Abstract
The minimal requirements to support protein import into mitochondria were investigated in the context of the phenomenon of ongoing gene transfer from the mitochondrion to the nucleus in plants. Ribosomal protein 10 of the small subunit is encoded in the mitochondrion in soybean and many other angiosperms, whereas in several other species it is nuclear encoded and thus must be imported into the mitochondrial matrix to function. When encoded by the nuclear genome, it has adopted different strategies for mitochondrial targeting and import. In lettuce (Lactuca sativa) and carrot (Daucus carota), Rps10 independently gained different N-terminal extensions from other genes, following transfer to the nucleus. (The designation of Rps10 follows the following convention. The gene is indicated in italics. If encoded in the mitochondrion, it is rps10; if encoded in the nucleus, it is Rps10.) Here, we show that the N-terminal extensions of Rps10 in lettuce and carrot are both essential for mitochondrial import. In maize (Zea mays), Rps10 has not acquired an extension upon transfer but can be readily imported into mitochondria. Deletion analysis located the mitochondrial targeting region to the first 20 amino acids. Using site directed mutagenesis, we changed residues in the first 20 amino acids of the mitochondrial encoded soybean (Glycine max) rps10 to the corresponding amino acids in the nuclear encoded maize Rps10 until import was achieved. Changes were required that altered charge, hydrophobicity, predicted ability to form an amphiphatic α-helix, and generation of a binding motif for the outer mitochondrial membrane receptor, translocase of the outer membrane 20. In addition to defining the changes required to achieve mitochondrial localization, the results demonstrate that even proteins that do not present barriers to import can require substantial changes to acquire a mitochondrial targeting signal.
Characterization of the mitochondrial proteome from yeast (Saccharomyces cerevisiae), mammals, and Arabidopsis (Arabidopsis thaliana) suggests that mitochondria contain from 2,000 to 3,000 proteins (Sickmann et al., 2003; Taylor et al., 2003; Heazlewood et al., 2004; Prokisch et al., 2004; Millar et al., 2005). As the mitochondrial coding capacity of these organisms varies from only seven to approximately 30 proteins, the majority of proteins are targeted to mitochondria from a cytosolic pool (Burger et al., 2003). Targeting to mitochondria is achieved by specific targeting signals (Schatz and Dobberstein, 1996). These signals may be located at the N terminus and, if removed from the protein after import into mitochondria, the signal is generally referred to as a presequence. Alternatively, targeting signals may be located within the protein itself, i.e. internal targeting signals that are not removed after import into mitochondria (Neupert, 1997; Pfanner and Geissler, 2001).
All targeting signals must be recognized by receptors on the mitochondrial surface to achieve targeting specificity. A single translocase of the outer membrane complex (TOM) is responsible for the recognition of all proteins destined to be located in mitochondria. The TOM complex plays three roles in the import of proteins into mitochondria: (1) recognition of mitochondrial targeting signals; (2) translocation of the unfolded polypeptide across the outer membrane; and finally (3) transfer to one of two translocases of the inner membrane (TIM; Pfanner and Chacinska, 2002). In yeast, two primary receptors, TOM20 and TOM70, which recognize N-terminal and internal targeting signals, respectively, have been characterized (Pfanner and Geissler, 2001). Characterization of the TOM complex from other organisms indicates that although there are some differences in composition compared to yeast, a single TOM complex plays an essential role in recognition on proteins destined to be imported into mitochondria (Hoogenraad et al., 2002; Werhahn et al., 2003; Macasev et al., 2004; Humphries et al., 2005).
Cleavable N-terminal mitochondrial targeting signals are well characterized. Analysis of sequences from a variety of organisms indicates that no primary amino acid sequence homology exists (von Heijne et al., 1989; Sjoling and Glaser, 1998). However, an enrichment of positively charged amino acids, Arg and Lys, and hydroxylated amino acids Ser and Thr are a feature of almost all N-terminal targeting signals. The ability to form an amphiphatic α-helix also appears to be a common feature of N-terminal mitochondrial targeting signals (Roise and Schatz, 1988). Detailed site directed mutagenesis studies of signals confirm these in silico predicted features, where both positive and hydrophobic residues have been confirmed to play a role in mitochondrial targeting. Changing these residues often, but not always, can reduce the amount of mitochondrial import (Hammen et al., 1996a, 1996b; Tanudji et al., 1999; Duby et al., 2001b; Zhang et al., 2001; Ambard-Bretteville et al., 2003).
