Abstract
The actin-related proteins (ARPs) that are localized to the nucleus are present as components of various chromatin-modifying complexes involved in chromatin dynamics and transcriptional regulation. Arabidopsis (Arabidopsis thaliana) ARP7 is a constitutively expressed nuclear protein belonging to a novel plant-specific ARP class. In this study, we demonstrate a vital role for ARP7 protein in embryogenesis and plant development. Knocking out the expression of ARP7 in an arp7-1 T-DNA mutant produced morphologically aberrant, homozygous embryos that were arrested at or before the torpedo stage of development. Hence, the arp7-1 null mutation is homozygous lethal. Knocking down the expression levels of ARP7 protein with RNA interference produced viable Arabidopsis lines affected in multiple developmental pathways and induced dosage-dependent, heritable defects in plant architecture. The transgenic plants containing greatly reduced levels of ARP7 in the nucleus were severely dwarfed with small rosette leaves that are defective in cell expansion and trichome morphology. Moreover, the ARP7-deficient RNA interference plants exhibited retarded root growth, altered flower development, delayed perianth abscission, and reduced fertility. These pleiotropic phenotypic changes suggest a critical role for the Arabidopsis ARP7 protein in the regulation of various phases of plant development through chromatin-mediated, global regulation of gene expression.
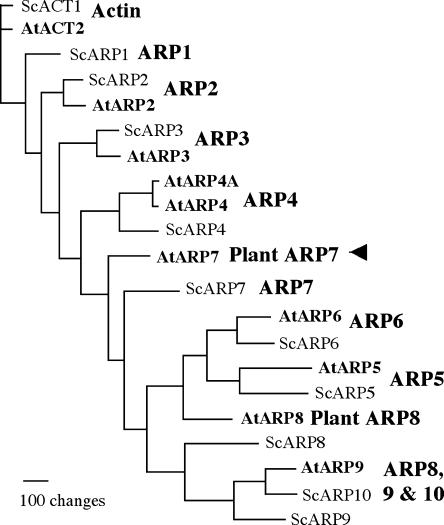
Eukaryotic cells encode a novel family of ancient and highly divergent actin-related proteins (ARPs) that share moderate sequence and structural homology with the conventional actins (Frankel and Mooseker, 1996; Schafer and Schroer, 1999). All ARPs contain the conserved ATP/ADP-binding pocket called the actin fold, but they have widely differing surface features caused by insertions, deletions, and point mutations, suggesting that they are functionally distinct from actins (Kabsch and Holmes, 1995; Mullins et al., 1996). Based on the degree of similarity to actin, ARPs have been grouped into several phylogenetic classes or subfamilies that are highly conserved among a wide range of eukaryotes, from yeast (Saccharomyces cerevisiae) to humans to plants (Goodson and Hawse, 2002). For example, the budding yeast contains 10 ARPs that are grouped into classes 1 to 10, where ARP1 is the most similar to and ARP10 is the most divergent from conventional actin (Poch and Winsor, 1997). The model plant Arabidopsis (Arabidopsis thaliana) contains nine ARP genes (ARP2–9 and ARP4A) and, based on the yeast classification system, they are grouped into eight classes (ARP2–9), as shown in Figure 1. Six of the Arabidopsis ARP classes have identifiable yeast orthologs, whereas two classes (ARP7 and 8) are distinct in plants and have no clear orthologs in yeast or other eukaryotes (McKinney et al., 2002). Moreover, all eight Arabidopsis ARP classes have orthologs in the monocot plant rice (Oryza sativa; Kandasamy et al., 2004), which has not shared a common ancestor with crucifers for more than 200 million years, suggesting that these eight classes are conserved in most higher plants.
Figure 1.
Phylogenetic relationships of Arabidopsis ARP7 to other Arabidopsis and yeast ARPs and actin. Arabidopsis (At) and yeast (Sc) sequences are compared in a neighbor-joining tree. Arabidopsis contains nine ARPs representing eight ancient ARP classes; six have homologs in yeast (ARP2-6 and ARP9), and two (ARP7 and ARP8) are unique to plants. ARP1 and ARP10 classes are missing in the Arabidopsis genome. ARP4 class in Arabidopsis has two ARP genes (ARP4 and ARP4A), whereas the remaining classes are represented each by a single ARP gene. The corresponding ARP classes are indicated to the right of each clade of ARPs. The ARP tree was constructed based on sequences cited in Kandasamy et al. (2004). The arrowhead points to Arabidopsis ARP7, the protein of interest in this study.
Orthologs of the more divergent yeast and human ARP classes 4 to 9 are localized predominantly to the nucleus (Harata et al., 2000; Goodson and Hawse, 2002). Functional and biochemical studies of the nuclear ARPs reveal that they, with various other proteins and sometimes actin, assemble into a wide variety of multiprotein complexes (e.g. SWI/SNF, RSC, INO80, SWR1, and NuA4 complexes) that are involved in the regulation of chromatin structure, transcription, and DNA repair (Cairns et al., 1998; Peterson et al., 1998; Olave et al., 2002; Shen et al., 2003; Mizuguchi et al., 2004). It has been proposed that a nucleotide-dependent, conformational change in ARP structure helps initiate the formation of these complexes (Peterson et al., 1998). Moreover, none of the nuclear ARPs are known to function outside such chromatin-modifying complexes. Thus, the biological functions of the nuclear ARPs should be considered in light of their roles as components of larger macromolecular machines. Significant advances have been made in identifying various ARP-containing multiprotein complexes and understanding the roles of their components in yeast (Cairns et al., 1998; Peterson et al., 1998; Galarneau et al., 2000; Shen et al., 2000; Mizuguchi et al., 2004). However, very little progress has been made in analyzing the functions of the nuclear ARPs or ARP complexes in multicellular eukaryotes.
Our molecular genetic characterization of Arabidopsis ARP7 provides an opportunity to address the role of a divergent ARP gene in multicellular plant development. The Arabidopsis ARP7 protein has a clear ortholog in rice that is 73% identical at the amino acid level, suggesting that orthologs to this ARP may be universally present in all higher plants (Kandasamy et al., 2004). Arabidopsis ARP7 shares 39% amino acid identity and the basal structure with a conventional actin. However, it is not clearly allied with any one yeast ARP, as it shares 23% to 27% identity with yeast ARP4, ARP5, ARP6, and ARP7. A similar relationship is seen for rice ARP7 (Kandasamy et al. 2004). Thus, ARP7 is considered to belong to a distinct, plant-specific ARP class (McKinney et al., 2002; Kandasamy et al., 2004). We have shown previously that the ARP7 protein is expressed ubiquitously in all organs and tissues and is localized predominantly to the nucleus in Arabidopsis (Kandasamy et al., 2003). In order to further uncover the function of this novel ARP protein, here we have analyzed a T-DNA null mutant and a series of RNA interference (RNAi) plant lines that showed significant and variable levels of silencing of ARP7 protein expression. Our studies demonstrate that ARP7 is an essential gene in Arabidopsis and is required for normal development of embryos and survival of plants. A substantial knockdown of its expression severely affected plant architecture and considerably delayed the abscission of floral organs. Silencing of ARP7 also induced several other morphological phenotypes, suggesting that its orthologs are involved in the regulation of multiple developmental pathways in plants. These results indicate a vital role for ARP7 protein in the control of Arabidopsis plant development, possibly through the epigenetic effects of ARP7-containing, multiprotein complexes on chromatin-mediated regulation of gene expression.
RESULTS
arp7-1 Mutation Results in Embryo Lethality
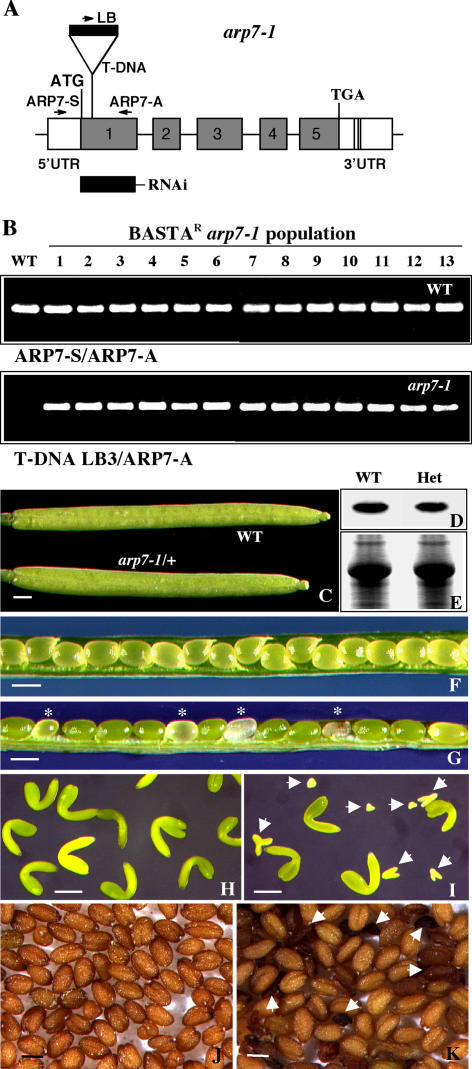
In order to understand the function of Arabidopsis ARP7, we have characterized a mutant allele, arp7-1, from Torrey Mesa Institute's T-DNA mutant collection. The arp7-1 mutation contains an insertion with a Basta resistance marker in the sixth codon of exon 1 of the ARP7 gene, as depicted in Figure 2A. PCR-based screening with the ARP7 gene-specific and mutant allele-specific primers revealed that all the Basta-resistant arp7-1 plants from T3 or T4 generation were heterozygous (Fig. 2B). Moreover, genetic analysis in a population of more than 200 plants derived from a self-pollinated heterozygous parent revealed that approximately one-third of the plants were wild type and Basta sensitive and two-thirds were heterozygous and Basta resistant. There were no homozygous arp7-1 plants in the population. All the heterozygous mutant plants were morphologically similar to wild-type plants under normal growth conditions. They developed normal roots, leaves, shoots, and flowers, which, after pollination and fertilization, produced siliques that were morphologically indistinguishable from wild-type siliques (Fig. 2C). Western-blot analysis with an ARP7-specific antibody, mAbARP7a, indicated that the heterozygous plants contained 85% to 90% of wild-type levels of ARP7 protein in the seedling (Fig. 2, D and E) or inflorescence samples (data not shown). However, examinations of seed development in heterozygous mutant siliques revealed abortion of a significant number of seeds (Fig. 2G, asterisks). Out of 1,210 seeds scored, 308 were small, white, or brown and shriveled, which comprised about 25% of the seed population. Those abnormal seeds contained small, rudimentary embryos that were arrested early during development (Fig. 2I). Siliques from wild-type plants, on the other hand, contained almost all normal seeds (only 8 out of 1,300 seeds were rudimentary) and the normal seeds contained similar-sized, fully developed green embryos (Fig. 2, F and H). Moreover, the mature dried seeds harvested from heterozygous mutants also showed a large number of shriveled, dark brown seeds (Fig. 2K). In contrast, the seeds from wild-type plants were all light brown and uniformly sized (Fig. 2J).
