Abstract
We report here on a novel transcription factor with a basic helix-loop-helix domain for tolerance to inorganic phosphate (Pi) starvation in rice (Oryza sativa). The gene is designated OsPTF1. The expression of OsPTF1 is Pi starvation induced in roots while constitutively expressed in shoots, as shown by northern-blot analysis. Overexpression of OsPTF1 enhanced tolerance to Pi starvation in transgenic rice. Tillering ability, root and shoot biomass, and phosphorus content of transgenic rice plants were about 30% higher than those of the wild-type plants in Pi-deficient conditions in hydroponic experiments. In soil pot and field experiments, more than 20% increase in tiller number, panicle weight, and phosphorus content was observed in transgenic plants compared to wild-type plants at low-Pi levels. In Pi-deficient conditions, transgenic rice plants showed significantly higher total root length and root surface area, which results in a higher instantaneous Pi uptake rate over their wild-type counterparts. Microarray analysis for transgenic plants overexpressing OsPTF1 has been performed to investigate the downstream regulation of OsPTF1.
Phosphorus (P) is an essential macronutrient for plant growth, development, and reproduction. Plants absorb P almost exclusively in the inorganic form (Pi). Pi concentration is limited in both low- and high-pH soils due to its propensity to form insoluble aluminum and iron phosphate in acidic soils and calcium and magnesium phosphate compounds in alkaline soils (Bar-Yosef, 1991). Pi starvation elicits developmental and biochemical adaptation in plants, including an increase in root hair length and number, root-to-shoot ratio, and induction of high-affinity Pi transporters, phosphatases, and RNases (Schachtman et al., 1998; Raghothama, 1999; Ma et al., 2001). Similar adaptive responses to Pi starvation also occur in bacteria and fungi. In Escherichia coli, the PHO regulon comprises at least 15 genes involved in the acquisition of Pi (Torriani, 1990). Yeast (Saccharomyces cerevisiae) PHO regulon is mainly controlled by the phosphorylation and dephosphorylation of the transcription factor PHO4. PHO4 is a basic helix-loop-helix (bHLH) transcription factor, which generally interacts with the second transcription factor, PHO2, to induce the expression of downstream genes, such as high-affinity phosphate transporters, during Pi starvation (O'Neill et al., 1996).
Physiological and molecular studies have identified several putative members of a Pi starvation-inducible rescue system in higher plants (for review, see Abel et al., 2002). The regulatory gene, PHR1, which participates in the Pi starvation response, has been isolated from Arabidopsis (Arabidopsis thaliana; Rubio et al., 2001). The PHR1 gene encodes a Myb transcription factor with homology to PSR1 from Chlamydomonas (Wykoff et al., 1999). The finding of PHR1 implies that a PHO-like regulation system may exist in higher plants, and PHR1 may be a central factor for controlling the Pi-signaling system in plants. However, more evidence is needed to elucidate the molecular pathways controlled by PHR1 for the adaptive responses to low-Pi regimes in plants. Increasing evidence has shown the existence of a complicated transcriptional regulation system that responds to Pi starvation in higher plants (for review, see Hammond et al., 2004). Two types of transcriptional regulations in response to Pi starvation have been reported, including the transient induction of response genes during early stages of stress and the high induction during later stages of stress (Wang et al., 2002; Hammond et al., 2003). The promoters of early response genes are enriched in two sequence motifs, the PHO-like (CDHGTGG; D:G, T, or A; H:C, T, or A) and the TATA box-like (TATAAATA) elements, which are different from the binding motif of PHR1 (Hammond et al., 2003). A systematic transcriptional change in response to Pi starvation was described by microarray analysis in Arabidopsis (Wu et al., 2003). In that study, several transcription factors were identified to be regulated by Pi starvation at different time points and in different plant parts.
Although the physiological responses to Pi starvation have become well understood, knowledge about the components of the Pi starvation signaling system and the mechanism of how those components control the plant's responses at the cellular and molecular level to Pi starvation was limited (Franco-Zorrilla et al., 2004; Ticconi and Abel, 2004).
In this study, a cDNA clone was isolated from a Pi starvation-induced rice (Oryza sativa) root cDNA library constructed using the suppression subtractive hybridization (SSH) method. The sequence analysis for the full-length cDNA clone indicates that the gene encodes a transcription factor with a bHLH domain, which is designated OsPTF1 (rice Pi starvation-induced transcription factor 1). The function and expression pattern of the transcription factor and its downstream regulating genes are investigated.
RESULTS
Cloning and Characterization of OsPTF1
A cDNA library of indica rice var. Kasalath was constructed using the SSH method. About 5,000 colonies from this library were transferred onto Hybond-N+ nylon membrane and screened by reverse northern-blot analysis using forward- and reverse-subtracted cDNA. The clones with positive signal by forward-subtracted screening and negative signal by reverse-subtracted screening were selected as the positive clones. A total of 150 positive clones were sequenced and analyzed. Among them, a clone with a bHLH domain was selected for further analysis.
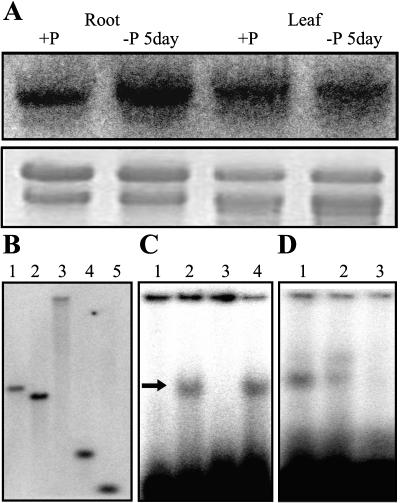
Northern-blot analysis, using the clone as a probe, showed that this gene was induced by Pi starvation in roots while constitutively expressed in shoots (Fig. 1A). The full-length cDNA encodes a protein of 478 amino acids with an estimated molecular mass of 50.7 kD (GenBank accession no. AY238991). A bHLH transcriptional regulatory domain was detected in this protein, and the basic region of the bHLH domain was predicted as a bipartite nuclear localization signal peptide by Motifscan. The high stringency of the genomic DNA gel-blot analysis, using full-length cDNA as a probe, indicated that the gene exists as a single copy in the rice genome (Fig. 1B). By screening a rice genomic library, the genomic sequence was obtained, which showed that the open reading frame of the gene was separated by seven introns. Similar to the most common pattern of the Arabidopsis bHLH domain gene family (Toledo-Ortiz et al., 2003), the coding region of the bHLH domain is separated by three introns. According to these results, the gene is designated OsPTF1.
Figure 1.