As mitochondria are endosymbiotic in origin, many proteins were once encoded within the organelle. However, soon after the establishment of the endosymbiosis, massive gene transfer meant that many proteins were encoded in the nucleus and subsequently had to be imported into the mitochondrion (Gray et al., 1999). Many other mitochondrial proteins came from other sources, i.e. not from the endosymbiont giving rise to mitochondria (Andersson et al., 2003). These proteins have also had to acquire mitochondrial targeting signals. Although gene transfer has ceased in fungi and animals, it is an ongoing process in plants (Adams and Palmer, 2003).
Examination of plant mitochondrial genomes from a variety of species indicates that some proteins are mitochondrially encoded in one species, and in others they are nuclear encoded. This process of ongoing gene transfer has allowed the opportunity to study the process and steps necessary for the successful transfer of a gene from the mitochondrion to the nucleus. Furthermore, it has yielded insights into how transferred genes have acquired mitochondrial targeting signals, and given clues as to why they have been transferred so late in evolution. For cases of mitochondrial proteins where active organelle and nuclear genes exist, a comparison of the proteins can point to the changes that were necessary for mitochondrial import. In the case of subunit 2 of cytochrome c oxidase in legumes, changes in the local hydrophobicity of the first transmembrane helix were required in addition to acquiring a cleavable mitochondrial targeting signal to facilitate gene transfer from the mitochondrion to the nucleus in soybean (Daley et al., 2002).
The Rps10 gene, coding for the small subunit ribosomal protein Rps10, is located in the mitochondrion of some flowering plants and in the nucleus of others because of recent and frequent transfers to the nucleus during flowering plant evolution (Knoop et al., 1995; Wischmann and Schuster, 1995; Adams et al., 2000; Kubo et al., 2000, 2003). This has resulted in a variety of gene structures for nuclear encoded Rps10 in various species (Fig. 1; Adams et al., 2000). Arabidopsis and carrot (Daucus carota) each have an N-terminal extension of 100 amino acids or more. The extension on carrot Rps10 is derived from the gene for mitochondrial hsp22 (Adams et al., 2000), whereas the extension on Arabidopsis Rps10 has similarity (39%) to a recently annotated gene encoding a DEAD-box RNA helicase. Lettuce (Lactuca sativa) Rps10 has an extension of 30 amino acids derived from a nonmitochondrial metalloprotease gene (Adams et al., 2000); Oxalis, spinach (Spinacia oleracea), and maize have gained no extension in comparison to the mitochondrially encoded rps10 from soybean (Glycine max).
Figure 1.
Amino acid sequence alignment of the predicted Rps10 proteins from several plant species. The mitochondrially encoded soybean [Gm rps10(m)] is shown on top with six nuclear encoded Rps10 proteins aligned below; n indicates a nuclear coding location and m indicates a mitochondrial coding location. Residues identical to soybean are white on a black background, and conservative changes are indicated with a gray box. Gaps introduced to align proteins are indicated by dots. The numbers refer to the number of amino acids. At = Arabidopsis, Dc = carrot, La = lettuce, Ox = Oxalis, So = spinach, and Zm = maize.
We have studied the minimal requirements for mitochondrial targeting of the nuclear encoded Rps10 proteins. We conclude that Rps10 either had to obtain a mitochondrial targeting presequence or alternatively undergo extensive sequence modification upon transfer to the nucleus to enable it to be retargeted back to the mitochondrion.