Figure 2.
Characterization of the arp7-1 mutant allele. A, Map of the Arabidopsis ARP7 gene indicating the location of T-DNA insertion in the arp7-1 mutant allele. The T-DNA is inserted into the first exon disrupting the sixth (Val) codon. The locations of primer sequences used for PCR genotyping of ARP7 (wild-type) and arp7-1 mutant alleles are marked with arrows. The solid rectangle (RNAi) indicates the location of the 300-bp target sequence used in a RNAi construct. B, PCR screening of the genotypes of wild-type (top) and mutant (bottom) alleles in a population of Basta-resistant arp7-1 T4 plants. The first lane represents wild-type control. C, Morphology of self-pollinated wild-type and mutant (arp7-1/+) siliques. D, Western-blot analysis of ARP7 protein in the wild-type and heterozygous mutant plants using mAbARP7a antibody. E, A duplicate gel stained with Coomassie Brilliant Blue to reveal equal loading of total protein. F, Wild-type silique with seeds. G, Heterozygous arp7-1 mutant silique with seeds. The aborting seeds are marked with asterisks. H, Embryos isolated from a wild-type silique. I, Embryos isolated from a heterozygous arp7-1 silique. Four normal embryos at the bent-curled cotyledon stage are shown along with homozygous mutant embryos (arrows) arresting at different stages of development. J and K, Mature wild-type (J) and heterozygous arp7-1 (K) mutant seeds. The arrows in K point to the aborted abnormal seeds. Scale bars = 1 mm (C, F, and G); 100 μm (H and I); and 500 μm (J and K). WT, Wild type; Het, heterozygous.
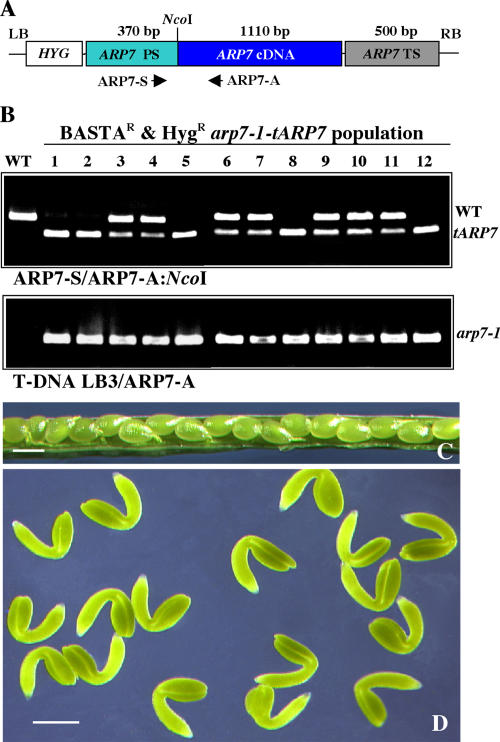
We performed genetic complementation to corroborate the homozygous lethality of the arp7-1 mutation. Plants heterozygous for the arp7-1 allele were transformed with the tARP7 transgene containing the Arabidopsis ARP7 cDNA controlled by native ARP7 regulatory sequences (Fig. 3A). Five independent T1 lines containing the tARP7 transgene were isolated, and PCR analysis indicated they were heterozygous for the arp7-1 allele (data not shown). Complementation was analyzed by two criteria. First, we used PCR assays to determine whether homozygous arp7-1/arp7-1 T2 plants could be recovered from the transgenic T1 plants. Second, we tested whether the siliques of transgenic T2 heterozygous plants contained a reduced number of aborted seeds. The sense (ARP7-S) and the antisense (ARP7-A) primers (see Fig. 3A) used in the PCR reaction amplified both wild-type ARP7 and transgene tARP7 sequences. These products were distinguished by digesting the PCR product with NcoI, a site found only in the tARP7 sequence. The results of the PCR analysis revealed that the T2 population of all the five transgenic lines contained plants homozygous for the arp7-1 mutant allele. For example, the genotypic screening of one of the complemented lines is presented in Figure 3B. Among a random sample of 12 Basta-resistant T2 mutant plants tested, five did not contain the chromosomal copy of wild-type ARP7 (1, 2, 5, 8, 12) and thus were homozygous for the mutant allele. The rest of the plants (seven) were heterozygous, having both the wild-type and transgene copies of ARP7. Moreover, the siliques of transformed heterozygous arp7-1 plants contained almost no shriveled brown seeds (Fig. 3C), and nearly all the seeds contained fully developed and uniformly sized embryos (Fig. 3D). Therefore, the homozygous recessive embryo-lethal phenotype of arp7-1 mutants can be fully complemented by the tARP7 transgene.
Figure 3.
Complementation of the homozygous arp7-1 embryo-lethal phenotype. A, Diagram of tARP7 transgene (not to scale). Homologous promoter (ARP7 PS) and terminator (ARP7 TS) sequences drive the full-length ARP7 cDNA in the transgene. Arrows indicate the orientation and position of primers. HYG, Hygromycin resistance gene; LB, left border; RB, right border. B, PCR-based screening of wild-type ARP7, tARP7 transgene, and arp7-1 mutant alleles in a Basta- and Hyg-resistant, transformed (T2) mutant population. The PCR product obtained with ARP7-S and ARP7-A primers was cut with NcoI to differentiate the chromosomal and transgene (cDNA) copies of ARP7 in the top panel. The mutant allele in the bottom image was amplified with T-DNA left border primer LB3 and the antisense primer, ARP7-A. C, Complemented heterozygous arp7-1 mutant silique with seeds. D, Mature green embryos from a transformed heterozygous arp7-1 mutant silique. Scale bars = 1 mm (C); 100 μm (D). WT, Wild type; Het, heterozygous.
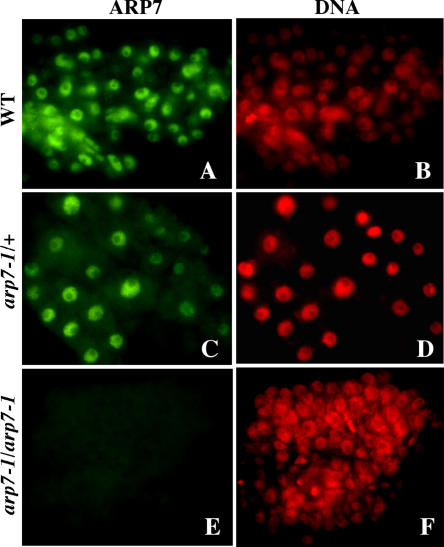
The biology of embryo lethality associated with the homozygous arp7-1 mutation was investigated more thoroughly by examining ARP7 protein expression and structural organization of normal and aberrant embryos from heterozygous siliques (Figs. 4 and 5, respectively). Immunocytochemical studies with the mAbARP7a antibody revealed that the cells of aberrant embryos did not contain detectable levels of ARP7 protein in the nucleus (Fig. 4, E and F). The cells of normal embryos in the heterozygous siliques (Fig. 4, C and D) or wild-type control embryos (Fig. 4, A and B), on the other hand, showed strong immunostaining for ARP7. The lack of ARP7 labeling in the nucleus of aborted embryos, which represent one-fourth of embryos from a heterozygous silique, suggested that they were the segregating homozygous mutant embryos.
Figure 4.
Immunolabeling of ARP7 protein in the arp7-1 mutant embryos. A and B, Wild-type control embryo. C and D, Normal embryo from a heterozygous mutant silique. E and F, Aborted embryo from a heterozygous mutant silique. Cells of embryos at the torpedo stage were used for labeling. ARP7 was labeled with mAbARP7a antibody and is shown in green (left) and DAPI staining of DNA is shown in red (right).
Figure 5.