A, RNA gel-blot analysis for the expression of OsPTF1 in roots and leaves of 15-d-old seedlings under P-sufficient (10 mg Pi L−1) and Pi-deficient (0.5 mg Pi L−1) hydroponic solutions. Twenty milligrams of total RNA from each sample were electrophoresed. A 32P-labeled cDNA fragment of OsPTF1 was used as probe for hybridization. An ethidium bromide-stained gel is shown at the bottom. B, Southern-blot analysis of rice genomic DNA from the Kasalath genotype. DNA samples were digested with DraI (1), EcoRI (2), EcoRV (3), HindIII (4), and XbaI (5). The blot was hybridized with a 32P-labeled cDNA fragment of OsPTF1. C, E-box (G-box)-specific DNA binding properties of OsPTF1. Arrow shows OsPTF1-bound DNA. Lane 1, 32P-labeled E-box probe without OsPTF1 protein; lane 2, 32P-labeled E-box probe with OsPTF1 protein; lane 3, 32P-labeled mutated G-box probe with OsPTF1 protein; and lane 4, 32P-labeled G-box probe with OsPTF1 protein. D, Sequence-specific DNA-binding properties of OsPTF1. Lane 1, 32P-labeled G-box probe with OsPTF1 protein; lane 2, 5-fold excess of no labeled G-box DNA added to the reaction mixture of lane1; and lane 3, 100-fold excess of no labeled G-box DNA added to the reaction mixture of lane1.
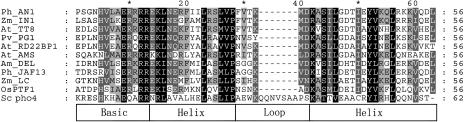
Alignment analysis using the ClustalW program indicates significant similarities between OsPTF1 and some bHLH proteins in the region of the bHLH domain (Fig. 2). Like the yeast PHO4 and other homologous bHLH transcription factors, His-5, Glu-9, Arg-12, and Arg-13 are present in the basic region of the OsPTF1 protein. These residues constitute the recognition motif for the G-box (CACGTG), which is one type of E-box (CANNTG; Atchley et al., 1999; Massari and Murre, 2000). The electrophoretic mobility shift assays showed that OsPTF1 is able to bind to the E-box and the G-box, but not to the mutated G-box DNA (CAATTG; Fig. 1C). To confirm the specificity of interaction between OsPTF1 and G-box DNA, competition experiments were performed. Unlabeled G-box DNA representing 5 to 100 times molar excess of the labeled probe was added to the interaction reaction mixture. A proportional decline in binding in response to increasing specific competitors indicates that the G-box-OsPTF1 interaction is specific (Fig. 1D).
Figure 2.
The bHLH domain of OsPTF1 shares highest identity with the bHLH motif of yeast PHO4. The accession numbers of these proteins in the GenBank database are as follows: PHO4 (Sc pho4), S56289; AMS (At_AMS), AC005167; ANTHROCYANIN 1 (Ph_AN1), AAG25928; PHASEOLIN G-BOX BINDING PROTEIN 1 (Pv_PG1), AAB00686; IN1 (Zm_IN1), AAB03841; TRANSPARENT TESTA 8 (At_TT8), CAC14865; RD22 BINDING PROTEIN 1 (At_RD22BP1), BAA25078; Lc (Zm_LC), AAA33504; DELILA (Am_DEL), AAA32663; and JAF13 (Ph_JAF13), AAC39455. The shading of the alignment presents as follows: identical residues in black, conserved residues in dark gray, and similar residues in light gray. The structure of the bHLH domain is shown in the box below.
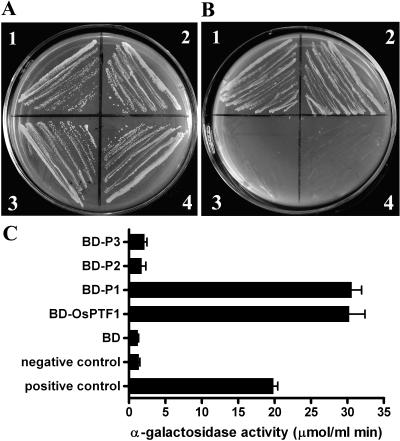
To analyze the transcription activation ability, OsPTF1 and its three subclones, the N-terminal 235 amino acids (P1), the bHLH domain from amino acid 236 to 392 (P2), and the last 86 amino acids in the C terminus (P3), respectively, were expressed in the yeast strain AH109 and fused to the DNA-binding domain (BD) of the yeast transcription factor GAL4. These four clones were designated BD-OsPTF1, BD-P1, BD-P2, and BD-P3, respectively. The activation of a MEL reporter gene (quantified by means of α-galactosidase assays) was tested. In the absence of Trp in the growth medium, all constructs grew well, indicating that the vector is present in the yeast strain AH109 (Fig. 3A). In the absence of histone (−His), −Trp, and adenine (−Ade), only the yeast with BD-OsPTF1 and BD-P1 grew well, indicating that full-length OsPTF1 and its N-terminal domain can activate the reporter genes for His and Ade (Fig. 3B). The OsPTF1 lacking the N-terminal domain can no longer activate the reporter gene. The reporter gene assay showed that the N-terminal peptide of OsPTF1 acts as the transcription activation domain (Fig. 3C).
Figure 3.
Detecting the activating domain of OsPTF1 using a yeast system. A, The transformed yeast strain AH109 with all four constructs grows well in synthetic dextrose growth medium without Trp. 1, BD-OsPTF1; 2, BD-P1, the N-terminal 270 amino acids not containing the bHLH domain; 3, BD-P2, from amino acids 272 to 403 containing the bHLH domain; and 4, BD-P3, the last 93 amino acids in the C terminus. B, The yeast with constructs of BD-OsPTF1 (1) and BD-P1 (2) grows well, but not with constructs of BD-P2 (3) and BD-P3 (4), in synthetic dextrose growth medium without His/Trp/Ade. Growth on −Trp means that the vector is present in yeast stain AH109, and growth on −His/−Trp/−Ade means that there is functional expression of His and Ade biosynthetic genes. C, Reporter gene activity (α-galactosidase activity, μmol mL−1 min−1) in yeast stain AH109 encoded by MEL1 with different constructs as above and the control (n = 8). Error bars = se.
Expression Pattern of OsPTF1
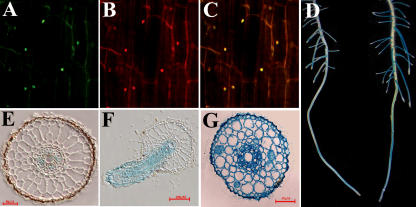
To determine the subcellular localization of OsPTF1, the reporter gene mGFP4 was fused to the 3′-end of OsPTF1. The cauliflower mosaic virus (CaMV) 35S promoter was used to drive the fusion protein. The results showed that OsPTF1::mGFP4 fusion protein was localized in the nuclei (Fig. 4, A–C).
Figure 4.
A, OsPTF1::mGFP green fluorescence shown in the nucleus. B, The same cells as in A stained with Pi. C, Merged image of A and B. D, OsPTF1 promoter-driven expression of the GUS reporter gene in primary roots and lateral roots of 15-d-old rice seedlings under Pi-sufficient (left) and Pi-deficient (right) conditions for 5 d. E, Transverse section of the primary root region above the elongation zone of a 15-d-old seedling after Pi starvation for 5 d. F, Vertical sections of a young lateral root formed on a primary root after Pi starvation for 5 d. G, Transverse section of the lateral root at 0.5 cm from root tip.