RESULTS
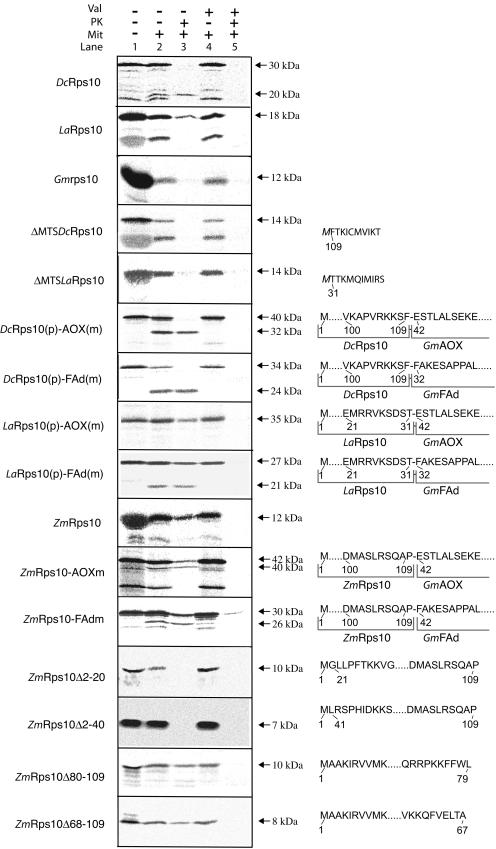
Upon transfer to the nucleus, the Rps10 gene from carrot was inserted into a gene for mitochondrial hsp22 and lettuce Rps10 was inserted into a gene for a nonmitochondrial gene encoding a metalloprotease (Adams et al., 2000). Each Rps10 gene acquired an N-terminal extension from its host gene (Fig. 1). Because the transferred Rps10 gene in some other plants, such as maize and spinach, contains no N-terminal extension, it is possible that the extensions in carrot and lettuce Rps10 are not needed for import. This hypothesis is particularly compelling for lettuce Rps10 whose extension is derived from a nonmitochondrial gene. To determine if the N-terminal extensions are necessary for mitochondrial targeting and import, we carried out import assays into purified potato (Solanum tuberosum) mitochondria with in vitro synthesized lettuce Rps10 and carrot Rps10. We used purified potato mitochondria where rps10 is encoded in the mitochondrion (Knoop et al., 1995), but essentially the same results were obtained when we used Arabidopsis mitochondria where Rps10 is nuclear encoded (data not shown; Wischmann and Schuster, 1995). Both Rps10 proteins were imported to a protease protected location in a membrane potential dependent manner into mitochondria. The 18-kD lettuce Rps10 protein was not processed upon import (Fig. 2A). Removal of the N-terminal extension abolished import (Fig. 2A), indicating that the extension, derived from the metalloprotease gene, is necessary for import. Carrot Rps10 was cleaved upon import to produce a mature product of 20 kD from the 30-kD precursor protein. However, removal of the N-terminal extension abolished import, indicating that the hsp22-derived extension is necessary for import. The mitochondrially encoded soybean rps10 was not imported into isolated mitochondria as evidenced by the lack of any protease protected product when incubated under identical conditions that supported the import of carrot and lettuce Rps10 (Fig. 2A).
Figure 2.
Import of Rps10 proteins into mitochondria. In vitro radiolabeled Rps10 proteins from carrot, lettuce, maize, and soybean were tested for import ability into purified mitochondria. Import of the various Rps10 wild-type proteins, carrot, and lettuce that have the N-terminal extension removed (ΔMTS) and the carrot and lettuce N-terminal extension placed in front of the alternative oxidase mature protein [DcRps10(p)-AOX(m) and LaRps10(p)-AOX(m)] and in front of the FAd subunit of mitochondrial ATP synthase [DcRps10(p)-FAd(m) and LaRps10(p)-FAd(m)]. The first 10 amino acids are shown for carrot and lettuce where the N-terminal extension has been removed; the M in italics indicates that this residue was added to provide a start codon. The N-terminal extension of carrot and lettuce place in front of the alternative oxidase and FAd subunit are shown with the 10 amino acids from each protein that border the junction point. Numbering is as from Figure 1, with dots indicating any number of amino acids. B, Localization of the mitochondrial targeting information in maize Rps10. The maize Rps10 protein was fused to the mature alternative oxidase and FAd subunit of mitochondrial ATP synthase to test if it could support import of other proteins [ZmRps10-AOX(m) and ZmRps10-FAd(m)]. Deletions were made from the N and C termini to determine the location of the mitochondrial targeting signal in the maize Rps10 protein. In the case of the chimeric proteins, the 10 amino acids boarding the junction are shown. For the deletions, the 10 amino acids nearest the deletion are shown. The numbering of the deletions is based on amino acids number as in Figure 1.