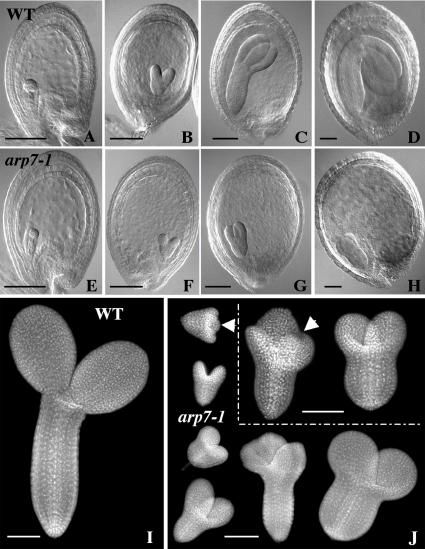
Embryo development in the arp7-1 mutant. A to H, Comparison of the development of retarded homozygous arp7-1 embryos (E–H) with normal, wild-type, or heterozygous sibling embryos (A–D). A and E, Globular stage. B and F, Heart stage. C and G, Torpedo stage. D, Bent-cotyledon stage. H, Abnormal mutant embryo arrested at the torpedo stage. Cleared seeds were examined with a light microscope equipped with Nomarski optics. I and J, Comparison of the organization of normal (I) and the aborted sibling embryos (J) from a self-pollinated, mature, green heterozygous mutant silique. Isolated embryos were stained with DAPI and observed with a fluorescence microscope. About one-fourth of seeds in each silique contained aborted embryos exhibiting abnormal morphology. The homozygous mutant embryos were arrested at various stages of development. The arrows in J point to embryos with the extra cotyledon. Scale bars = 100 μm.
To further examine the growth and morphology of mutant embryos during seed maturation, we cleared developing seeds from heterozygous plants and observed them by light microscopy using Nomarski optics, as shown in Figure 5, A to H. Morphologically abnormal seeds were compared to the normal seeds from the same silique. Observation of embryos at the globular (Fig. 5E) and heart (Fig. 5F) stages showed no significant morphological difference, except that about one-fourth of them were slightly smaller than the rest at the corresponding developmental stages (Fig. 5, A and B). However, at torpedo (Fig. 5, C and G) and bent-cotyledon (Fig. 5, D and H) stages, drastic differences in size were observed between the 75% normal, heterozygous mutant or wild-type embryos and the remaining 25% aborted, homozygous mutant embryos. The homozygous embryos were rudimentary and arrested at or before the torpedo stage of development. Despite aberrant embryo development, endosperm development, and enlargement of seeds, harboring small mutant embryos appeared normal until the late torpedo stage, after which the endosperm and the integument layers started to disintegrate. Eventually the seeds turned brownish and dried out prematurely. To study the cellular organization of these aborted embryos, we performed fluorescence microscopic analysis of 4′,6-diamino-phenylindole (DAPI)-stained embryos isolated from abnormal seeds of mature, but still green, heterozygous siliques. The results revealed that abnormal embryos were arrested at different developmental time points well before the cotyledon stage (Fig. 5J). Moreover, they exhibited uneven polar growth, resulting in irregularly shaped embryos. The aberrant embryos often contained two or three rudimentary cotyledons and stunted radicles, with either fewer or more files of cells than normal wild-type embryos, which contain two equally sized cotyledons and a cylindrical radicle with regular arrays of cells (Fig. 5, I and J).
Generation and Molecular Characterization of ARP7 RNAi Plants
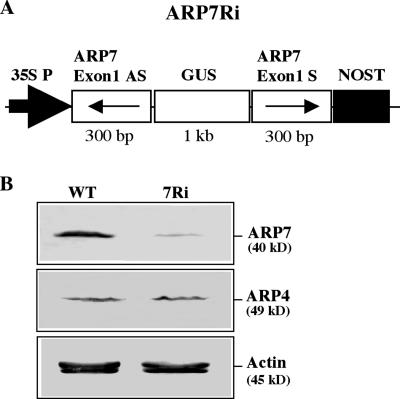
Because the only mutant allele (arp7-1) we isolated was null for ARP7 expression and embryo lethal in the homozygous state, we used RNAi to generate multiple knockdown lines that could survive and be useful in characterizing the role of this ARP protein in developmental pathways beyond early embryo development. Moreover, we used RNAi because of its high specificity, heritability, and ability to generate a phenotypic series (Chuang and Meyerowitz, 2000). ARP7 is ubiquitously expressed in all organs and cell types; therefore, we expected that a knockdown of ARP7 activity might lead to defects in overall plant development or phenotypes in various plant organs. An ARP7Ri construct was designed to express 300 bp of an ARP7-specific sequence from exon 1 in a double-stranded inverted repeat stem-loop structure to silence the expression of ARP7 mRNA (Fig. 6A). We introduced the ARP7Ri construct into wild-type Arabidopsis plants and generated 58 independent plant lines resistant to kanamycin. After transferring to the soil, the transformants were examined for silencing of ARP7 expression by western-blot analysis with ARP7-specific antibody mAbARP7a (Kandasamy et al., 2003). The results revealed that the ARP7Ri construct drastically reduced the expression of ARP7 protein in transgenic plants exhibiting visible phenotypic changes, as compared to wild type (Fig. 6B, top). On the other hand, the levels of actin or the most closely related actin-related protein, ARP4, were unaffected in the RNAi plants (Fig. 6B, bottom and middle, respectively). Thus, there was a specific knockdown of ARP7 expression in the ARP7Ri plants.
Figure 6.
RNAi-mediated silencing of ARP7 expression. A, The ARP7Ri construct contains an inverted repeat of 300 bp of ARP7-exon 1 (see Fig. 2A) as the target sequence and a 1-kb partial GUS gene as the linker sequence. The cauliflower mosaic virus-35S promoter (35S P) and nopaline synthase terminator (NOST) control expression from this construct. B, Western-blot analysis of selective knockdown of ARP7 expression. Top, middle, and bottom images were probed with mAbARP7a, mAbARP4, and mAbGPa, respectively. WT, Wild-type; 7Ri, an ARP7Ri line exhibiting distinct morphological phenotypes. Equal amounts of total protein from inflorescence samples were loaded per each lane. Note the drastic reduction in the level of ARP7, but not ARP4 or actin proteins, in the ARP7Ri plant.
The T1 generation plant lines exhibited a range of phenotypic changes that were proportional to the levels of silencing of ARP7 expression. Based on the severity of morphological phenotypes, the ARP7Ri plant lines were arranged into three groups: normal (N), moderate (M), and strong (S) RNAi lines (7Ri-N, 7Ri-M, 7Ri-S, respectively, in Fig. 7A). Out of the 58 T1 plants isolated, 15 were normal and appeared morphologically indistinguishable from the wild-type plants (Fig. 7A). Twenty-three had moderate phenotypes, with slightly reduced size, altered floral characteristics, and reduced fertility relative to wild type (Fig. 7, A and B). The remaining 20 plants were grouped as strong RNAi lines because they were severely dwarfed, highly sterile, and contained abnormal flowers and leaves (Fig. 7, A and B).
Figure 7.
Phenotypic and molecular characterization of T1 ARP7Ri plants. A, Six-week-old wild-type and ARP7Ri plants. Based on morphology, the transgenic RNAi plant lines were grouped as normal (7Ri-N), moderate (7Ri-M), or strong (7Ri-S). WT, Wild type. B to E, Dosage-dependent effects of silencing of ARP7 on flower development in ARP7Ri plants. Wild-type and strong, moderate, and normal RNAi plants were examined and are shown in the same sequence in all the figures. B, Morphology of inflorescences. C, Immunolabeling of ARP7 in pistil cells. D, Western-blot analysis of ARP7 in inflorescence samples with mAbARP7a antibody. The relative levels of ARP7 protein are indicated at the bottom of each band (see Table I for details). E, Western-blot analysis of actin on a duplicate blot probed with mAbGPa, a general anti-actin antibody.
To show that there is a correlation between the severity of the phenotypes exhibited by the RNAi plants and the levels of ARP7 protein expression, we performed western-blot analysis of protein samples from inflorescences (Fig. 7D) of different groups of T1 RNAi lines with mAbARP7a antibody. In addition, we immunolabeled chemically fixed cells from flower buds (Fig. 7C) with mAbARP7a to further verify the knockdown of ARP7 protein expression in the nuclei. The results from representative samples of wild-type and strong, moderate, and normal RNAi lines are arranged in four columns, respectively (from left), in Figure 7, B to E. The strong group of RNAi plants, which were dwarfed (Fig. 7A) and exhibited severe defects in the flower morphology (Fig. 7B), showed an 80% or greater reduction in ARP7 protein expression (Fig. 7D). Moreover, the nuclei of floral cells did not reveal any detectable labeling (Fig. 7C; second column from left). The moderate plants that showed less severe flower (Fig. 7B) phenotypes contained 25% to 50% of wild-type levels of ARP7 protein (Fig. 7D). The immunolabeling was also poor in the nuclei of flowers of moderate plants (Fig. 7C; third column from left). On the other hand, the RNAi plants that were normal and did not exhibit any noticeable plant or flower phenotype (Fig. 7, A and B) contained ARP7 protein at similar levels as the wild type (Fig. 7D). The floral cells of normal RNAi plants (fourth column) also showed strong staining for ARP7 protein in the nuclei, as did the wild-type cells (Fig. 7C; first column). Probing of duplicate blots with the general actin antibody mAbGPa, which reacts with all plant actin isovariants (Kandasamy et al., 1999), showed similar levels of total actin in all the samples (Fig. 7E). Thus, expression of the ARP7Ri transgene did not affect the levels of actin protein expression, and all the lanes were loaded with an equal amount of total protein.