The putative promoter, a 2.4-kb upstream sequence of OsPTF1, was fused to the GUS reporter gene to explore the expression pattern of OsPTF1. Histochemical GUS assays were carried out on the roots of 2-week-old seedlings of T2 plants under Pi starvation and Pi-sufficient conditions. Strong GUS staining was observed in lateral roots and in the elongation zone of primary roots in the Pi starvation condition (Fig. 4D). The GUS staining of transverse and vertical sections showed that the fusion gene was expressed in phloem cells in the primary root (Fig. 4E), while in all cells of newly developed lateral roots (Fig. 4, F and G). GUS staining was also observed in leaves (data not shown).
Overexpression of OsPTF1 Enhanced Tolerance to Pi Starvation in Transgenic Rice
To investigate the function of OsPTF1 tolerance to Pi starvation, the CaMV 35S promoter-driven OsPTF1 was introduced into rice (Nipponbare) via Agrobacterium tumefaciens-mediated gene transformation. T2 plants of two independent transgenic lines with overexpression of OsPTF1 (35S:OsPTF1-1, 35S:OsPTF1-2; Fig. 5, A–C) were tested for tolerance to Pi starvation. In hydroponic culture solution with a high-Pi level, no significant differences in tiller number, root and shoot biomass, root length, and P content were observed between wild-type and transgenic plants. With a low-Pi level, however, tiller number of transgenic plants was about 40% higher than that of wild-type plants. Similarly, root and shoot biomass and Pi content of transgenic plants were more than 30% higher than those of wild-type plants (Fig. 5D; Table I). The number of rice tillers is considered an indicator of the P nutrient status in plants (IRRI, 1996). Therefore, the higher tiller number and biomass yield in transgenic plants implies that the overexpression of OsPTF1 enhanced tolerance to Pi deficiency.
Figure 5.
Growth performances of 4-week-old wild-type (W) and two transgenic lines (35S1 and 35S2) under hydroponic and soil pot experiments. A, Southern-blot analysis for the wild-type (W; Nipponbare) and two transgenic lines (35S1 and 35S2) using the hygromycin gene as probe. Five-microgram genomic DNA of wild type, 35S:OsPTF1-1, and 35S:OsPTF1-2 was digested by EcoRI and separated on an agarose gel. B, Northern-blot analysis of OsPTF1 for wild type (W) and two transgenic lines. Ethidium bromide-stained gel is shown at the bottom. C, Western blot of OsPTF1 in the roots of wild-type (W) and two transgenic plants (35S1 and 35S2) under Pi-supplied conditions. Each lane was loaded with 50 μg of total protein extracted from the roots. D, Growth performance of wild-type (W) and two transgenic lines (35S1 and 35S2) under low-P solution (0.5 mg Pi L−1) for 28 d. E, Growth performance of a 45-d-old wild-type plant and one transgenic line in a pot experiment using acidic red soil (pH 4.3; water/soil, 1:1) under low-P conditions (15 mg P/kg Bray-1, P = 3.25 mg P/kg). Right, Wild-type (W); left, 35S:OsPTF1-1 line (35S1).
Table I.
Plant growth parameters and total P content (mg P plant−1) of transgenic and wild-type plants (Nipponbare) from solution culture experiments
Four-week-old seedlings were sampled for the measurements. The value represents the mean ± se of three experiments with 10 plants each.
| Nip | 35S:OsPTF1-1 | 35S:OsPTF1-2 | |
|---|---|---|---|
| 10 mg P/L | |||
| Tiller number | 3.80 ± 0.31 | 3.20 ± 0.14 | 3.60 ± 0.17 |
| Plant height (cm) | 75.67 ± 1.11 | 76.00 ± 0.70 | 75.40 ± 1.26 |
| Root biomass (mg plant−1) | 0.26 ± 0.00 | 0.28 ± 0.02 | 0.27 ± 0.01 |
| Shoot biomass (mg plant−1) | 1.40 ± 0.04 | 1.57 ± 0.09 | 1.46 ± 0.03 |
| Root P content (mg P plant−1) | 1.85 ± 0.08 | 2.07 ± 0.13 | 2.09 ± 0.08 |
| Shoot P content (mg P plant−1) | 10.71 ± 0.21 | 11.38 ± 0.45 | 10.98 ± 0.38 |
| 0.5 mg P/L | |||
| Tiller number | 1.80 ± 0.13 | 2.50 ± 0.16a | 2.80 ± 0.13a |
| Plant height (cm) | 67.11 ± 0.87 | 74.60 ± 1.02a | 73.40 ± 0.95a |
| Root biomass (mg plant−1) | 0.25 ± 0.01 | 0.33 ± 0.02a | 0.34 ± 0.01a |
| Shoot biomass (mg plant−1) | 0.66 ± 0.02 | 0.92 ± 0.03a | 0.88 ± 0.03a |
| Root P content (mg P plant−1) | 0.26 ± 0.01 | 0.35 ± 0.01a | 0.36 ± 0.02a |
| Shoot P content (mg P plant−1) | 0.63 ± 0.03 | 0.89 ± 0.03a | 0.84 ± 0.02a |
Most significant (P < 0.01) difference in means between wild-type and transgenic plants.
Two-pot experiments using an acid clay soil (pH 4.3) with two rates of Pi level were carried out in a greenhouse. High-Pi level contained 60 mg Pi kg−1 soil (Bray-1, P = 13.4 mg P kg−1), while low-Pi level contained 15 mg Pi kg−1 soil (Bray-1, P = 3.25 mg P kg−1). Fifteen-day-old rice seedlings of wild-type and 35S:OsPTF1-1 lines were transplanted into pots individually. In the high-Pi condition, no significant differences in tiller number, shoot biomass, panicle weight, and P content of the plants were observed between wild-type and transgenic plants in both experiments. In the low-Pi condition, tiller number, shoot biomass, panicle weight, and P content of transgenic plants were more than about 40%, 30%, 20%, and 25% higher than in wild-type plants, respectively (Fig. 5E; Table II). A field test for a representative transgenic line (35S:OsPTF1-1) and wild-type plants was carried out as described in “Materials and Methods.” The average of tiller number and panicle weight of the transgenic plants was 14.3 and 14.4 g per plant, respectively, which is significantly higher (P < 0.01) than those of wild-type plants (11.0 and 12.5) in the low-Pi level. In the high-Pi level, however, no significant differences in the above parameters were observed between transgenic and wild-type plants.
Table II.
