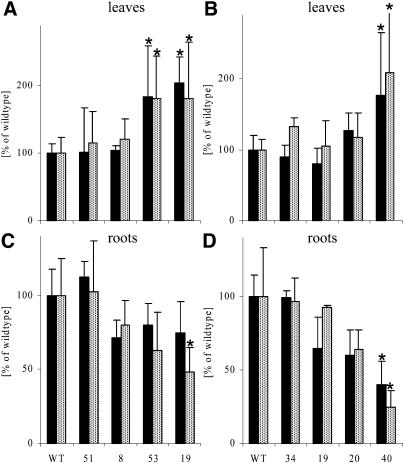
One of the marked changes in plants with reduced amounts of protein of enzymes involved in pyrimidine de novo synthesis was a reduction in growth (Fig. 4) and the decreased levels of sugars (Fig. 5) and almost unchanged levels of amino acids (Table IV) in roots. Figure 6 shows root growth in more detail in these transformants. Shoot cuttings (homogeneous shoot tips of approximately 1 to 1.5 cm length with four leaves) were grown for 7 d on vertical plates in tissue culture on Murashige and Skoog medium containing 2% Suc. Root number and root elongation rate were monitored every 12 h for at least 3 d after the first two roots had been established. A summary of the averaged elongation rates and root number at the end of the observation period is shown in Figure 6 for RBPa and ROSa transformants. Significant decreases in root elongation rates were found when <25% of the ATCase or DHOase protein remained in the transformants. When elongation rates were reduced, the total amount of roots per plant also decreased. Significant reduction in the number of roots was observed when the root elongation rate was reduced by >50%. As already found for the above-ground organs, where a reduction of ATCase or DHOase protein led to a decrease in the overall plant height, accompanied by a reduction in total area and number of leaves (Fig. 4), the size of the roots was also strongly reduced, followed by a decrease in total root number per plant.
Figure 6.
Root growth analysis of plants with reduced expression of either ATCase (RBPa) or DHOase (ROSa). A, Root elongation rate (black bars) and number of roots per plant (black bars with white dots) of RBPa plants. B, Root elongation rate (black bars) and number of roots per plant (black bars with white dots) of ROSa plants. Homogeneously looking sterile plantlets (shoot tips of approximately 1–1.5 cm length with four leaves) were transferred to 12 × 12-cm petri dishes containing Murashige and Skoog medium with 2% Suc. Six randomized plantlets were explanted in one row 2.5 cm from the top and grown vertically in dark boxes to minimize light exposure of the root system. Every 12 h the root elongation and the number of roots per plant were monitored. For better comparison, all values are given as means and sd of 15 replicates (RBPa) or six replicates (ROSa) in percentage of the wild-type amount. For assignment of the lines, please refer to Table II and Figure 3. Significant differences (P < 0.05) are marked with an asterisk. WT, Wild type.
Detailed Analysis of Leaves Reveals That Organ Size and Number Were Reduced Due to a Lower Cell Number but without a Decrease in Cell Size
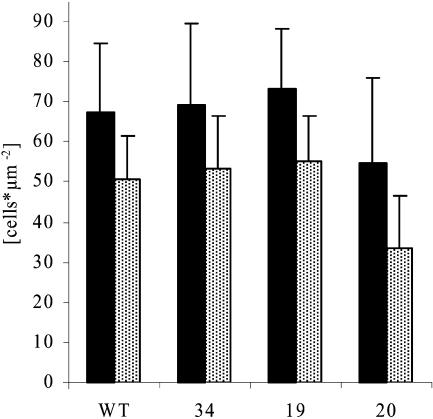
In order to look for growth differences at a cellular level, the total cell numbers and cell sizes were analyzed in the tips and bases of source leaves of ROSa transformants. To obtain standardized material, the plants were cultivated in a growth chamber on sand and watered daily with nutrient solution. After 3 weeks, when growth differences were fully established (see above), the terminal leaflet of the fourth leaf from the tip of the plant (first leaf is defined as being longer than 1 cm) was harvested, and discs were cut from the apical and basal part. As shown in Figure 7, the cell number per square centimeter was higher in the leaf base than in the leaf tip, as expected because the leaf base is a growing zone while the leaf tip is more expanded. When DHOase protein was reduced to approximately 20% of wild-type levels, the cell sizes in the tip and the base of the leaves increased, which means that the cells were slightly bigger in both regions. Together with the finding that total leaf size was also reduced (Fig. 4D), growth reduction was also due to smaller organ size because of fewer cells per organ. Comparable results were found for RBPa plants (data not shown).