Effect of ARP7 Knockdown on Vegetative Development
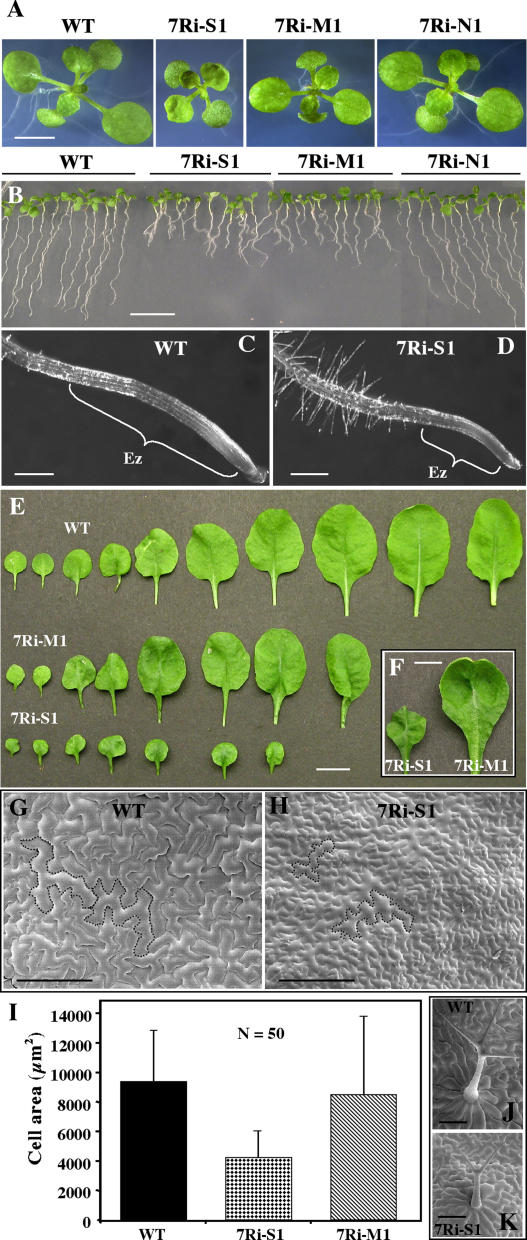
Because of extreme dwarfism and poor fertility, several of the strong RNAi lines that expressed drastically low levels of ARP7 protein were difficult to propagate and were lost during the T1 generation. However, the morphological phenotypes of the moderate and a few surviving strong lines continued into T2 and T3 generations. To explore the effect of ARP7 knockdown on different developmental pathways, we analyzed in detail the growth and morphology of T2 plants from eight of these RNAi lines representing the full range of phenotypic classes and summarized the results in Table I. Transgenic T2 plants were selected briefly on kanamycin and transferred to soil and observed for ARP7Ri-induced phenotypic changes. There was no substantial difference between the kanamycin-selected plants and those grown directly on soil and selected based on phenotype and ARP7 protein expression. Depending upon the level of reduction in the expression of ARP7 protein, as determined by quantification of protein blots analyzed with mAbARP7a, the RNAi plants showed defects in plant size and architecture from seedling to adult stages of plant development (Fig. 8A; Table I). The plants belonging to the strong group with significantly reduced ARP7 expression were dwarfed, as in the T1 generation, with only 25% to 35% of the height of wild-type plants (Table I). These RNAi plants produced severely retarded roots (Fig. 8B) with highly reduced elongation zones compared to the wild type (Fig. 8, C and D). Moreover, the strong RNAi plants contained fewer (7–8) rosette leaves than the wild type (10–12; Fig. 8E) and the leaves were small, leathery, and severely curled (Fig. 8, E and F). Scanning electron microscopic (SEM) observations revealed that the smaller leaves had a similar number of cells, but the cells were smaller with highly reduced surface area, about one-half of the wild type (Fig. 8, G–I). The leaves of dwarf plants also contained trichomes with two branches compared to three in the wild type (Fig. 8, J and K). In spite of their dwarf, altered architecture, the strong plant lines flowered at the same time as wild-type plants.
Table I.
Phenotypic analysis of the effect of knocking down Arabidopsis ARP7 expression in T2 RNAi plants
| Line | Phenotype Class | ARP7 Levela | Leaf No.b | Leaf Length ±sdc | Root Length ±sdd | Plant Height ±sdecm | Silique Length ±sd | Seed No. per Silique ±sd | Fertilityf |
|---|---|---|---|---|---|---|---|---|---|
| mm | mm | mm | |||||||
| WT | Normal | +++++ | 10–12 | 50 ± 4 | 27 ± 3 | 24.5 ± 0.8 | 13.3 ± 0.8 | 59 ± 4 | Fertile (99%) |
| 7Ri-S1 | Strong | + | 6–8 | 15 ± 3 | 9 ± 2 | 6.2 ± 1.0 | 7.4 ± 1.9 | 29 ± 9 | Highly sterile (22%) |
| 7Ri-S2 | Strong | + | 6–8 | 15 ± 3 | 10 ± 1 | 8.2 ± 1.4 | 7.7 ± 1.5 | 30 ± 10 | Highly sterile (20%) |
| 7Ri-S3 | Strong | + | 7–8 | 17 ± 3 | 11 ± 1 | 9.9 ± 1.7 | 7.8 ± 1.7 | 27 ± 13 | Highly sterile (16%) |
| 7Ri-S4 | Strong | ++ | 7–9 | 19 ± 3 | 12 ± 1 | 11.2 ± 1.6 | 7.6 ± 1.2 | 35 ± 10 | Partly sterile (32%) |
| 7Ri-M1 | Moderate | +++ | 8–10 | 24 ± 5 | 13 ± 2 | 14.8 ± 0.6 | 9.1 ± 1.0 | 37 ± 10 | Partly sterile (40%) |
| 7Ri-M2 | Moderate | +++ | 9–11 | 25 ± 4 | 13 ± 2 | 19.1 ± 1.1 | 10.6 ± 1.2 | 42 ± 9 | Partly sterile (59%) |
| 7Ri-N1 | Normal | ++++ | 10–12 | 50 ± 4 | 26 ± 2 | 23.5 ± 2.2 | 12.0 ± 0.6 | 57 ± 4 | Fertile (96%) |
| 7Ri-N2 | Normal | +++++ | 10–12 | 50 ± 3 | 28 ± 2 | 23.8 ± 1.6 | 11.9 ± 0.7 | 58 ± 5 | Fertile (98%) |
+++++, Wild-type ARP7 level; ++++, approximately 80% of wild-type ARP7 level; +++, approximately 60% of wild-type ARP7 level; ++, approximately 40% of wild-type ARP7 level; +, approximately 20% of wild-type ARP7 level, based on Western-blot analysis.
Number of rosette leaves at the time of bolting.
Average length of the two largest rosette leaves from 15 plants at the time of flowering.
Average length of roots from 15 vertically grown 9-d-old seedlings.
Average height (the tallest point of plant) of 15 plants, 5 weeks after germination.
Numbers in parentheses represent the percentage of flowers that produced siliques.
Figure 8.
Characterization of root growth and leaf phenotypes of T2 ARP7Ri plants. A, Fourteen-day-old wild-type and ARP7Ri seedlings. 7Ri-S1, 7Ri-M1, and 7Ri-N1 represent strong, moderate, and normal RNAi lines, respectively. WT, Wild type. B, Nine-day-old vertically grown seedlings. C and D, Enlarged view of root tips from wild-type (C) and a strong ARP7Ri (D) plant line. Note the difference in the length of their elongation zones (Ez). E, Rosette leaf series of wild-type and ARP7Ri plants at the time of bolting. All the leaves (except the cotyledons) from each representative plant line are shown. F, Curled rosette leaves from strong and moderate RNAi plants. G and H, SEMs of adaxial leaf surfaces of a wild-type and a strong, dwarf ARP7Ri plant. The perimeters of two representative cells are marked for each sample. I, Average area of leaf epidermal cells. Bars represent sds. J and K, Wild-type (J) and ARP7Ri (K) leaf trichomes. Scale bars = 5 mm (A, F); 10 mm (B, E); 100 μm (C, D, J, K), and 200 μm (G, H).
Plants with moderate reduction in ARP7 expression had only a modest decrease in overall plant size (Table I), but still showed severely stunted root growth (Fig. 8B) and abnormal, curved leaf morphology (Fig. 8, E and F). The moderate plants, which also bolted at the same time as the wild type, produced eight to nine rosette leaves before bolting, compared to 10 to 12 leaves for the wild type (Fig. 8E; Table I). Although the average surface area of leaf cells was only marginally reduced in the moderate plants compared to the wild type (Fig. 8I), these RNAi plants contained mixed areas of large and small cells (data not shown), which probably caused the curved leaf morphology. Transgenic T2 plants with normal levels of ARP7 expression were indistinguishable in size from wild-type plants; their architecture, leaf development, and fertility were also unaffected (see Table I).
Effect of ARP7Ri Expression on Reproductive Development
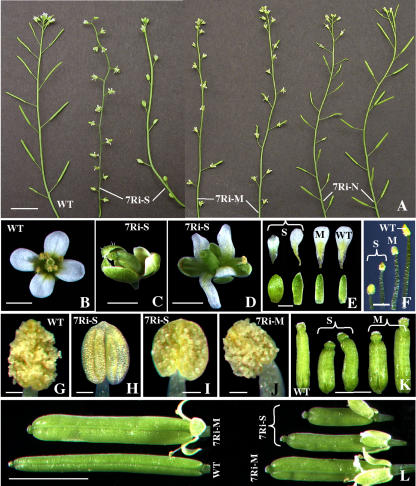
Depending upon the level of reduction in the expression of ARP7 protein, which is abundant in floral tissues (Kandasamy et al., 2003), the RNAi plants displayed a series of reproductive phenotypes as depicted in Figure 9 and Table I. In plants with strong reduction in ARP7 expression, the defects in flower organization and fertility were quite distinct and severe. In the extreme cases of the strong class of dwarf plants, their flowers never opened completely (Figs. 7B and 9C), their anthers never dehisced (Fig. 9H), and their pistils remained stunted (Fig. 9K). In addition, their petals were rudimentary and the sepals were stunted and broader (Fig. 9E, first column from left) and the stamens were shorter than the wild type (Fig. 9F). In those dwarf lines where the flowers did open, the petals were narrower and twisted compared to the wild type (Fig. 9, B, D, and E, second column from left) and the anthers produced less pollen than the wild type (compare Fig. 9, G and I). Defects in stamen morphology, along with poor pollen content, resulted in lack of efficient self-pollination, poor silique development, and reduced fertility or complete sterility (Fig. 9A). Thus, in most of the strong plant lines, only a few flowers (15%–30%) produced siliques and these ranged in size from 10% to 50% of the wild type (Fig. 9L) and contained 10 to 30 seeds per silique (Table I). The wild-type plants, on the other hand, had about 60 seeds per silique. Even severe knockdown of ARP7 in the strong RNAi plants did not affect floral organ number.