Tiller number, dry shoot biomass, panicle weight, and P content in shoot and panicle of each plant at harvest stage of the wild-type (WT) and 35S-driven OsPTF1 transgenic plants (35S:OsPTF1) in two-pot experiments
Two P fertilizer levels were employed at 60 mg P/kg (Bray-1, Pi = 13.70 mg P/kg) and 15 mg P/kg (Bray-1, Pi = 3.25 mg P/kg). Each pot contained 7.5 kg soil and individual plants were transplanted into each pot (n = 6). The value represents the mean ± se.
| First Experiment
|
Second Experiment
|
|||
|---|---|---|---|---|
| WT | 35S:OsPTF1-1 | WT | 35S:OsPTF1-1 | |
| 60 mg Pi kg soil−1 | ||||
| Tiller number | 25.00 ± 0.57 | 25.33 ± 0.84 | 20.31 ± 0.80 | 21.17 ± 1.17 |
| Shoot biomass (g) | 40.72 ± 1.79 | 42.20 ± 0.86 | 35.75 ± 1.97 | 36.64 ± 2.03 |
| Panicle weight (g) | 24.65 ± 2.03 | 25.24 ± 1.40 | 28.31 ± 3.10 | 29.46 ± 2.56 |
| P content (mg) | 37.75 ± 2.17 | 39.10 ± 1.91 | 34.16 ± 3.35 | 35.44 ± 3.05 |
| 15 mg Pi kg soil−1 | ||||
| Tiller number | 5.17 ± 0.17 | 7.50 ± 0.22a | 5.54 ± 0.24 | 7.58 ± 0.34a |
| Shoot biomass (g) | 13.80 ± 0.49 | 18.54 ± 0.18a | 10.88 ± 0.51 | 14.16 ± 0.33a |
| Panicle weight (g) | 11.38 ± 0.84 | 14.14 ± 1.03a | 10.52 ± 0.93 | 12.85 ± 1.60a |
| P content (mg) | 14.17 ± 1.02 | 18.08 ± 0.91a | 13.07 ± 1.14 | 16.30 ± 1.04a |
Significant differences in means between transgenic lines and wild-type plants at the P < 0.01 level.
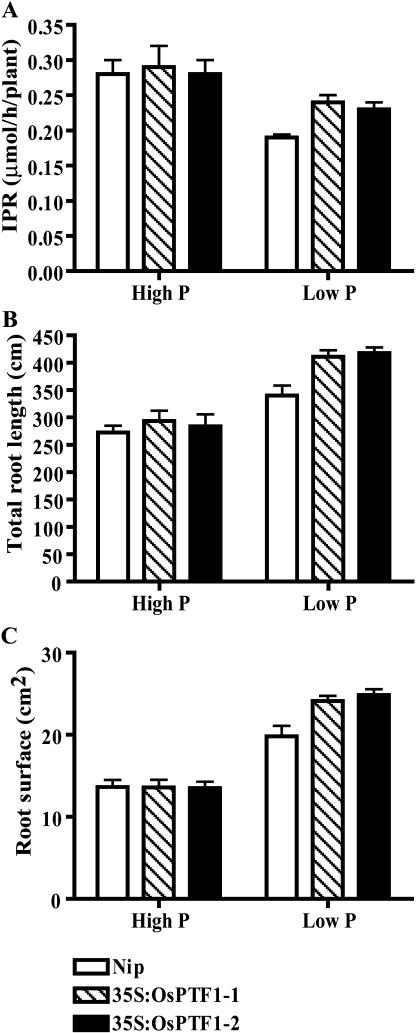
To elucidate the mechanism of high-Pi uptake ability in transgenic plants in Pi-deficient conditions, the instantaneous Pi uptake rate and root parameters were measured. In the high-Pi condition, no significant differences were observed in the instantaneous Pi uptake rates (μmol h−1 per seedling), root total length, and root surface area between wild-type and transgenic seedlings. In the low-Pi condition, however, the instantaneous Pi uptake rate of transgenic seedlings was significantly higher (P < 0.01) than that of wild-type seedlings (Fig. 6A), and the total root length and root surface area of the transgenic seedlings were about 20% higher than those of the wild-type seedlings (Fig. 6, B and C). The results indicate that the higher Pi uptake ability of transgenic plants in Pi-deficient conditions may be attributable, at least partially, to the larger root surface area.
Figure 6.
Instantaneous Pi uptake rate (μmol h−1 seedling−1), root surface area (cm2), and total root length (cm) of 21-d-old seedlings of wild type (Nipponbare) and two transgenic lines with overexpression of OsPTF1 (35S:OsPTF1-1/2) under high-Pi (10 mg Pi L−1) and low-Pi levels (0.5 mg Pi L−1) from the hydroponic experiment. A, Instantaneous Pi uptake rate. B, Root surface area. C, Total root length. Values represent the mean of 10 seedlings ± se.
Downstream Regulation of OsPTF1
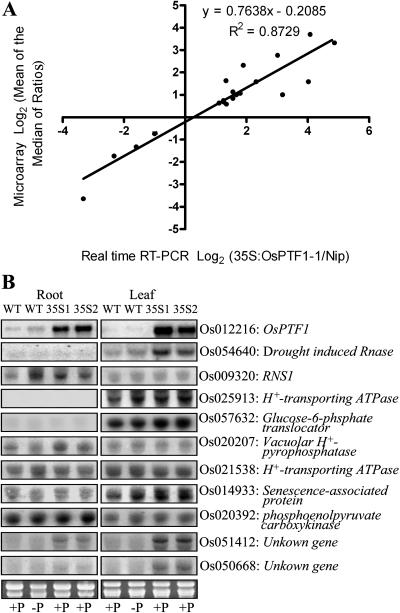
To investigate the downstream genes regulated by OsPTF1, microarray analysis was performed using rice whole-genome oligo chips provided from the DNA microarray laboratory at Yale University. Total RNA was extracted from 15-d-old seedlings of transgenic lines and wild-type seedlings in the Pi-sufficient condition (10 mg Pi L−1). To confirm the differentially expressed genes identified by the microarray analysis, quantitative PCR was used to examine the confidence level. A high degree of concordance (r = 0.87) was observed between the results generated by the two methods (Fig. 7A).
Figure 7.
A, Correlation between microarray data and real-time RT-PCR data. Log-transformed fold changes (base 2) for 20 differentially expressed genes (root, Os009320; Os009602; Os012216; Os014603; Os020207; Os021935; Os042773; Os050668; Os050997; Os051412; leaf, Os012216; Os020392; Os021538; Os022118; Os024733; Os025913; Os050668; Os051412; Os054640; Os057632). B, Northern-blot analysis for selected genes based on microarray assay results (see supplemental data). Total RNA (20 μg) samples from leaves and roots of 15-d-old wild-type seedlings (Nipponbare) and two transgenic lines with 35S-driven overexpression of OsPTF1 (35S1 and 35S2) were hybridized with 32P-labeled probes of the selected genes. Ethidium bromide-stained gel is shown at the bottom.