Figure 7.
Cell densities of leaves from plants with reduced expression of DHOase (ROSa). Plants were grown for 3 weeks on sand and watered daily with nutrient solution in a growth chamber. Discs were cut from the apical and basal part of the terminal leaflet of the fourth leaf from the tip of the plant. All cell types per square millimeter of the leaf base (black bars) and the leaf tip (black bars with white dots) were counted after bleaching the tissue and staining of the nuclei. WT, Wild type.
DISCUSSION
Plants Expressing Antisense- or Sense-Specific cDNAs Are an Appropriate Tool for a Functional Analysis of Nucleotide de Novo Synthesis
A functional analysis of the whole pyrimidine de novo synthesis pathway was performed by using antisense and cosuppression strategies to create plants with a stepwise reduction in the expression of each single gene involved in this pathway. As discussed in the introduction, each step in this pathway is encoded by a single gene. This may explain why no homozygous Arabidopsis T-DNA insertion line in pyrimidine de novo synthesis has been described yet. So far, only the establishment of tobacco cell cultures with reduced UMPSase activity has been reported (Santoso and Thornburg, 1992, 2000). We conclude that pyrimidine de novo synthesis is an essential process, where a complete loss of one function will lead to death of the plant. For this reason, sense or antisense constructs were stably transformed into tobacco or potato plants to obtain a decrease in expression of each gene required for the pathway.
We found reduced steady-state mRNA levels of 50% to 80% in 16% to 20% of the transformants and a reduction of >80% in 2% of the transformants. Similar results were obtained, irrespective of whether the sense or antisense strategy was used. In cases where full-length constructs were used, the values for reductions caused by cosuppression using the sense strategy might be underestimated because we have not analyzed the appearance of small interfering RNAs to visualize that posttranscriptional gene silencing has already started although high levels of steady-state mRNAs are still present. The analysis of inhibition on mRNA level is also imprecise as no clear correlation of the remainder of steady-state mRNA and the remaining protein amounts exist when posttranscriptional gene silencing has started (Schubert et al., 2004). At the protein level, a reduction of 50% to 80% was found in 7% of all transformants, and in 2% of the plants, a reduction in protein level of >80% was found. These values match results from comparable studies with enzymes in pathways (Stitt and Sonnewald, 1995; Zrenner et al., 1995, 1996; Haake et al., 1998). In the case of the enzymes of the pyrimidine pathway, the relatively small reduction of transcript and protein is clearly due to an incomplete inhibition of expression because each of these genes is encoded by a single gene. The antisense strategy gave us a whole range of transformants, including both moderately and severely affected lines, and was therefore more useful than RNA interference techniques (Waterhouse et al., 1998), which usually only give plants with strong repressed expression. Plants showing intermediate reduction in expression with a progressive increase in phenotypic differences are a better tool for a more detailed analysis of physiological and developmental consequences of reduced nucleotide biosynthesis because fewer secondary or pleiotropic responses are expected to occur than in severely affected plants.
Reduced Steady-State mRNA Levels Do Not Necessarily Lead to Phenotypic Differences When Compared to Plants without Reduced Expression
A strong reduction in steady-state mRNA level could be found for all of the genes, with the exception of UMPSase. However, the impact on the plant phenotype depended upon which enzyme was inhibited. Plants with 80% reduction of transcript for both CPSase subunits and DHODH did not show any obvious phenotypic change with respect to plant growth and development. Although reduced steady-state mRNA levels are not necessarily reflected in lower protein amounts, in the case of both subunits of CPSase, reduced expression could also be monitored at the protein level. When <20% of the CPSase-specific steady-state mRNA was present, there was a reduction of 60% to 75% in CPSase-specific protein, without any visible phenotypic differences. It has been argued that expression of enzymes that are subject to complex posttranslational or allosteric regulation can often be strongly reduced without a change in pathway flux because feedback loops that decrease in vivo enzyme activity are relaxed when less protein is present (Stitt and Sonnewald, 1995; Stitt, 1999).