Figure 9.
Flower phenotypes of ARP7Ri plants. A, Inflorescences of wild-type and strong (7Ri-S), moderate (7Ri-M), and normal (7Ri-N) group of ARP7Ri plant lines. WT, wild type. B to D, Flowers of wild-type (B) and strong ARP7Ri (C and D) lines. E, Sepals (bottom) and petals (top) of wild-type and strong (S) and moderate (M) ARP7Ri lines. F, Stamens from wild-type and strong and moderate ARP7Ri lines at the time of anthesis. G to J, Mature anthers from just-opened flowers of wild-type (G), strong (H and I), and moderate (J) ARP7Ri lines. K, Pistils of wild-type, strong (S), and moderate (M) RNAi lines at the time of anthesis. L, Mature green siliques of wild-type and ARP7Ri plants. Each silique represents an independent RNAi line. Scale bars = 10 mm (A); 1 mm (B–E, K); 0.5 mm (F); 0.1 mm (G–J); and 5 mm (L).
The moderate group of plants displayed less severe flower phenotypes than the dwarf plants and showed partial sterility (Figs. 7B and 9A). The flowers of these plants opened normally, but only 40% to 60% of the flowers produced siliques compared to 99% in wild-type plants. The siliques on the moderate plants were wider and slightly (15%–20%) shorter than the wild type (Fig. 9L), and they contained a 30% to 40% reduction in the number of seeds per silique compared to the wild type (Table I). The morphology of the flowers, development of siliques, and fertility of the normal group of RNAi plants resembled very much that of wild-type plants (Figs. 7B and 9A; Table I).
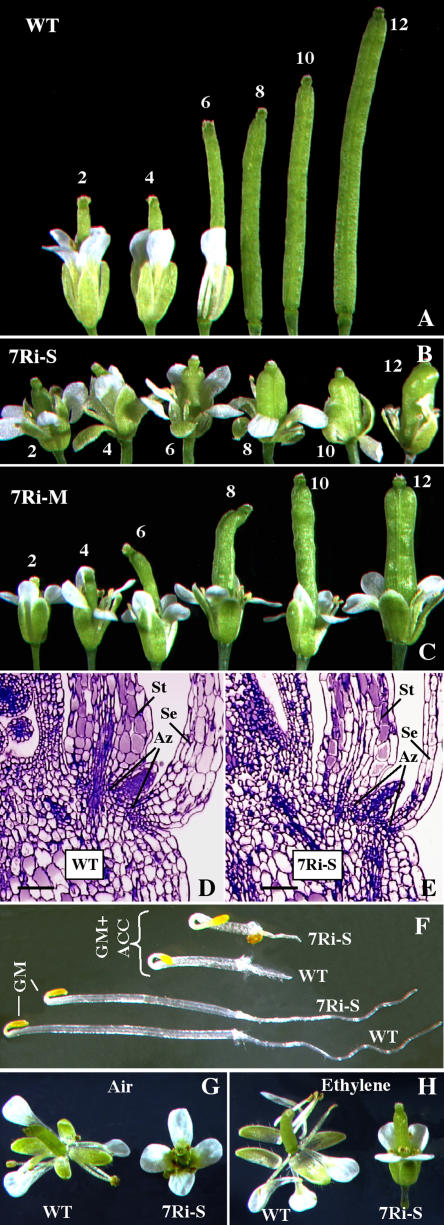
Delayed Floral Organ Abscission and Ethylene Response
In wild-type Arabidopsis, shedding of turgid floral organs, such as petals, sepals, and stamens, which no longer served a functional role, generally followed pollination and fertilization. As a consequence, in a wild-type inflorescence, only four to five opened flowers retained intact sepals and petals (Figs. 9A and 10A). On the other hand, in the ARP7 RNAi plants, especially in the strong and medium lines, there was a significant delay in the abscission of floral organs. The inflorescences of these RNAi plants typically had 12 or more flowers with intact sepals and petals regardless of whether they produce siliques following successful pollination and fertilization (Figs. 9A and 10C) or they produce no siliques due to poor pollination (Figs. 9A and 10B). Thus, in most of the RNAi plants with significant reduction in ARP7 expression, even the mature siliques retained the sepals (Fig. 9L). On the other hand, the floral organ abscission in normal plants with only minor reduction or no reduction in ARP7 expression was indistinguishable from the wild type (Fig. 9A).
Figure 10.
Delayed perianth senescence and abscission in ARP7Ri plants. A, Wild-type flowers/siliques that are consecutively numbered from fully opened flower toward the base of the inflorescence. WT, Wild type. B and C, Retention of sepals and petals in ARP7Ri flowers that are consecutively numbered from fully opened flower toward the base of the inflorescence. B and C represent strong (7Ri-S) and moderate (7Ri-M) RNAi lines, respectively. D, Longitudinal section of a wild-type flower showing the abscission zones (Az) associated with the sepal (Se) and stamen (St). Scale bar = 100 μm. E, Longitudinal section of a strong ARP7Ri flower showing abscission zones associated with the sepal and stamen. Scale bar = 100 μm. F, Triple response in wild-type and ARP7Ri seedlings. Seeds were germinated and grown for 3 d in the dark on growth medium (GM) with or without ACC (20 μm). G and H, Effect of ethylene on senescence of floral organs. Isolated just-opened wild-type and ARP7Ri flowers were exposed to 100 ppm ethylene (G) or air (H) overnight, incubated further for 66 to 72 h in growth chambers, and photographed.
To examine whether the ARP7 RNAi-induced delayed abscission is due to structural defects in abscission zones, we performed light microscopic analysis of newly opened flowers and buds just before anthesis. The size and organization of cells in the abscission zones at the base of sepals and stamens in a wild-type flower are shown in Figure 10D. Abscission zones were present at the base of sepals and stamens in strong RNAi plants and they were anatomically indistinguishable from the wild type (Fig. 10E). Petals also contained distinct abscission zones at the base (data not shown) and, in addition, moderate plants revealed similar results.
Because the hormone ethylene has been implicated in the regulation of abscission in many plants (Bleecker and Patterson, 1997; Klee, 2002), we tested whether the ARP7 knockdown plants were sensitive and responded to ethylene and whether the delayed floral organ abscission phenotype could be suppressed by exogenous application of ethylene. First, we conducted the triple-response assay by vertically growing seedlings in the dark on growth medium supplemented with 20 μm 1-aminocyclopropane-1-carboxylic acid (ACC), the natural precursor of ethylene. The results depicted in Figure 10F reveal that the ARP7 RNAi plants displayed similar morphological changes (inhibition of hypocotyl and root elongation, radial swelling of hypocotyls, and more pronounced curvature of the apical hook) to the wild type. Thus, the ARP7 RNAi seedlings are capable of perceiving and responding to ethylene. We then incubated wild-type and transgenic flowers in chambers with air or ethylene at 10 or 100 μg mL−1. The perianth organs of wild-type flowers (left) abscised in the presence of both air (Fig. 10G) and ethylene (Fig. 10H), although there was an accelerated response when exposed to exogenous ethylene. On the contrary, the perianth organs of the strong transgenic flowers were retained even after ethylene treatment at 100 μg mL−1 (Fig. 10H). Similarly, incubation of mature plants in pots containing fresh flowers with ethylene at 10 or 100 μg mL−1 also did not result in accelerated abscission of floral organs (data not shown). Thus, ARP7 RNAi plants have an ethylene-independent block of flower abscission. This suggests that ARP7 controls the floral abscission process either by an ethylene-independent developmental pathway or by acting at some point downstream of ethylene perception.
DISCUSSION
Arabidopsis ARP7 Controls Multiple Pathways of Plant Development
ARP7 is a novel and highly divergent member of the Arabidopsis ARP family. Our previous molecular and immunocytochemical studies demonstrate that it is a nuclear protein expressed ubiquitously in all cell types throughout the plant (Kandasamy et al., 2003). Here, we have shown through genetic analysis of a null mutant allele arp7-1 that ARP7 is an essential gene and is required for normal embryogenesis in Arabidopsis. The growth of homozygous mutant embryos that contained no ARP7 protein in the nucleus was blocked at the heart or torpedo stage of development. Therefore, one-fourth of the seeds in the siliques of self-fertilized heterozygous mutants aborted before maturity and no homozygous arp7-1 plant survived in a large mutant population raised from the seeds of heterozygous parents. We also found that the homozygous embryo-lethal phenotype of arp7-1 mutant plants could be rescued by complementation with a transgene containing the ARP7 cDNA sequence. Thus, in the presence of the tARP7 transgene, self-pollinated heterozygous mutant plants produced siliques fully filled with fertile seeds, and the complemented homozygous mutant embryos developed into healthy adult plants that are indistinguishable from the wild type. This confirms that ARP7 is crucial for regular development of embryos and survival of plants.