Microarray data showed that 158 genes, with a ratio greater than 2-fold in roots and/or in shoots, were found to be regulated by overexpression of OsPTF1 (see supplemental data). Among those, 14 genes were up-regulated in both leaves and roots. Forty-three genes were up-regulated in either leaves or roots. Twenty-seven and 30 genes were down-regulated in leaves and roots. Ninety-eight genes were classified based on a database search from the 158 genes. The function-classified genes include nutrient transporters and metabolism, carbon metabolism, transcription factors, ATP-binding protein, oxidoreductase, protease, disease resistance protein, RNase, H+-transporting ATPase, vacuolar H+-pyrophosphatase, senescence-associated protein, receptor-like kinase, and several cytochrome P450 genes (see supplemental data). Many function-unknown or putative genes were strongly up- and down-regulated by overexpression of OsPTF1. Some of them did not respond to Pi starvation.
The element analysis of the promoter region of these genes showed that almost all of the regulated genes have the E-box elements, and about 20% of the genes have at least one copy of the G-box element (see supplemental data). Ten genes up-regulated by overexpression of OsPTF1 from more than 2-fold to about 16-fold revealed by microarray analysis (see supplemental data) were also subjected to RNA gel-blot analysis. These genes include plant PHO genes and two function-unknown genes. The RNA gel-blot results are consistent with the results from microarray analysis (Fig. 7B).
DISCUSSION
In this study, we cloned and characterized a bHLH transcription factor, OsPTF1, which is responsible for tolerance to Pi starvation in rice. Overexpression of OsPTF1 can enhance tolerance to Pi deficiency. It was reported that PHO4, which was also a bHLH protein, acts as a key regulator of PHO regulon in yeast. In Pi starvation conditions, PHO4 combines with its partner, PHO2, and induces the expression of PHO5 and PHO84, the Pi starvation-induced acid phosphatase and the high-affinity Pi transporter (Johnston and Carlson, 1992; Oshima, 1997). Microarray analysis was performed to test whether OsPTF1 regulates downstream genes as PHO4. Unexpectedly, no high-affinity phosphate transporter genes or acid phosphatase genes were found to be controlled by OsPTF1 (see supplemental data). In addition, OsPTF1 is located in the nucleus in both high- and low-Pi conditions, which is different from PHO4, which is imported into or exported out of the nucleus under Pi-sufficient or starvation conditions (Kaman et al., 1998). These results support the conclusion that the Pi regulation system in higher plants is much more complicated than that in yeast, and perhaps there are some other bHLH proteins that act as the PHO4 in plants.
A MYB transcription factor, PHR1, has been isolated in Arabidopsis and shown to be involved in a regulation system responsive to Pi starvation (Rubio et al., 2001). In the Pi starvation condition, the phr1 mutant did not show the typical Pi starvation phenotypes (accumulation of anthocyanins and reduced root-to-shoot ratio) and changes in the expression of Pi starvation marker genes. Because the expression of PHR1 is not sensitively affected by the status of Pi supply levels and the PHR1 is located in the nucleus independent of Pi status, it was proposed that either PHR1 activity is regulated posttranscriptionally or a second Pi starvation regulatory protein is needed to mount a proper Pi starvation response (Franco-Zorrilla et al., 2004). The OsPTF1 is located in the nucleus independent of the Pi status of the plant (Fig. 4A) and it is induced in the rice roots under Pi-deficient conditions (Fig. 1A), which is different from the response of PHR1 to the Pi status of the plant. It indicates that the regulation system of OsPTF1 may work through a different mechanism than the Arabidopsis MYB-containing transcription factor, PHR1. To investigate whether a PHR1-like system exists in rice, we searched rice genome data for the homologies of PHR1. Two homologous genes, OsPHR1 and OsPHR2, were identified. Transformation of these two homologous genes is ongoing to investigate their functions and their possible relationship with OsPTF1.
Many genes have been presumed to play a role in the Pi starvation rescue system, such as RNases (Green, 1994), high-affinity Pi transporters (Raghothama, 2000), H+-transporting ATPase and vacuolar H+-pyrophosphatase (H+-PPase), phosphoenolpyruvate (PEP) carboxylase (PEPC), and pyrophosphate-dependent phosphofructokinase (Plaxton and Carswell, 1999). Microarray analysis revealed that some Pi starvation-induced genes, such as RNS1, H+-transporting ATPase, vacuolar H+-pyrophosphatase, and one senescence-associated gene, were expressed in transgenic plants with overexpression of OsPTF1 in the Pi-supplied condition (Fig. 7B; see supplemental data). The results strongly suggest that the overexpressed OsPTF1 can trigger the expression of plant PHO genes involved in the efficient utilization of absorbed Pi in plants as a Pi starvation signal. Several genes showed coexpression with overexpression of OsPTF1 but not with response to Pi starvation, including a gene for RNS2 and some function-unknown genes (Fig. 7B; see supplemental data). These function-unknown genes might be important components of the regulatory system controlled by OsPTF1.
No high-affinity Pi transporter genes were found to be up-regulated in the overexpression lines from microarray analysis. Transgenic lines with an overexpression of OsPTF1 showed longer total root length and larger root surface area under Pi-deficient conditions than those of wild-type plants (Fig. 6). It has been reported that small changes in root growth-related parameters had big effects on P uptake (Itoh and Barber, 1983; Wissuwa, 2003). In this case, the total root length and root surface area of 3-week-old transgenic plants were increased about 20% compared to wild-type plants, which results in the higher instantaneous Pi uptake rates of transgenic lines under Pi-deficient conditions and makes a contribution to tolerance for Pi deficiency.
PEP plays a central role in the modification of carbon and energy metabolism in response to Pi starvation. In the cytosol, PEP can be converted to pyruvate catalyzed by pyruvate kinase or to oxaloacetate catalyzed by PEPC. The latter has been suggested to be a Pi starvation-induced bypass to preserve Pi (Bieleski, 1973). In Arabidopsis and rice, however, this is not the case (Wu et al., 2003; Shinano et al., 2005). In this study, we did not find the gene for PEPC to be regulated, but a gene for PEP carboxykinase was up-regulated by Pi starvation and overexpression of OsPTF1 in the shoots (Fig. 7B). PEP carboxykinase catalyzes the conversion of phosphate and oxaloacetate to PEP and CO2. The results suggest that the stimulation of the original metabolic pathways should be important in alleviating P-deficient conditions, which were enhanced by overexpression of OsPTF1. It is noted in transgenic plants with overexpression of OsPTF1 that the phosphohydrolases, the translocator of Glc-6-P, and the H+-transporting ATPase in the shoot were up-regulated, which suggests that overexpression of OsPTF1 in the shoot may also contribute to tolerance to Pi starvation.
Together, our results confirm that there must be several key regulators in the control of the PHO system in plants, and the OsPTF1 is an important one that is involved in higher root growth and, consequently, higher uptake rates of Pi. It is also involved in the efficient utilization of P in plants. Therefore, OsPTF1 has potential in engineering plants with higher Pi use efficiency.