It is known that CPSase enzymes from various sources are regulated allosterically (Delannay et al., 1999; Simmons et al., 1999), and one might speculate that relaxation of this feedback loop compensates for the reduced amount of CPSase protein. In the case of plant DHODH, nothing is known about posttranscriptional regulation. The plant enzyme has only recently become accessible for in vitro characterization, and a significant structural difference to the animal enzymes has been reported (Ullrich et al., 2002). A suitable antibody was not available to further characterize the reduction on protein level. When 90% of the DHODH expression was abolished, no phenotypic consequences were obvious at the whole plant level. Since our previous studies strongly suggest the absence of isoforms with high homology (Giermann et al., 2002), we conclude that only 10% or less of the steady-state mRNA levels of DHODH are necessary for adequate pyrimidine de novo biosynthesis in plants grown under standard conditions. This might be due to posttranscriptional or posttranslational adjustments or to the presence of isoforms with a related dehydrogenase activity but with low sequence homology.
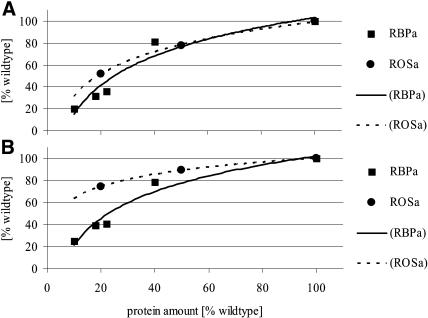
A 50% to 60% reduction in protein amount of the enzymes catalyzing the second and third steps in the pyrimidine de novo synthesis pathway (ATCase and DHOase reaction) led to a clearly visible inhibition of plant growth. Transformants were usually maintained in tissue culture for up to 3 weeks, before transferring plantlets of near identical size and appearance to sand or soil culture. After a further 3 weeks on sand culture, lines with reduced ATCase and DHOase protein abundance showed reduced fresh and dry weight accumulation, reduced height, reduced number and size of leaves, and reduced tuber yield. The qualitative effects of reduced ATCase and DHOase were essentially the same. For both enzymes, a stepwise decrease in protein level was accompanied by a concomitant appearance of phenotypical differences. When the growth rates were plotted against ATCase or DHOase protein amount, a steeper decrease in growth parameters was found for ATCase (Fig. 8). This is the enzyme that catalyzes the first unique step for pyrimidine synthesis. Interestingly, decreased expression of the enzyme that catalyzes the next step, DHOase, led to almost the same consequences for plant growth. This is another example of a protein lacking strong regulatory properties (Washabaugh and Collins, 1984) but not being expressed in large excess (Haake et al., 1998) and exerting high control over the pathway when expression is slightly reduced.
Figure 8.
Growth rate analysis of plants in relation to either ATCase-specific (RBPa) or DHOase-specific (ROSa) amount of protein. A, Total plant fresh weight increase per day. B, Leaf area increase per day. Values were calculated from the measurements shown in Figures 3 and 4. Values are given in percentage of the wild type.
Based on biochemical assays and mutant cell lines, it has previously been proposed that ATCase and UMPSase are the rate limiting steps for pyrimidine biosynthesis (Lovatt and Cheng, 1984; Santoso and Thornburg, 1998). Our results demonstrate that ATCase is not expressed in large excess, as halving of the amount of protein already limits flux to nucleotide biosynthesis, resulting in a progressive decrease in growth rates. No phenotypical differences were achieved for UMPSase, but the changes in transcript levels were also smaller for this gene than the others targeted in this study. An RNA interference strategy might be useful to obtain larger reductions in UMPSase steady-state mRNA or protein levels.