Moreover, we applied the technique of RNAi to specifically knockdown the expression of ARP7 protein, and showed that an adequate level of this protein was essential for multiple aspects of plant growth and development. Consistent with the constitutive expression pattern of ARP7 (Kandasamy et al., 2003), silencing of its expression altered the overall organization of the plants, affected the development of different organs such as leaves, flowers, and siliques, and, in addition, greatly reduced cell expansion, root growth, and fertility of the plants. All these developmental processes, which are affected in the RNAi plants deficient in ARP7 protein, require the regulated expression of many genes. In plants, epigenetic modifications of chromatin, which regulates transcription, have been suggested to play a key role in the control of many of the above-mentioned developmental pathways (Berger and Gaudin, 2003). Because the nuclear ARP-containing multiprotein complexes have been implicated in chromatin remodeling, knocking down ARP7 expression might have affected various aspects of plant development through the regulation of chromatin dynamics and global regulation of gene expression.
It is striking that knocking down ARP7 expression also affected the floral abscission process in Arabidopsis. RNAi plants with markedly reduced levels of ARP7 showed a significant delay in the abscission of sepals and petals, even though they contained normal abscission zones at their base. The delayed abscission often resulted in inflorescences that were fully covered with flowers, even in lines that showed moderate silique development following pollination and fertilization. Flower senescence, like whole-plant senescence, is an active process that is hormone mediated and executed via a defined genetic program (van Doorn and Stead, 1997; Chang et al., 2003). The hormone ethylene has been suggested to play a central role in the regulation of flower senescence and abscission of floral organs in many plants (Bleecker and Patterson, 1997; Roberts et al., 2000; Klee, 2002). However, exogenous application of ethylene did not suppress the delayed floral organ abscission in the Arabidopsis ARP7 RNAi plants, which are otherwise sensitive to ethylene, as demonstrated by the triple response of seedlings. This suggests that ARP7 is involved in an ethylene-independent pathway controlling floral organ abscission in Arabidopsis. This pathway should involve proteins that influence abscission, but not ethylene sensitivity. Recent reports indicate that misregulation or silencing of proteins like AGL15, a MADS domain factor (Fang and Fernandez, 2002), HAESA, a receptor-like protein kinase (Jinn et al., 2000), and INFLORESCENCE DEFICIENT ABSCISSION (IDA) in a putative ligand (Butenko et al. 2003), also affects abscission of floral organs without affecting ethylene response, like ARP7. It will be interesting to see whether the expression of any of these proteins is altered in the ARP7 knockdown plants. On the contrary, ARP7 may be required for some developmental steps in floral senescence acting downstream of ethylene perception. Our future analysis of transcriptional arrays should reveal the target genes regulated by ARP7.
Possible Functions of ARP7 in Chromatin-Mediated Gene Regulation and Plant Development
In Arabidopsis, distinct facets of embryogenesis and plant growth and development are regulated through the highly coordinated expression of interconnected networks of genes that include different chromatin-remodeling factors (Wagner, 2003). The aberrant development of homozygous arp7-1 embryos due to a null mutation in the ARP7 gene and the pleiotropic effects of knocking down ARP7 protein in several independent RNAi plant lines suggest that this protein controls multiple gene networks affecting plant development. In yeast, several of the nuclear ARPs are essential genes, and their encoded proteins are present as components of various multiprotein complexes that are implicated in key roles in the regulation of chromatin structure and transcription (see Blessing et al., 2004). For example, yeast ARP7 and ARP9 are important subunits of ATP-dependent chromatin-remodeling complexes SWI/SNF and RSC (Cairns et al., 1998; Peterson et al., 1998). Also, the human SWI/SNF complex contains BAF53, which is a homolog of yeast ARP4 (Zhao et al., 1998), a close relative of the yeast ARP7. Moreover, the other yeast chromatin-modifying complexes INO80 and SWR1 contain three (ARP4, ARP5, and ARP8; Shen et al., 2000), and two different (ARP4 and ARP6) ARPs (Mizuguchi et al., 2004), respectively. ARP4 is also present as an integral component of histone acetyltransferase complexes such as yeast NuA4 (Galarneau et al., 2000) and human TIP60 (Ikura et al., 2000), which both acetylate histones. Therefore, all the nuclear ARPs studied in detail so far in yeast and humans are present as components of one or more multiprotein chromatin-remodeling complexes. Although we understand little about the function of the nuclear ARPs within various complexes, genetic and biochemical studies in yeast point to structural and enzymatic roles for ARPs in chromatin-mediated gene regulation (Jiang and Stillman, 1996; Cairns et al., 1998; Galarneau et al., 2000; Shen et al., 2003; Szerlong et al., 2003). Mutations in several plant genes with predicted roles in chromatin remodeling produce alterations in multiple developmental pathways (Verbsky and Richards, 2001; Goodrich and Tweedie, 2002; Wagner, 2003). Because of its localization to the nucleus at the transcriptionally active interphase stage of the cell cycle (Kandasamy et al., 2003), Arabidopsis ARP7 is likely to participate as a component of chromatin-remodeling complexes and to play a role in transcriptional regulation similar to the ARP counterparts in other kingdoms. In fact, our size exclusion chromatographic analysis revealed that most of ARP7 protein present in leaf and inflorescence samples exists as part of macromolecular assemblies of greater than 500 kD (R.B. Deal, M.K. Kandasalmy, and R.B. Meagher, unpublished data). Considering the pleiotropic effects of silencing ARP7 in Arabidopsis, it is reasonable to propose that knocking down ARP7 protein has a global effect on transcription through its activity in several multiprotein complexes. Thus, we suggest that silencing ARP7 interferes with the expression of genes controlling various developmental pathways and thereby effects changes in the morphology of different organs throughout the life of plants.
Although the characterization of the arp7-1 null mutant facilitated our understanding of the vital role of ARP7 during embryogenesis, analysis of an RNAi series, which was viable and showed dosage-dependent pleiotropic phenotypes, helped us to demonstrate the importance of this ARP in the regulation of various aspects of plant development. Even in unicellular yeast, some of the nuclear ARPs are identified as a component of more than one chromatin-modifying complex (e.g. ARP4 in INO80, SWR1, NuA4, and TIP60 complexes). From our studies, it seems logical to propose that plant ARP7 proteins may be part of several distinct chromatin-remodeling complexes, which act in various developmental pathways via global changes in gene regulation. Genomic studies reveal that plants contain several dozen homologs of SWI/SNF ATPases (e.g. PKL [Ogas et al., 1999]; DDM1 [Jeddeloh et al., 1999]; SYD [Wagner and Meyerowitz, 2002]) that are thought to be involved in the chromatin-mediated control of gene expression during plant development (Verbsky and Richards, 2001). Therefore, the presence of multiple forms of SWI/SNF-like complexes containing ARP7 in different tissues and cell types in plants is highly possible. Our recent analysis of RNAi-induced knockdown of Arabidopsis ARP4 revealed morphological phenotypes both similar to and entirely different from those of ARP7. For example, silencing the expression of ARP4, but not ARP7, induced early flowering. On the other hand, knocking down the expression of either ARP4 or ARP7 proteins delayed senescence and abscission of sepals and petals (Kandasamy et al., 2005). These results suggest that both of these ARPs may exist as parts of the same and different chromatin-modifying complexes in plants. However, very little progress has been made in the characterization of the SWI/SNF and other chromatin-remodeling complexes in plants. Future functional studies, such as identification of the proteins within various ARP-containing complexes and the target genes that are altered in their expression by these complexes, will help us understand the crucial role of ARP7 orthologs in plant multicellular development.
MATERIALS AND METHODS
Plant Materials
The arp7-1 mutant allele of Arabidopsis (Arabidopsis thaliana) in a Columbia genetic background was isolated by searching the sequences of the T-DNA insertion lines provided by Torrey Mesa Research Institute. Wild-type and mutant seeds or seeds from transgenic plant lines expressing the ARP7Ri construct were surface sterilized, plated on plant growth medium (Murashige and Skoog, 1962) supplemented with 1% Suc, incubated at 4°C for 2 d, and grown at 22°C under a 16-h photoperiod. For examining the root phenotype, seedlings were grown vertically in plates without any selection. Seeds for soil-grown plants were sown on moist soil, chilled at 4°C for 2 d, and then germinated and grown in growth chambers maintained under the same growth conditions described above. The Basta-resistant mutant plants were selected by spraying them with the herbicide Finale (AgrEvo, Montvale, NJ) diluted with water at 240 μg/mL concentration.
Ethylene Responses
To examine the triple response, seeds from wild-type and ARP7Ri plant lines were surface sterilized, planted on growth medium supplemented with or without the ethylene precursor ACC (20 μm; Sigma, St. Louis) and cold treated at 4°C in the dark for 3 to 4 d. The seeds were then germinated and grown vertically in the dark at 22°C and the responses of the seedlings were observed after 72 h. To study the effect of ethylene on abscission of floral organs, whole plants or isolated flowers placed on growth medium were exposed to 10 or 100 μg mL−1 of ethylene for 16 h in Mason jars as described in Weigel and Glazebrook (2002). Following further incubation in growth chambers, the flowers on plates were observed after 3 to 4 d and the flowers attached to plants were monitored periodically up to 10 d.