To determine if the N-terminal extensions of Rps10 from carrot and lettuce possess generic mitochondrial import information, the extensions were fused to two other mitochondrial proteins: alternative oxidase (AOX) and the FAd subunit of mitochondrial ATP synthase (FAd). Previously, we have demonstrated that both AOX and FAd mature proteins have no mitochondrial targeting ability (Tanudji et al., 2001). The extensions from both Rps10 proteins could support the import of two other mitochondrial proteins (Fig. 2A), indicating that they contained generic mitochondrial import information. The carrot extension was cleaved from both proteins upon import to generate a lower molecular mass mature protein. In the case of the lettuce protein, no processing was observed with AOX as a passenger but some processing was observed with the FAd as a passenger. The inefficient processing seen in the latter case may have resulted due to the generation of a cryptic processing site as has been previously observed with chimeric constructs upon import into mitochondria (Duby et al., 2001a). The results of these experiments show that a region of the nonmitochondrial gene encoding a metalloprotease that is immediately upstream of the transferred Rps10 gene in lettuce either possesses mitochondrial targeting capability or has evolved it upon association with Rps10. The metalloprotease sequence underwent numerous amino acid substitutions (up to 16 out of 29 amino acids were changed) after association with Rps10 that may have facilitated creation of a mitochondrial targeting sequence.
In contrast to Rps10 from lettuce and carrot, maize Rps10 has no N-terminal extension, but the maize Rps10 can be imported into isolated mitochondria (Fig. 2B; Adams et al., 2000). The entire maize Rps10 protein could support import of passenger proteins, as both AOX and FAd were imported into mitochondria under the direction of maize Rps10 (Fig. 2B). To define the location of the mitochondrial targeting signals in maize Rps10, we made deletions of the N and C termini. Deletion of 20 or 40 amino acids from the N terminus completely abolished mitochondrial import, whereas deletion of 20 or 40 amino acids from the C terminus had no affect on mitochondrial uptake (Fig. 2B).
With the various fusion proteins containing AOX or FAd mature proteins it appears that processing was not always at the junction of the fusion proteins. The AOX mature protein has an apparent molecular mass of 32 kD and the mature FAd protein has an apparent molecular mass of 22 kD (Tanudji et al., 2001). As these sized products were not always generated when processing was observed, it indicates that processing was unlikely to be at the fusion site (Fig. 2). However, this does not affect targeting ability and we concluded that the N-terminal region of the maize Rps10 had mutated following gene transfer, to contain mitochondrial targeting information.
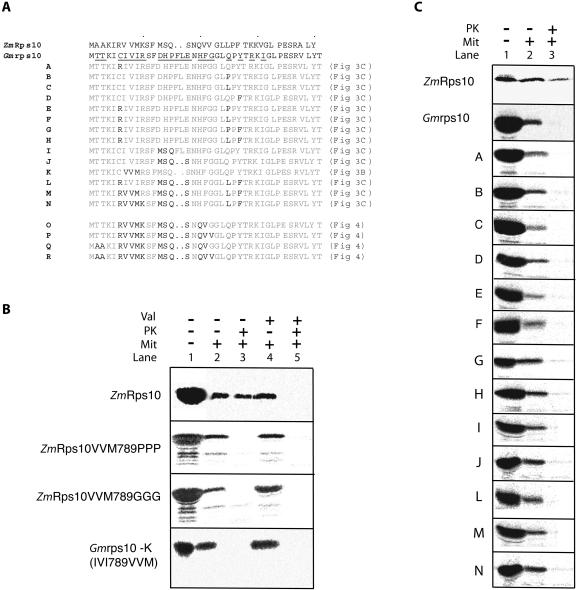
To recreate this evolutionary process, the N-terminal region of the soybean rps10 protein was mutagenized to resemble the maize Rps10. A previous study also identified the targeting information of rice Rps10 to be located in the N-terminal region and mutated three residues to inhibit import (Kubo et al., 2003). The three residues mutated in the previous study correspond to seven to nine of the maize Rps10 and soybean rps10 protein (Fig. 3A). Mutating residues 7 to 9 (VVM) of maize Rps10 to Gly or Pro inhibited import in agreement with previous findings (Fig. 3B). To test the role of these residues in supporting mitochondrial import, we mutated residues 7 to 9 of soybean rps10 (IVI, which was not imported) to what they are in maize Rps10 (VVM, which was imported; Fig. 3A). Changing these residues in soybean rps10 did not result in import (Fig. 3B). This illustrates the limitations of mutating residues as changes may simply inhibit import and thus not identify the features required to support mitochondrial uptake.