Reduced Growth Is Established as Slower Growth Rates and Is Neither Accompanied by Pleiotropic Effects nor by Changes in the Developmental Program
A gradual reduction in the ATCase or DHOase protein abundance below 50% in source leaves was linked to a consecutive reduction in plant size, total plant fresh and dry weight, and leaf area. When growth rates were calculated as fresh weight increase per day and leaf area increase per day and were normalized as percentage of the wild type, a steeper decrease in growth parameters was found with reduced abundance of ATCase protein. Tuberization of potato plants started at approximately the same time, but tuber yield per plant was reduced with decreasing abundance of ATCase or DHOase protein. The reduced growth rates with reduced final organ sizes were accompanied by a decrease in organ number but no big changes in cell size. It is known from cellular studies of tobacco that the final organ and cell size is dependent on nutrient availability (Walter and Schurr, 2005). In contrast to our plants, the reduction of final leaf size is always pronounced as a reduction of final cell size and, depending on the stage of development when the nitrogen limitation occurs, a concomitant reduction of the numbers of cells (Roggatz et al., 1999).
At the metabolite level, there were no major changes, except that carbohydrate levels in leaves were increased and carbohydrate levels in roots decreased with reduction in ATCase or DHOase protein abundance. Despite this, no obvious pleiotropic effects like chlorosis or necrosis were visible. The extractable amounts of free nucleotides in mature leaves were almost unchanged, with only a small decrease in the leaves from the strongest inhibited plants. In roots, there was an increase in nucleotide levels in the intermediate lines and a decrease in the strongest inhibited line. This change occurs for both adenine and uridine nucleotides. As purines and pyrimidines are affected in the same way, it is presumably not a direct effect of the inhibition of pyrimidine nucleotide biosynthesis but an indirect effect due to adjustments in growth and physiology. This explanation is further supported by comparable changes that were also found for amino acid levels in leaves and roots, excluding a general nitrogen limitation for growth.
The rather unaffected free nucleotide pools in fully grown tissues clearly shows that nucleotide levels are maintained in mature tissues. This finding leads to the suggestion that in mature tissue, nucleotide recycling is sufficient to run basic metabolism. It is already known that the expression of genes involved in nucleotide de novo synthesis is under a certain developmental control (Giermann et al., 2002). In addition, under standardized growth conditions, the nucleotide pools seem to be coordinated because changes occur for both adenine and uridine nucleotides. In contrast, this adjustment can be disturbed when feeding high concentrations of nucleotide precursors to mature potato tuber slices (Loef et al., 1999), but these changes might be caused by the rather unphysiological conditions.
It was reported for Arabidopsis seedlings grown in the presence of the potent ATCase transition-state analog inhibitor PALA (Bassett et al., 2003) that there is a general reduction in growth and development, accompanied by chlorosis. These findings were linked to a strong decrease in the extractable amount of total RNA and protein. In our plants with a strong reduction in the amount of ATCase protein, we did not find comparable effects on total extractable RNA or protein amounts (data not shown) and did not observe any chlorosis. Whether these differences are due to the different plant material or due to additional effects of the PALA treatment needs to be investigated further.
In summary, root growth seems to be limited by the decreased supply of carbohydrates rather than nucleotides, whereas in leaves, a small decrease in nucleotide pools is accompanied by an increase in carbohydrate content. Leaves are the organs where carbohydrate synthesis occurs and the decreased leaf growth rate might be limited by nucleotide supply. To further test this hypothesis, a more cell-specific determination of nucleotide pools is needed, especially from dividing rather than elongating or fully grown tissue. The approach should be further confirmed by looking at expression patterns of cell cycle markers, for example, but this more detailed analysis will be further complicated by the fact that nucleotide pools are strongly dependent on the developmental stage of the tissue (Meyer and Wagner, 1986).
CONCLUSION
Each single step of the pyrimidine de novo synthesis pathway exerts different control strength over the whole process. Plants with reduced pyrimidine de novo synthesis compensate for the restricted nucleotide availability with reduced growth rates, organ number, and size. In fully grown tissues, almost no reduction in nucleotide pools could be detected, indicating that nucleotide recycling is sufficient to run basic metabolism.