Plasmid Constructs and Plant Transformation
RNAi
To generate plant lines that exhibit varying levels of silencing of ARP7 expression, an ARP7Ri construct was produced using an inverted repeat-PCR gene assembly technique (L. Pawloski and R.B. Meagher, unpublished data). This construct consisted of a 300-bp segment of ARP7 exon 1 in the antisense orientation, a 1-kb partial GUS gene as the linker sequence, and the same 300-bp segment of ARP7 in the sense orientation (Fig. 6A). The exon 1 nucleotide sequence used is unique in the Arabidopsis genome, as it contains no sequence homology of more than 20 nucleotides with any other gene. Therefore, the ARP7Ri construct is predicted to specifically target the ARP7 mRNA. The ARP7 sequence was amplified with two separate primer sets such that one product contained a region homologous to the upstream end of the GUS linker while the other contained a region homologous to the downstream end of the linker. The two products were assembled with the GUS linker in an overlap-extension PCR to generate the inverted repeat structure shown in Figure 6A. The ARP7 inverted repeat assembly was then placed under the control of the cauliflower mosaic virus-35S promoter and nopaline synthase terminator in pBIN19 containing the kanamycin resistance marker to make the ARP7Ri gene construct for plant transformation.
Complementation
To complement the embryo-lethal phenotype of the arp7-1 mutant, we made a transgene construct containing homologous ARP7 regulatory and coding sequences. A 370-bp 5′-untranslated region (UTR) and promoter sequence of ARP7 was PCR amplified from the bacterial artificial chromosome clone T4C21 (Arabidopsis Biological Resource Center) and cloned into the SalI-EcoRV restriction sites of the Bluescript KS (Stratagene, La Jolla, CA) multilinker. A 500-bp region containing the 3′-UTR and terminator sequences of ARP7 was then PCR amplified from bacterial artificial chromosome T4C21 and cloned into the BamHI-SacI restriction sites of the ARP7 promoter-Bluescript KS construct to generate the ARP7pt expression vector. In the next step, the 1,100-bp cDNA of ARP7 was amplified from an existing ARP7cDNA clone (McKinney et al., 2002) and inserted into the NcoI-BamHI sites of the ARP7pt vector. The complete 1,970-bp ARP7pt:ARP7 cDNA assembly in Bluescript KS was then cut with SalI-SacI and cloned into the replacement region of pCambia binary vector carrying a hygromycin (Hyg) resistance marker (Hajdukiewicz et al., 1994). The resulting construct, named tARP7, was transformed into the Agrobacterium strain C58C1 and used for the transformation of arp7-1 mutant plants.
Plant Transformation
To generate transgenic plant lines expressing the ARP7Ri construct, wild-type plants were transformed by the Agrobacterium-mediated vacuum infiltration method as described in Weigel and Glazebrook (2002). Transgenic plants were selected on 0.5× Murashige and Skoog medium (Murashige and Skoog, 1962) containing 35 mg/L kanamycin. After brief selection, the kanamycin-resistant T1 plants were transferred to soil and grown in growth chambers at 22°C with 16-h light and 8-h dark photoperiods. To check complementation of the embryo-lethality phenotype, heterozygous arp7-1 mutant plants were transformed with the tARP7 construct. The transformed arp7-1 mutant plants (T1) were selected by plating the seeds harvested from vacuum-infiltrated plants on Murashige and Skoog medium containing 50 mg/L Hyg. After transferring to the soil, the Hyg seedlings were sprayed with herbicide to identify transgenic plants that contained the mutant allele. These T1 plants were allowed to self and produce T2 seeds that were then germinated and selected as above to isolate Hyg-Basta mutant transgenic plants. PCR was performed on those T2 plants to determine whether homozygous mutants with the complementing transgene exist in the population.
Determination of Genotypes of the Mutants by PCR
Mutant and wild-type allele-specific primers were designed to PCR amplify regions of the ARP7 (wild type), arp7-1, or tARP7 transgene. A sense primer ARP7-S (5′-TCACTGACTTGCTGCTACCACTAA-3′) and an antisense primer ARP7-A (5′-TTAGGAGTACAAAGTGGATCTGTA-3′) were used to amplify approximately a 400-bp fragment of the wild-type allele. Approximately a 400-bp fragment of the mutant allele was amplified with a left-border T-DNA primer LB3 (5′-TAGCATCTGAATTTCATAACCAATCTCGATACAC-3′) and the antisense primer ARP7-A. The PCR product of the mutant allele was sequenced to confirm the exact location of T-DNA insertion. More than 100 plants each from T3 and T4 generations and a population of more than 200 F1 plants derived from selfed heterozygous parents were genotyped by PCR to verify the genetics and homozygous lethal phenotype of the mutant. While screening the complemented mutants, the PCR product of the wild-type allele was cut with Ncol to differentiate the native gene product from that of the transgene, which has an Nco1 site at the junction between the 5′-UTR and the coding sequence (Fig. 3A). Therefore, after digestion, the heterozygous mutant plants are predicted to have wild-type and tARP7 gene products, and the homozygous mutants have only the transgene product (Fig. 3B). The DNA used as template for genotype screening was prepared by a modified rapid alkali DNA prep method described previously (Gilliland et al., 1998).
Protein-Blot Analysis
Total proteins from inflorescence or leaf samples of Arabidopsis arp7-1 mutants or ARP7Ri plants were prepared as described previously (Kandasamy et al., 1999). Proteins were separated on 10% SDS-PAGE gels and transferred to Immobilon polyvinylidene difluoride membrane (Millipore, Bedford, MA) or nitrocellulose (Bio-Rad, Hercules, CA) by semidry blotting (Hofer, San Francisco, CA). Duplicate gels were stained with Coomassie Brilliant Blue to check equal loading. After blocking for 30 min to 1 h in Tris-buffered saline plus Tween (TBST; 10 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.05% Tween 20) containing 10% goat serum and 5% dry milk, the membrane was probed with the primary antibody (0.5 μg/mL) diluted in the blocking solution for 1 h. The membrane was then washed in TBST and probed with horseradish peroxidase or alkaline phosphatase-conjugated secondary antibody (Amersham, Piscataway, NJ) diluted in blocking solution at 1:2,000 or 1:7,000, respectively, for 30 min. The blots were washed again, treated with ECL detection solution (Amersham) for 2 min, and exposed to Hyperfilm ECL (Amersham) to visualize the protein bands on the film, or treated with the alkaline phosphatase substrate buffer (Kandasamy et al., 1999) containing nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate (Sigma) to detect protein bands directly on the membrane. Identical blots were probed with mAbARP7a, an ARP7-specific monoclonal antibody, mAbARP4, an ARP4-specific antibody (Kandasamy et al., 2003), or mAbGPa, a general actin monoclonal antibody (Kandasamy et al., 1999), to detect the expression of ARP7, ARP4, and actin proteins, respectively. After scanning the processed film or membrane, protein bands were quantified using the National Institutes of Health (NIH) SCI Image program.
Microscopy
For Nomarski microscopy of developing seeds, wild-type and heterozygous mutant siliques were fixed in 4% paraformaldehyde in 50 mm PIPES buffer, pH 7.0, for 1 h and cleared with 80% (w/v) chloral hydrate in 10% glycerol overnight. The cleared seeds were then observed with a Zeiss (Jena, Germany) microscope, equipped with Nomarski optics, and photographed using a Hamamatsu (Shizuoka, Japan) digital camera (C-4742-95).
To analyze the cellular organization, normal and aborted mutant embryos were isolated from developing seeds, fixed for 30 min to 1 h in 4% paraformaldehyde containing traces of Triton X-100, stained with DAPI, and observed with a Zeiss fluorescence microscope. Morphological phenotypes of flowers, embryos, and other plant parts were characterized using a stereomicroscope fitted with a Hamamatsu color-chilled 3CCD camera (C-5810).
Light microscopic observations of the floral abscission zone were made on wild-type and ARP7Ri flower buds just before anthesis or newly opened flowers that were fixed in 3% glutaraldehyde, postfixed in 1% osmium tetroxide, and embedded in Spurr's resin as described before (Kandasamy and Kristen, 1987). One-micron-thick sections were stained with toluidine blue and observed with a Zeiss light microscope.
SEM of the adaxial leaf surfaces of wild-type and ARP7Ri plant lines were made on cryopreserved samples from at least two independent plants each. Identical leaf pieces were placed on a cryotransfer holder covered with a thin layer of Tissue Tek mixed with conductive carbon dust and rapidly plunged into liquid nitrogen slush (approximately 206°C) to avoid ice crystallization. The samples were then transferred to a Gatan Alto 2500 Cryoprep chamber (Gatan, Pleasanton, CA) kept at −110°C, and sublimated for 15 min at −90°C to remove any remaining surface water. After coating with gold-palladium (approximately 25-nm thickness), the samples were transferred to the cryostage in the field emission SEM (LEO Electron Microscopy, Thornwood, NY) and kept at −110°C while viewing. The surface area of 50 epidermal cells for each sample was measured from the SEM images using the ImageJ program (NIH Image), and the average values were calculated and plotted using Microsoft Excel.