Figure 3.
Converting mitochondrially encoded soybean rps10 to a protein that can be imported into mitochondria. A, Amino acid residues that differed in soybean rps10 were changed to the corresponding amino acid in maize Rps10. The mutants made are labeled A to R and the location of the import assay for each mutant is indicated in brackets. B, The inhibitory affect of changing residues 7, 8, and 9 to Gly or Pro in maize Rps10. However, changing residues 7, 8, and 9 in soybean to the corresponding amino acids in maize Rps10 does not support import. C, Import assays of various soybean rps10 mutants into isolated mitochondria. The mutants are designated as outlined in part A.
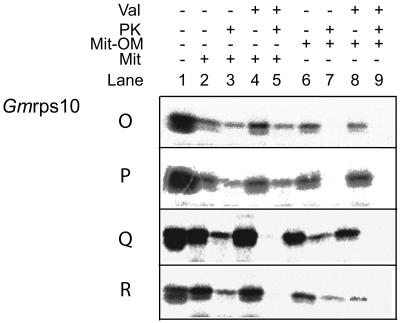
Changing single or double residues of soybean rps10 was also not sufficient to support mitochondrial uptake. Even changing up to nine residues in the first 20 amino acids of soybean rps10, out of the 15 that differed between maize and soybean, did not result in mitochondrial import (Fig. 3, A and C). Of the 15 residues that differed, five would be considered to be in the same physicochemical grouping, i.e. hydrophobic, aliphatic, or charged (Fig. 3A). Changes at positions 18 to 20 did result in mitochondrial import (Fig. 3A, mutants O and P; Fig. 4). However, it was evident that this import was not sensitive to the addition of valinomycin, which abolishes the membrane potential (Fig. 4, mutants O and P, lanes 4 and 5). As all proteins imported into or across the inner membrane require a membrane potential, this suggested that import had only taken place across the outer membrane (Neupert, 1997; Glaser et al., 1998). To test this possibility, we carried out import assays into outer membrane ruptured mitochondria. Rupture of the outer membrane by osmotic shock, followed by washing, removes intermembrane space components. Import into outer membrane ruptured mitochondria is still possible at a reduced efficiency, but it allowed assessment of whether import takes place across the inner mitochondrial membrane (Lister et al., 2002; Murcha et al., 2004). Import assays carried out with soybean rps10 mutants O and P into outer membrane ruptured mitochondria resulted in no protease protected products (Fig. 4, lanes 6 and 7), confirming that these proteins were only imported across the outer membrane in intact mitochondria. We then changed the only two remaining residues that differed in the first 20 amino acids between soybean rps10 and maize Rps10, Thr at positions 2 and 3 to Ala (Fig. 3A, mutants Q and R). Import assays with these two precursors resulted in a protease protected product that was sensitive to the addition of valinomycin (Fig. 4, mutants Q and R, lanes 1–5). To confirm that these proteins were imported across the inner membrane, we carried out import assays into outer membrane ruptured mitochondria. Again, a protease protected valinomycin sensitive product was generated indicating import across the inner membrane (Fig. 4, lanes 6–9).