MATERIALS AND METHODS
Plant Materials, Bacterial Strains, and Reagents
Nicotiana tabacum L. cv SNN plants were grown in the greenhouse with supplementary light (200 μmol photons·m−2·s−1 irradiance, 23°C) under a 14-h/10-h day/night regime at 60% relative humidity in 1.5-L pots of soil or in 1.5-L pots of quartz sand culture (a 1:3 mixture of 0.3–0.8 mm and 0.6–1.2 mm diameter sand grains; Dorsolit, Mannheim, Germany). Solanum tuberosum L. cv Desirée and cv Solara plants were grown in growth chambers (350 μmol photons·m−2·s−1 irradiance, 20°C) under a 14-h/10-h day/night regime at 60% relative humidity either in 2.5-L pots of soil or in 1.5-L pots of quartz sand culture (as above). The soil mixture contained three parts Torfkultursubstrat 2 (Floragard, Oldenburg, Germany), two parts Fruhstorfer Typ T (Achut, Lauterbach-Wallenrod, Germany), and one part quartz sand (a 1:1 mixture of 0.3–0.8 mm and 0.6–1.2 mm diameter). Plants grown in soil were watered daily and fertilized once a week with 0.3% (v/v) Wuxal Super (Aglukon, Düsseldorf, Germany); plants grown in sand were watered daily with nutrient solution containing 2.5 mm Ca(NO3)2, 1 mm KNO3, 0.5 mm KH2PO4/K2HPO4 at pH 5.8, 0.5 mm MgSO4, 90 μm H3BO3, 20 μm Fe-EDTA, 20 μm MnSO4, 1.5 μm ZnSO4, 0.9 μm CuSO4, 0.6 μm NiCl, 0.45 μm HMoO4, and 30 nm CoCl2. Plants in tissue culture were grown at 100 μmol photons·m−2·s−1 at 20°C under a 16-h/8-h day/night regime at 50% relative humidity on Murashige and Skoog medium (Duchefa Biochemie, Haarlem, The Netherlands) containing 2% Suc.
Escherichia coli strain XL-1 Blue and Agrobacterium tumefaciens strain C58C1 containing pGV2260 were cultivated using standard procedures (Sambrock and Russel, 2001).
Agrobacterium-mediated gene transfer was performed as by Rosahl et al. (1989) for tobacco plants and as by Rocha-Sosa et al. (1989) for potato plants.
DNA restriction and modification enzymes were obtained from MBI Fermentas (Vilnius, Lithuania), and all other chemicals were from Sigma-Aldrich Chemical (St. Louis) or Merck (Darmstadt, Germany).
Northern Analysis
RNA was isolated, fractioned by electrophoresis, and blotted on DuralonUV (Stratagene, La Jolla, CA) as by Zrenner et al. (1995). Probes were labeled using the PCR DIG probe synthesis kit and the DIG luminescent detection kit (Roche Diagnostics, Mannheim, Germany). Hybridization was performed at 50°C overnight and filters were washed 3 × 20 min in 2× SSC, 0.1% SDS at 50°C, 55°C, and 60°C and 1 × 20 min in 0.2× SSC, 0.1% SDS at 65°C. Northern blots were normalized on total RNA amount and quantified using spot-density measurements (E.A.S.Y. Enhanced Analysis System; Herolab, Wiesloch, Germany). The sizes of some mRNAs are in the range of the ribosomal RNAs, and the signals appear more or less in two bands (Fig. 2A). For densitometric quantitation, the whole signal was measured. For the comparative analysis within one blot, pixel densities of areas of identical sizes were measured and background was subtracted. Afterward, everything was normalized on percentage of the wild-type signal.
SDS-PAGE and Immunoblotting
SDS-PAGE was performed using standard procedures. Proteins were transferred electrophoretically to Immobilon-P membrane (Millipore, Billerica, MA) and visualized using an ECL chemiluminescent kit (Amersham Bioscience, Uppsala) or SIGMA-FAST (Sigma-Aldrich Chemical). Western blots were normalized on total protein amount and quantified using spot-density measurements (E.A.S.Y. Enhanced Analysis System).