MATERIALS AND METHODS
Hydroponic, Soil Pot, and Field Experiments
Hydroponic experiments were conducted using normal rice (Oryza sativa) culture solution with 10 mg Pi L−1 (Yoshida et al., 1976) and Pi-deficient solution (0.5 mg Pi L−1), based on our previous studies under a photosynthetic photon flux density of approximately 200 μmol photons m−2 s−1 with a 16-h-light (28°C)/8-h-dark (22°C) photoperiod and 70% to 80% relative humidity. Three biological replications for hydroponic experiments were carried out under the same growth conditions for wild-type rice (japonica var. Nipponbare) and transgenic lines overexpressing OsPTF1. Soil pot experiments were conducted using fine-clay acid red soil (pH 4.3; water/soil, 1:1, fine-clay kaolinitic thermic typic plinthudults; Soil Survey Staff, 1996), with two Pi-supplied levels: 60 mg Pi kg−1 soil as KH2PO4 (Bray-1, P = 13.70 mg P kg−1) and 15 mg Pi kg−1 soil (Bray-1, P = 3.25 mg P kg−1). Each pot contained 7.5 kg of air-dried soil with the individual plant. In 2003, two independent experiments were carried out in a greenhouse. The pots were randomly arranged with six replications in each experiment. A field experiment was carried out in the Field Experiment Station of the China National Rice Research Institute (CNRRI; Hangzhou, China) from June to October 2004. Twenty-day-old seedlings of a transgenic line (35S:OsPTF1-1) overexpressing OsPTF1 and wild type were transplanted at 15- to 20-cm spacing. Nitrogen (N) and potassium (K) were applied before transplanting at 60 kg ha−1 N as urea and 40 kg ha−1 K as KCl. At midtillering and panicle initiation, 45 kg ha−1 N and 60 kg ha−1 N were added, respectively. Two levels of Pi were used. For low-Pi levels in soil (Bray-1, Pi = 6.02 mg P kg−1, total P = 370 mg P kg−1), no Pi fertilizer was applied. For high-Pi levels, 60 kg Pi ha−1, P (Bray-1, Pi = 20.11 mg P kg−1) were applied to the soil before transplanting. Three plots each of 3 × 2 m size were designed for each Pi level. At harvest, 15 plants were randomly sampled from each plot to determine the number of tillers and air-dried panicle weight.
Gene Cloning
The root mRNAs of Kasalath (indica rice) in normal and Pi-deficient cultures were used to construct a cDNA library by the SSH method using the PCR-select cDNA subtraction kit (CLONTECH Laboratories, Palo Alto, CA) and the PCR cloning kit (CLONTECH). A cDNA clone for OsPTF1 was isolated from this cDNA library. Full-length cDNA of the gene was obtained by the RACE method using the Smart RACE cDNA amplification kit (CLONTECH), according to the manufacturer's instructions.
DNA and RNA Gel-Blot Analysis
Genomic DNA was isolated from young leaves using the previously described method (Murray and Thompson, 1980) and was digested using five enzymes, DraI, EcoRI, EcoRV, HindIII, and XbaI. Total RNA was extracted using the Trizol D0410 reagent, according to the manufacturer's instructions (Invitrogen, Carlsbad, CA). Five micrograms of digested DNA and 20 μg of total RNA were used for DNA and RNA gel-blot analysis, respectively.
Shifting Assay Analysis
The coding region of OsPTF1 was amplified using primers 5′-GCTGAATTCGATGGACTACTCCGCTGGTTCCT-3′ (added EcoRI site underlined) and 5′-TGTGTCGACTTTTTCAGGAGGGATTGCAGCAG-3′ (with added SalI site underlined) and subcloned into a His × 6 expression vector pET29b (Promega, Madison, WI) with EcoRI and SalI. Recombinant protein produced in Escherichia coli was purified using a Ni-NTA column, according to the manufacturer's specifications (Qiagen, Valencia, CA). Purified protein was dialyzed against a DNA-binding reaction buffer (15.0 mm Tris, pH 8.0, 65.0 mm NaCl, 7.5% glycerol, 1.25 mm dithiothreitol [DTT], 1.8 mm EDTA, and 100 ng PolyIPolyC). 32P-end-labeled oligonucleotides were mixed with purified protein in DNA-binding reaction buffer. Samples were incubated at room temperature for 30 min, electrophoresed on a 4% polyacrylamide gel for 2 h at 15 V cm−1 at 4°C, and subsequently visualized with a Typhoon 8600 (Amersham-Pharmacia Biotech, Piscataway, NJ).
Western-Blot Analysis
For antibody of OsPTF1 preparation, purified recombinant OsPTF1 protein was mixed with Freund's complete adjuvant and injected intramuscularly into New Zealand rabbit. Blood samples were taken at 2-week intervals after the third immunization. For western-blot analysis, proteins were separated by 7.5% SDS-PAGE and electroblotted onto a polyvinylidene difluoride membrane (Amresco, Solon, OH), and visualized using rabbit anti-OsPTF1 antisera followed by alkaline phosphatase-conjugated goat anti-rabbit serum (Santa Cruz Biotechnologies, Santa Cruz, CA).
Detecting Activation Domain of OsPTF1
The Matchmaker yeast (Saccharomyces cerevisiae) two-hybrid system (K1615-1; CLONTECH) was used to detect the activation domain in OsPTF1 in accordance with the manufacturer's protocol. The deduced amino acid sequence of OsPTF1 was divided into three fragments: P1 (N-terminal 235 amino acids not containing the bHLH domain), P2 (from amino acid 236–392 containing the bHLH domain), and P3 (the last 86 amino acids in the C terminus), and inserted into pGBKT7, resulting in fusions with the GAL4 DNA BD. Quantitative galactosidase assays were performed using the method described in the CLONTECH protocol. The interaction between the BD-p53 and the AD-SV40 large T-antigen (provided by Matchmaker) served as a positive control, while the pGBKT7 vector was used as a negative control.
Construction of GUS Fusion and Overexpression Vectors and Development of Transgenic Plants
To construct an OsPTF1 promoter-GUS chimeric gene, the pCAMBIA 1391Z plasmid was digested using HindIII/NcoI and blunt-ended using T4 DNA polymerase. The promoter region of the OsPTF1 was obtained by PCR amplification using the primers 5′-GAGCAAGAAAAGCGGCAACTAAAG-3′ and 5′-TACGAAATACCCTCTGACGCTGAA-3′, and then subcloned into pCAMBIA 1391Z. The overexpression vector was constructed as follows. First, the CaMV 35S promoter was subcloned between the EcoRI and SacI sites of pCAMBIA1301. Second, the poly(A) addition sequence of pea small subunit of Rubisco E9 (Coruzzi et al., 1984) was inserted into the site between the HindIII and PstI. The resulting plasmid was named 35S-pCAMBIA1301. Third, the open reading frame of OsPTF1 was introduced into 35S-pCAMBIA1301 using the SmaI site. The Agrobacterium strain (EHA105) harboring the constructs was used to transform the rice cv Nipponbare as previously described (Chen et al., 2003).