For immunocytochemistry, embryos or root samples were fixed with 4% paraformaldehyde in pectin methylesterase (50 mm PIPES buffer, pH 7.0, 5 mm EGTA, 1 mm MgSO4, 0.5% casein) containing a protease inhibitor cocktail (Roche, Mannheim, Germany) for 1 h at room temperature. Following washing in pectin methylesterase, the samples were permeabilized and partially dissociated by treating with 1% cellulysin (Calbiochem, San Diego) and 0.1% pectolyase (Sigma) for 60 min. Further processing of the tissue for immunolabeling was done as described earlier (Kandasamy et al., 1999). In brief, the slides containing dissociated tissue were blocked with TBST-bovine serum albumin-glutamine synthetase (5% bovine serum albumin, 20% goat serum) for 1 to 2 h, incubated in the primary antibody mAbARP7a (2.5 μg/mL) overnight, and in fluorescein isothiocyanate-conjugated (Sigma) anti-mouse secondary antibody (1:100) for 3 to 4 h. Labeling was done at room temperature and the antibodies were diluted with the blocking solution. After washing in phosphate-buffered saline (PBS), the slides were incubated in DAPI (0.1 μg/mL PBS; Sigma) for 10 min, washed again in PBS, and mounted with 80% glycerol in PBS containing 1 mg/mL p-phenylenediamine (Sigma). Observations were made with a Zeiss fluorescence microscope equipped with Improvision Openlab software (Improvision, Lexington, MA).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number BK000423.
Acknowledgments
We thank Dr. Stanley J. Kays and Ms. Betty P. Schroeder of the University of Georgia (UGA) Horticulture Department for their help with ethylene treatment, Ms. Elizabeth A. Richardson of the UGA Plant Biology Electron Microscopy Lab for the use of the light microscopy facility, Dr. Donald R. Roberts of the UGA Center for Ultrastructural Research for his help with scanning electron microscopy, and Ms. Gay Gragson for critical reading of the manuscript.
This work was supported by funds from the National Institutes of Health (GM 36397–18).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.065326.
References
- Berger F, Gaudin V (2003) Chromatin dynamics and Arabidopsis development. Chromosome Res 11: 277–304 [DOI] [PubMed] [Google Scholar]
- Bleecker A, Patterson SE (1997) Last exit: senescence, abscission, and meristem arrest in Arabidopsis. Plant Cell 9: 1169–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blessing CA, Ugrinova GT, Goodson HV (2004) Actin and ARPs: action in the nucleus. Trends Cell Biol 14: 435–442 [DOI] [PubMed] [Google Scholar]
- Butenko MA, Patterson SE, Grini PE, Stenvik G-S, Amundsen SS, Mandal A, Aalen RB (2003) INFLORESCENCE DEFICIENT IN ABSCISSION controls floral organ abscission in Arabidopsis and identifies a novel family of putative ligands in plants. Plant Cell 15: 2296–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns BR, Erdjument-Bromage H, Tempst P, Winston F, Kornberg RD (1998) Two actin-related proteins are shared functional components of the chromatin-remodeling complexes RSC and SWI/SNF. Mol Cell 2: 639–651 [DOI] [PubMed] [Google Scholar]
- Chang H, Jones ML, Banowetz GM, Clark DG (2003) Overproduction of cytokinins in petunia flowers transformed with P(SAG12)-IPT delays corolla senescence and decreases sensitivity to ethylene. Plant Physiol 132: 2174–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang CF, Meyerowitz EM (2000) Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc Natl Acad Sci USA 97: 4985–4990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang SC, Fernandez DE (2002) Effect of regulated overexpression of the MADS domain factor AGL15 on flower senescence and fruit maturation. Plant Physiol 130: 78–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel S, Mooseker MS (1996) The actin-related proteins. Curr Opin Cell Biol 8: 30–37 [DOI] [PubMed] [Google Scholar]
- Galarneau L, Nourani A, Boudreault AA, Zhang Y, Heliot L, Allard S, Savard J, Lane WS, Stillman DJ, Cote J (2000) Multiple links between the NuA4 histone acetyltransferase complex and epigenetic control of transcription. Mol Cell 5: 927–937 [DOI] [PubMed] [Google Scholar]
- Gilliland LU, McKinney EC, Asmussen MA, Meagher RB (1998) Detection of deleterious genotypes in multigenerational studies. I. Disruptions in individual Arabidopsis actin genes. Genetics 149: 717–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich J, Tweedie S (2002) Remembrance of things past: chromatin remodeling in plant development. Annu Rev Cell Dev Biol 18: 707–746 [DOI] [PubMed] [Google Scholar]
- Goodson HV, Hawse WF (2002) Molecular evolution of the actin family. J Cell Sci 115: 2619–2622 [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25: 989–994 [DOI] [PubMed] [Google Scholar]
- Harata M, Oma Y, Tabuchi T, Zhang Y, Stillman DJ, Mizuno S (2000) Multiple actin-related proteins of Saccharomyces cerevisiae are present in the nucleus. J Biochem (Tokyo) 128: 665–671 [DOI] [PubMed] [Google Scholar]
- Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y (2000) Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102: 463–473 [DOI] [PubMed] [Google Scholar]
- Jeddeloh JA, Stokes TL, Richards EJ (1999) Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat Genet 22: 94–97 [DOI] [PubMed] [Google Scholar]
- Jiang YW, Stillman DJ (1996) Epigenetic effects on yeast transcription caused by mutations in an actin-related protein present in the nucleus. Genes Dev 10: 604–619 [DOI] [PubMed] [Google Scholar]
- Jinn TL, Stone JM, Walker JC (2000) HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes Dev 14: 108–117 [PMC free article] [PubMed] [Google Scholar]
- Kabsch W, Holmes KC (1995) The actin fold. FASEB J 9: 167–174 [DOI] [PubMed] [Google Scholar]
- Kandasamy MK, Deal RB, McKinney EC, Meagher RB (2004) Plant actin-related proteins. Trends Plant Sci 9: 196–202 [DOI] [PubMed] [Google Scholar]
- Kandasamy MK, Deal RB, McKinney EC, Meagher RB (2005) Silencing the nuclear actin-related protein AtARP4 in Arabidopsis has multiple effects on plant development, including early flowering and delayed floral senescence. Plant J 41: 845–858 [DOI] [PubMed] [Google Scholar]
- Kandasamy MK, Kristen U (1987) Developmental aspects of ultrastructure, histochemistry and receptivity of the stigma of Nicotiana sylvestris. Ann Bot (Lond) 60: 427–437 [Google Scholar]
- Kandasamy MK, McKinney EC, Meagher RB (1999) The late pollen-specific actins in angiosperms. Plant J 18: 681–691 [DOI] [PubMed] [Google Scholar]
- Kandasamy MK, McKinney EC, Meagher RB (2003) Cell cycle-dependent association of Arabidopsis actin-related proteins AtARP4 and AtARP7 with the nucleus. Plant J 33: 939–948 [DOI] [PubMed] [Google Scholar]
- Klee HJ (2002) Control of ethylene-mediated processes in tomato at the level of receptors. J Exp Bot 53: 2057–2063 [DOI] [PubMed] [Google Scholar]
- McKinney EC, Kandasamy MK, Meagher RB (2002) Arabidopsis contains ancient classes of differentially expressed actin-related protein genes. Plant Physiol 128: 997–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C (2004) ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303: 343–348 [DOI] [PubMed] [Google Scholar]
- Mullins RD, Kelleher JF, Pollard TD (1996) Actin' like actin? Trends Cell Biol 6: 208–212 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15: 473–497 [Google Scholar]
- Ogas J, Kaufmann S, Henderson J, Somerville C (1999) PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc Natl Acad Sci USA 96: 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olave IA, Reck-Peterson SL, Crabtree GR (2002) Nuclear actin and actin-related proteins in chromatin remodeling. Annu Rev Biochem 71: 755–781 [DOI] [PubMed] [Google Scholar]
- Peterson CL, Zhao Y, Chait BT (1998) Subunits of the yeast SWI/SNF complex are members of the actin-related protein (ARP) family. J Biol Chem 273: 23641–23644 [DOI] [PubMed] [Google Scholar]
- Poch O, Winsor B (1997) Who's who among the Saccharomyces cerevisiae actin-related proteins? A classification and nomenclature proposal for a large family. Yeast 13: 1053–1058 [DOI] [PubMed] [Google Scholar]
- Roberts JA, Whitelaw CA, Gonzalez-Carranza ZH, McManus MT (2000) Cell separation processes in plants: models, mechanisms, and manipulation. Ann Bot (Lond) 86: 223–235 [Google Scholar]
- Schafer DA, Schroer TA (1999) Actin-related proteins. Annu Rev Cell Dev Biol 15: 341–363 [DOI] [PubMed] [Google Scholar]
- Shen X, Mizuguchi G, Hamiche A, Wu C (2000) A chromatin remodelling complex involved in transcription and DNA processing. Nature 406: 541–544 [DOI] [PubMed] [Google Scholar]
- Shen X, Ranallo R, Choi E, Wu C (2003) Involvement of actin-related proteins in ATP-dependent chromatin remodeling. Mol Cell 12: 147–155 [DOI] [PubMed] [Google Scholar]
- Szerlong H, Saha A, Cairns BR (2003) The nuclear actin-related proteins Arp7 and Arp9: a dimeric module that cooperates with architectural proteins for chromatin remodeling. EMBO J 22: 3175–3187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorn WJ, Stead AD (1997) Abscission of flowers and floral parts. J Exp Bot 48: 821–837 [Google Scholar]
- Verbsky ML, Richards EJ (2001) Chromatin remodeling in plants. Curr Opin Plant Biol 4: 494–500 [DOI] [PubMed] [Google Scholar]
- Wagner D (2003) Chromatin regulation of plant development. Curr Opin Plant Biol 6: 20–28 [DOI] [PubMed] [Google Scholar]
- Wagner D, Meyerowitz EM (2002) SPLAYED, a novel SWI/SNF ATPase homolog, controls reproductive development in Arabidopsis. Curr Biol 12: 85–94 [DOI] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J (2002) Arabidopsis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Zhao K, Wang W, Rando OJ, Xue Y, Swiderek K, Kuo A, Crabtree GR (1998) Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell 95: 625–636 [DOI] [PubMed] [Google Scholar]