Antibody Production
Overexpression of heterologous proteins in E. coli was done with the QIAexpress system followed by His-tagged purification of the recombinant proteins using a standard procedure (Diagen, Hilden, Germany). The following protein fragments were expressed: CPSaseSU from amino acids 45 to 432, CPSaseLU from amino acids 73 to 335, and ATCase from amino acids 1 to 405 of the full-length protein. Peptide antibodies were made against the peptide 245-LGTDSAPHDRRRKEC specific for DHOase. All antibodies were produced in rabbits by Eurogentec (Liege, Belgium).
Metabolite Analysis
Sugars and starch were determined in the soluble and residual fractions of an ethanol-water extract (Jellito et al., 1992). Phosphorylated intermediates and nucleotides were extracted in trichloroacetic acid (Jellito et al., 1992) and measured as described by Stitt et al. (1989). Amino acids were determined by fluorescence after derivatization with orthophthaldialdehyde and separation by HPLC as by Geiger et al. (1998). Nucleotides were measured by HPLC according to Geigenberger et al. (1997). Amino acids and nucleotides were identified by cochromatography with authentic standards. Unpaired two-tailed t tests were used to compare pooled data from the different types of transgenic material.
Cell Densities
Leaf discs were fixed with phosphate buffer (140 mm NaCl, 2.6 mm KCl, 8 mm Na2HPO4, and 1.4 mm KH2PO4, pH 7.2) containing 10% dimethyl sulfoxide and 2% formaldehyde for 1 h at room temperature and subsequently washed three times with phosphate buffer. Bleaching of the leaf discs was performed with 90% water saturated phenol overnight at room temperature. Leaf discs were washed three times with 4 m KOH and three times with phosphate buffer before vacuum infiltration with the DNA-intercalating dye 4′,6-diamidino-2-phenylindole (in a dilution of 1:105) was performed. Detection and manual counting of the nuclei were done by image analysis with fluorescence microscopy using an Olympus BX 40 light microscope (Tokyo) with UV detection and a CCD camera (Sony XC 75; Tokyo) by sequential analysis of focal series.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AJ319873, AJ319872, AJ319874, AX093580, AX093582, and U22260.
Acknowledgments
We wish to thank Mark Stitt (Max Planck Institute of Molecular Plant Physiology, Potsdam, Germany) for his encouragement and support. We are grateful to Ralf-Michael Schmidt (BASF Plant Science, Ludwigshafen, Germany), Thomas Ehrhardt (BASF-AG), and Jens Lerchl (BASF Plant Science) for their support, Emilia Sancho-Vargas and Jolanta Pniewski (Botanisches Institut, Universität Heidelberg, Heidelberg, Germany) for technical assistance, Tina Ritter (Botanisches Institut, Universität Heidelberg) for her help with plant screening, Jana Halamova (Botanisches Institut, Universität Heidelberg) for her help with the growth analysis, Uwe Sonnewald and Andrea Knospe (Institute of Plant Genetics and Crop Plant Research, Gatersleben, Germany) for plant transformation, Josef Bergstein (Max Planck Institute of Molecular Plant Physiology) for photographic work, and Renate Schmidt and John Lunn (Max Planck Institute of Molecular Plant Physiology) for critical reading of the manuscript.
This work was supported within the framework of BioRegio by the German Federal Ministry of Education and Research (grant no. 0311626) and BASF-AG.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.063693.