Subcellular Localization of OsPTF1
The coding region of the OsPTF1 with the eliminated stop codon was inserted in-frame after the initiator ATG of the cDNA of a soluble modified green fluorescent protein (smGFP4). The OsPTF1::GFP fusion gene was subcloned into the binary vector 35S-pCAMBIA1301. Agrobacterium tumefaciens (EHA105) harboring the construct was used to transform the rice cv Nipponbare as previously described (Chen et al., 2003). The GFP was visualized using a LSM 510 laser-scanning microscope (Zeiss, Jena, Germany).
Histochemical Analysis and GUS Assay of Transient Expression
Histochemical GUS analysis was performed according to Jefferson et al. (1987). The stained materials were visualized using light or fluorescent microscopy. To investigate subcellular expression patterns, the stained tissues were rinsed and fixed in FAA (formalin:acetic acid:70% ethanol (1:1:18)) at 4°C for 24 h, embedded in Spurr resin, and then sectioned. Sections (5 μm) were mounted on slides and photographed.
Computational Analysis
The sequence of full-length cDNA was analyzed using BLAST and Motifscan (http://hits.isb-sib.ch/cgi-bin/PFSCAN). Sequence alignment was generated by ClustalW analysis (http://www.ebi.ac.uk/clustalw/index.html). For element analysis, 1 kb upstream of the ATG start site for the rice genome pseudomolecules was obtained from The Institute for Genomic Research (TIGR) (http://www.tigr.org/tdb/e2k1/osa1/pseudomolecules/info.shtml), and the PLACE signal scan search (http://www.dna.affrc.go.jp/PLACE/signalscan.html) was used to count the presence and copy number of the E- and G-box elements.
Measurement of P Concentration in Plants and Soils
Shoots and roots of wild-type and the transgenic seedlings under Pi-sufficient and Pi-deficient cultures were separately sampled and oven dried at 80°C for 3 d to determine the biomass dry weight. P concentrations of plant samples were analyzed with the phosphomolybdenum blue reaction using a Spectroquant NOVA60 spectrophotometer and Spectroquant phosphate test kit (Merck, Darmstadt, Germany). Total soil P content and Bray-1 Pi concentrations were determined according to the previously described method (Bray and Kurtz, 1945). P content was calculated as the concentration of the sample dry weight.
Measurements of Instantaneous Pi Uptake Rate, Root Total Length, and Root Surface Area
For instantaneous Pi uptake rate measurement, 15-d-old seedlings were divided into two groups. One group was cultured in the normal culture solution with 10 mg P L−1 (Yoshida et al., 1976) and another group was cultured in Pi-deficient culture solution with 0.5 mg P L−1. After 7 d of growth, plants were transferred to 50-mL flasks containing the normal culture solution. Each flask contained two plants with 10 replications, and the solution was sampled at 0, 5, 10, 15, 30, 40, 50, 60, 90, 120, 150, 180, 210, and 240 min, respectively. Phosphate concentration of the samples was determined using malachite green reagent. The instantaneous Pi uptake rate was calculated in micromoles per hour per seedling using the method described by Ning and Cumming (2001). The total root length and root surface composed of the primary root, adventitious roots, and lateral roots of each seedling were measured with WinRhizo (MAC STD1600; Regent Instruments, Quebec City, Canada).
Statistical Analysis of Data
Student's t test was performed for mean comparisons of plant growth parameters and total P content between wild-type and transgenic plants using the algorithm incorporated into Microsoft Excel (Microsoft, Redmond, WA).
Oligo Microarray Analysis
The rice oligo microarray analysis was performed for two transgenic lines (35S:OsPTF1-1 and 35S:OsPTF1-2) versus wild-type plants under Pi-sufficient hydroponic conditions (10 mg P L−1) with two replicates. The chips with 60,727 oligo spots on two slides were used, which were developed from the DNA microarray laboratory at Yale University using the Rice Genome Oligo Set Version 1.0 from Qiagen (http://omad.qiagen.com/download/storage/rice_V1.0.2_genelist_s-.xls.gz). All oligos were designed from cDNAs, expressed sequence tag sequence predicted genes of the Beijing Genomics Institute (BGI) rice genome database, and other public resources. Fifteen plants of 15-d-old transgenic lines and Nipponbare were used for total RNA extraction, and the mRNA was prepared using the Dynabeads mRNA purification kit (Dynal Biotech, Oslo). mRNA labeling, purification of labeled cDNA, and prehybridization were performed according to the method described by Ma et al. (2002), and the hybridization was followed as in Lee et al. (2004). Separate TIFF images for Cy-3 and Cy-5 channels were obtained by scanning with an Axon GenePix 4100 scanner (Axon Instruments, Foster City, CA) at a resolution of 10 μm. Laser and photomultiplier tube voltages were adjusted manually to minimize background. Normalization of the two channels of signal intensity was obtained by adjusting the photomultiplier tube and laser power settings. Spot intensities were quantified using Axon GenePix image analysis software. The channel ratio was determined using the GenePix median-of-ratio method and normalized using the corresponding GenePix Pro 5.1 default normalization method (the data can be downloaded from http://www.genomics.zju.edu.cn/ricemicroarray.html). Spots flagged Bad or Not Found by GenePix software were removed from the final data analysis, the mean of the median of ratios from the two independent lines was used for further analysis, only the genes with a mean >2 or <0.5 were used for further restricted screening criteria (in the two independent lines, at least 1-fold variation of >2.0 and the other of >1.8 for up-regulated genes; at least 1-fold of <0.5 and the other of <0.56 for down-regulated genes). These genes were picked up for the annotation using BLASTn to the dataset from rice full-length cDNA (http://cdna01.dna.affrc.go.jp/cDNA) and rice TGI (http://www.tigr.org/docs/tigr-scripts/tgi/tc_report) database. After data analysis, quantitative PCR and northern blotting were used to determine the confidence level of the microarray analysis.
For quantitative PCR analysis, 5 μg of DNase I-treated total RNA from the 35S:OsPTF1-1 and Nipponbare were used for RT using SuperScript II (Invitrogen). The cDNA samples were diluted to 1, 0.5, and 0.1 ng/μL. Triplicate quantitative assays were performed on each cDNA dilution with the SYBR Green Master Mix with an ABI 7000 sequence detection system, according to the manufacturer's protocol (Applied Biosystems, Foster, City, CA); the gene-specific primers were designed by using PRIMEREXPRESS software (Applied Biosystems). The relative quantification method was used to evaluate quantitative variation between replicates examined. The amplification of OsRAc1 (actin) was used as an internal control to normalize all data. Probes for northern blotting were obtained by RT-PCR amplification using specific primers for 10 selected genes (see supplemental data).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AY238991.
Supplementary Material
This work was supported by the Key Basic Research Special Foundation of China (G1999011700), the Special Program of Rice Functional Genomics of China (2002AAZZ1003), the National Education Ministry of China, and the Science and Technology Bureau of Zhejiang Province.
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.063115.