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Bassett EV, Bouchet BY, Carr JM, Williamson CL, Slocum RD (2003) PALA-mediated pyrimidine starvation increases expression of aspartate transcarbamoylase (pyrB) in Arabidopsis seedlings. Plant Physiol Biochem 41: 695–703 [Google Scholar]
- Boldt R, Zrenner R (2003) Purine and pyrimidine biosynthesis in higher plants. Physiol Plant 117: 297–304 [DOI] [PubMed] [Google Scholar]
- Cervera J, Conejero-Lara F, Ruiz-Sanz J, Galisteo ML, Mateo PL, Lusty CJ, Rubio V (1993) The influence of effectors and subunit interactions on Escherichia coli carbamoyl-phosphate synthetase studied by differential scanning calorimetry. J Biol Chem 268: 12504–12511 [PubMed] [Google Scholar]
- Delannay S, Charlier D, Tricot C, Villeret V, Pierard A, Stalon V (1999) Serine 948 and threonine 1042 are crucial residues for allosteric regulation of Escherichia coli carbamoylphosphate synthetase and illustrate coupling effects of activation and inhibition pathways. J Mol Biol 286: 1217–1228 [DOI] [PubMed] [Google Scholar]
- Geigenberger P, Reimholz R, Geiger M, Merlo L, Canale V, Stitt M (1997) Regulation of sucrose and starch metabolism in potato tubers in response to short-term water deficit. Planta 201: 502–518 [Google Scholar]
- Geiger M, Walch-Liu P, Engels C, Harnecker J, Schulze E-D, Ludewig F, Sonnewald U, Scheible W-R, Stitt M (1998) Enhanced carbon dioxide leads to a modified diurnal rhythm of nitrate reductase activity in older plants, and a large stimulation of nitrate reductase activity and higher levels of amino acids in young tobacco plants. Plant Cell Environ 21: 253–268 [Google Scholar]
- Giermann N, Schröder M, Ritter T, Zrenner R (2002) Molecular analysis of de novo pyrimidine synthesis in solanaceous species. Plant Mol Biol 50: 393–403 [DOI] [PubMed] [Google Scholar]
- Guillou F, Rubino SD, Markovitz RS, Kinney DM, Lusty CJ (1989) Escherichia coli carbamoyl-phosphate synthetase: domains of glutaminase and synthetase subunit interaction. Proc Natl Acad Sci USA 86: 8304–8308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haake V, Zrenner R, Sonnewald U, Stitt M (1998) A moderate decrease of plastid aldolase activity inhibits photosynthesis, alters the levels of sugars and starch, and inhibits growth of potato plants. Plant J 14: 147–157 [DOI] [PubMed] [Google Scholar]
- Höfgen R, Willmitzer L (1990) Biochemical and genetic analysis of different patatin isoforms expressed in various organs of potato (Solanum tuberosum L.). Plant Sci 66: 221–230 [Google Scholar]
- Jellito T, Sonnewald U, Willmitzer L, Hajirezeai M, Stitt M (1992) Inorganic pyrophosphate content and metabolites in potato and tobacco plants expressing E. coli pyrophosphatase in their cytosol. Planta 188: 238–244 [DOI] [PubMed] [Google Scholar]
- Kafer C, Zhou L, Santoso D, Guirgis A, Weers B, Park S, Thornburg R (2004) Regulation of pyrimidine metabolism in plants. Front Biosci 9: 1611–1625 [DOI] [PubMed] [Google Scholar]
- Loef I, Geigenberger P, Stitt M (1999) Orotate leads to a specific increase in uridine nucleotide levels and a stimulation of sucrose degradation and starch synthesis in discs from growing potato tubers. Planta 209: 314–323 [DOI] [PubMed] [Google Scholar]
- Lovatt CJ, Cheng AH (1984) Aspartate carbamyltransferase site of end-product inhibition of the orotate pathway in intact cells of Cucurbita pepo. Plant Physiol 75: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer R, Wagner KG (1986) Nucleotide pools in leaf and root tissue of tobacco plants: influence of leaf senescence. Physiol Plant 67: 666–672 [Google Scholar]
- Moffatt BA, Ashihara H (2002) Purine and pyrimidine nucleotide synthesis and metabolism. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD. doi: 10.1199/tab.0018, http://www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed]
- Rocha-Sosa M, Sonnewald U, Frommer W, Stratmann M, Schell J, Willmitzer L (1989) Both developmental and metabolic signals activate the promoter of a class I patatin gene. EMBO J 8: 23–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roggatz U, McDonald AJS, Stadenberg I, Schurr U (1999) Effects of nitrogen deprivation on cell division and expansion in leaves of Ricinus communis L. Plant Cell Environ 22: 81–89 [Google Scholar]
- Rosahl S, Schell J, Willmitzer L (1989) Expression of a tuber-specific storage protein in transgenic tobacco plants: Demonstration of an esterase activity. EMBO J 6: 1155–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrock J, Russel DW (2001) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York
- Santoso D, Thornburg RW (1992) Isolation and characterization of UMP synthase mutants from haploid cell suspensions of Nicotiana tabacum. Plant Physiol 99: 1216–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoso D, Thornburg RW (1998) Uridine 5′-monophosphate synthase is transcriptionally regulated by pyrimidine levels in Nicotiana plumbaginifolia. Plant Physiol 116: 815–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoso D, Thornburg RW (2000) Fluoroorotic acid-selected Nicotiana plumbaginifolia cell lines with a stable thymine starvation phenotype have lost the thymine-regulated transcriptional program. Plant Physiol 123: 1517–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert D, Lechtenberg B, Forsbach A, Gils M, Bahadur S, Schmidt R (2004) Silencing in Arabidopsis T-DNA transformants: the predominant role of a gene-specific RNA sensing mechanism versus position effects. Plant Cell 16: 2561–2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons AJ, Rawls JM, Piskur J, Davidson JN (1999) A mutation that uncouples allosteric regulation of carbamyl phosphate synthetase in Drosophila. J Mol Biol 287: 277–285 [DOI] [PubMed] [Google Scholar]
- Stasolla C, Katahira R, Thorpe TA, Ashihara H (2003) Purine and pyrimidine nucleotide metabolism in higher plants. J Plant Physiol 160: 1271–1295 [DOI] [PubMed] [Google Scholar]
- Stitt M (1999) The first will be last and the last will be first: non-regulated enzymes call the tune? In JA Burrell, MM Bryant, and NJ Kruger, eds, Plant Carbohydrate Biochemistry. Bioscience Scientific Publishing, Oxford, pp. 1–16
- Stitt M, Lilley RM, Gerhardt R, Heldt HW (1989) Determination of metabolite levels in specific cells and subcellular compartments of plant leaves. Methods Enzymol 174: 518–552 [Google Scholar]
- Stitt M, Sonnewald U (1995) Regulation of metabolism in transgenic plants. Annu Rev Plant Physiol Plant Mol Biol 46: 341–368 [Google Scholar]
- Strand A, Zrenner R, Trevanion S, Stitt M, Gustafsson P, Gardeström P (2000) Decreased expression of two key enzymes in the sucrose biosynthesis pathway, cytosolic fructose-1,6-bisphosphatase and sucrose phosphate synthase, has remarkably different consequences for photosynthetic carbon metabolism in transgenic Arabidopsis thaliana. Plant J 23: 759–770 [DOI] [PubMed] [Google Scholar]
- Ullrich A, Knecht W, Piskur J, Löffler M (2002) Plant dihydroorotate dehydrogenase differs significantly in substrate specificity and inhibition from the animal enzymes. FEBS Lett 529: 346–350 [DOI] [PubMed] [Google Scholar]
- Walter A, Schurr U (2005) Dynamics of leaf and root growth: endogenous control versus environmental impact. Ann Bot (Lond) 95: 891–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washabaugh MW, Collins KD (1984) Dihydroorotase from Escherichia coli. J Biol Chem 259: 3293–3298 [PubMed] [Google Scholar]
- Waterhouse PM, Graham MW, Wang MB (1998) Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc Natl Acad Sci USA 95: 13959–13964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson CL, Slocum RD (1994) Molecular cloning and characterization of the pyrB1 and pyrB2 genes encoding aspartate transcarbamoylase in pea (Pisum sativum L.). Plant Physiol 105: 377–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zrenner R, Krause K-P, Apel P, Sonnewald U (1996) Reduction of the cytosolic fructose-1,6-bisphosphatase in transgenic potato plants limits photosynthetic sucrose biosynthesis with no impact on plant growth and tuber yield. Plant J 9: 671–681 [DOI] [PubMed] [Google Scholar]
- Zrenner R, Salanoubat M, Willmitzer L, Sonnewald U (1995) Evidence of the crucial role of sucrose synthase for sink strength using transgenic potato plants (Solanum tuberosum L.). Plant J 7: 97–107 [DOI] [PubMed] [Google Scholar]