References
- Abel S, Ticconl CA, Delatorre CA (2002) Phosphate sensing in higher plants. Physiol Plant 115: 1–8 [DOI] [PubMed] [Google Scholar]
- Atchley WR, Therhalle W, Dress A (1999) Positional dependence, cliques and predictive motifs in the bHLH protein domain. J Mol Evol 48: 501–516 [DOI] [PubMed] [Google Scholar]
- Bar-Yosef B (1991) Root excretion and their environmental effects: influence on availability of phosphorus. In Y Weisel, A Eschel, V Kafkafi, eds, Plant Roots, the Hidden Half. Marcel Dekker, New York, pp 529–557
- Bieleski RL (1973) Phosphate pools, phosphate transport, and phosphate availability. Annu Rev Plant Physiol 24: 225–252 [Google Scholar]
- Bray RH, Kurtz LT (1945) Determination of total organic and available forms of phosphorus in soils. Soil Sci 59: 39–45 [Google Scholar]
- Chen SY, Jing WZ, Wang MY, Zhang F, Zhou J, Jia QJ, Wu YR, Liu FY, Wu P (2003) Distribution and characterization of over 1000 T-DNA tags in rice genome. Plant J 36: 105–113 [DOI] [PubMed] [Google Scholar]
- Coruzzi G, Broglie R, Edwards C, Chua NH (1984) Tissue-specific and light-regulated expression of a pea nuclear gene encoding the small subunit of ribulose-1,5-bisphosphate carboxylase. EMBO J 3: 1671–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Gonzalez E, Bustos R, Linhares F, Leyva A, Paz-Ares J (2004) The transcriptional control of plant responses to phosphate limitation. J Exp Bot 396: 285–293 [DOI] [PubMed] [Google Scholar]
- Green PJ (1994) The ribonucleases of higher plants. Annu Rev Plant Physiol Plant Mol Biol 45: 421–445 [Google Scholar]
- Hammond JP, Bennett MJ, Bowen HC, Broadley MR, Eastwood DC, May ST, Ralm C, Swarup R, Woolaway KE, White PJ (2003) Changes in genes expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiol 132: 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JP, Broadley MR, White PJ (2004) Genetic responses to phosphorus deficiency. Ann Bot (Lond) 94: 323–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- IRRI (1996) Annual Report for 1995. International Rice Research Institute. Los Banos, The Philippines
- Itoh S, Barber SA (1983) A numerical solution of whole plant nutrient uptake for soil-root systems with root hairs. Plant Soil 70: 403–413 [Google Scholar]
- Jefferson RD, Kavanagh TA, Bevan MW (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in highter plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M, Carlson M (1992) Regulation of carbon and phosphate utilization. In EW Jones, JR Pringle, JR Broach, eds, The Molecular and Cellular Biology of the Saccharomyces Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 193–281
- Kaman A, Rank NM, O'Neill EM, Huang LS, O'Shea EK (1998) The receptor Msn5 exports the phosphorylated transcription factor Pho4 out of the nucleus. Nature 396: 482–486 [DOI] [PubMed] [Google Scholar]
- Lee HS, Wang J, Tian L, Jiang H, Black MA, Madlung A, Watson B, Lukens L, Pires JC, Wang JJ, et al (2004) Sensitivity of 70-mer oligonucleotides and cDNAs for microarray analysis of gene expression in Arabidopsis and its related species. Plant Biotech J 2: 45–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Gao Y, Qu LJ, Chen ZL, Li JM, Zhao HY, Deng XW (2002) Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13: 2589–2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Bielenberg DG, Brown KM, Lynch JP (2001) Regulation of root hair density by phosphorus availability in Arabidopsis thaliana. Plant Cell Environ 24: 459–467 [Google Scholar]
- Massari ME, Murre C (2000) Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol 20: 429–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8: 4321–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning J, Cumming JR (2001) Arbuscular mycorrhizal fungi alter phosphorus relations of broomsedge (Andropogon virginicus L.) plants. J Exp Bot 52: 1883–1891 [DOI] [PubMed] [Google Scholar]
- O'Neill EM, Kaman A, Jolly ER, Shea EK (1996) Regulation of PHO4 nuclear localization by the PHO80-PHO85 cyclin-CDK complex. Science 271: 209–212 [DOI] [PubMed] [Google Scholar]
- Oshima Y (1997) The phosphatase system in Saccharomyces cerevisiae. Genes Genet Syst 72: 323–334 [DOI] [PubMed] [Google Scholar]
- Plaxton WC, Carswell MC (1999) Metabolic aspects of the phosphate starvation response in plants. In HR Lerner, ed, Plant Responses to Environmental Stresses: From Phytohormones to Genome Reorganization. Marcel Dekker, New York, pp 349–372
- Raghothama KG (1999) Phosphate acquisition. Annu Rev Plant Physiol 50: 665–693 [DOI] [PubMed] [Google Scholar]
- Raghothama KG (2000) Phosphate transport and signaling. Curr Opin Plant Biol 3: 182–187 [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martin AC, Iglesias J, Leyva A, Paz-Ares J (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15: 2122–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachtman DP, Reid RJ, Ayling SM (1998) Phosphorus uptake by plants: from soil to cell. Plant Physiol 116: 447–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinano T, Nanamori M, Dohi M, Wasaki J, Osaki M (2005) Evaluation of phosphorus starvation inducible genes relating to efficient phosphorus utilization in rice. Plant Soil 269: 81–87 [Google Scholar]
- Soil Survey Staff (1996) Keys to Soil Taxonomy, Ed 8. USDA-NRCS, Washington, DC
- Ticconi CA, Abel S (2004) Short on phosphate: plant surveillance and countermeasures. Trends Plant Sci 9: 548–555 [DOI] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Huq E, Quail PH (2003) The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torriani A (1990) From cell membrane to nucleotides: the phosphate regulon in Escherichia coli. Bioessays 12: 371–376 [DOI] [PubMed] [Google Scholar]
- Wang YH, Garvin DF, Kochian LV (2002) Rapid induction of regulatory and transporter genes in response to phosphorus, potassium, and iron deficiencies in tomato roots. Evidence for cross talk and root/rhizosphere-mediated signals. Plant Physiol 130: 1361–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissuwa M (2003) How do plants achieve tolerance to phosphorus deficiency? Small causes with big effects. Plant Physiol 133: 1947–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Ma LG, Hou XL, Wang MY, Wu YR, Liu FY, Deng XW (2003) Phosphate starvation triggers distinct alterations of genome expression in Arabidopsis roots and leaves. Plant Physiol 132: 1260–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wykoff DD, Grossman AR, Weeks DP, Usuda H, Shimogawara K (1999) Psr1, a nuclear localized protein that regulates phosphorus metabolism in Chlamydomonas. Proc Natl Acad Sci USA 96: 15336–15341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Forno DA, Cock JH, Gomez KA (1976) Laboratory Manual for Physiological Studies of Rice, Ed 3. The International Rice Research Institute, Manila, The Philippines
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.