Figure 4.
Import of soybean rps10 mutants into mitochondria and outer membrane ruptured mitochondria. The import of four mutants was tested into intact mitochondria and outer membrane ruptured mitochondria. Mutants O and P were imported into intact mitochondria as they were protease protected (lanes 1–3). This import was not sensitive to addition of valinomycin (lanes 4 and 5). Mutants O and P were not imported across or into the inner membrane in outer membrane ruptured mitochondria (lanes 6–9). Mutants Q and R were imported into intact mitochondria in a valinomycin sensitive manner (lanes 1–5) and were imported into outer membrane ruptured mitochondria in a membrane potential dependent manner (lanes 6–9).
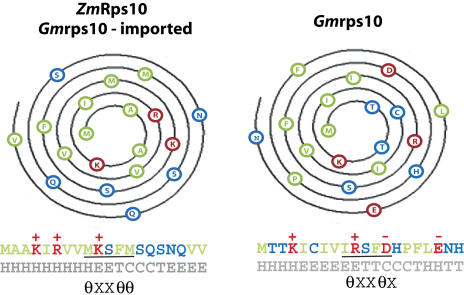
Converting soybean rps10 to an imported protein allows us to examine the requirements for import. The changes required can be summarized as follows (Fig. 5): abolition of negative residues and addition of one positive residue, change of hydrophobic residues to form an α-helical amphiphatic structure, and formation of a core binding motif defined for TOM20 (Abe et al., 2000; Muto et al., 2001).
Figure 5.
Analysis of the requirements to support import of soybean rps10 into mitochondria. Helical wheel projections of maize Rps10 protein, imported soybean rps10 protein, and unimportable soybean rps10 proteins are shown. The first amino acid is at the interior with successive amino acids moving outwards on the helix in a clockwise manner. The amino acids are colored according to chemical properties, of side chain Red = charged, Green = hydrophobic, and Blue = polar. The first 20 amino acids of each protein are depicted below the helical projection with coloring as described above. Structural prediction of secondary is indicated below in θXXθθ.
Overall, the changes resulted in features that combine to achieve mitochondrial import. Individually, none of these changes appeared to be sufficient to support import. Soybean rps10 mutant N for instance has the changes that incorporate both the charge and TOM20 binding motif but still fails to be imported into mitochondria. Thus, altering the hydrophobicity is also an essential feature to support mitochondrial import. Hydrophobic moment analysis indicates that both soybean rps10 and maize Rps10 display highest predicted hydrophobic moment at residues 5 (Supplemental Table I). However, from residues 5 to 10, the hydrophobicity differs between the two proteins, with soybean rps10 maintaining relatively high hydrophobicity. Overall, there are only two differences in the number of hydrophobic residues between maize Rps10 and soybean rps10: 11 compared to 9 residues. However, the changes have resulted in a change in the position of these hydrophobic residues, combined with the changes in charge to achieve a predicted amphiphatic structure. Helical wheel projections indicate that for the imported Rps10 proteins, eight hydrophobic residues are on one face and the hydrophilic and positive residues are on the other face (Fig. 5, ZmRps10 and Gmrps10-imported). The unimportable soybean rps10 does appear to have a hydrophobic face when plotted on a helical wheel. However, predictions indicate that overall it is less helical forming than the imported Rps10 proteins and negative residues are also on the more polar face (Fig. 5, Gmrps10). The changes also resulted in the generation of a motif that has been defined for TOM20 binding, θXXθθ, where θ is any hydrophobic amino acid and X is aliphatic with a preference for a long side chain.
DISCUSSION
Following transfer to the nucleus, the Rps10 gene gained a mitochondrial targeting sequence in different ways as a result of separate transfer events. Rps10 in carrot acquired a long N-terminal extension from the mitochondrial hsp22 gene and this extension is essential for import of the protein into mitochondria. Rps10 in lettuce acquired a short N-terminal extension from a gene encoding a nonmitochondrial metalloprotease. The extension of lettuce Rps10 is also essential for mitochondrial import. Thus, a region of a nonmitochondrial gene became a mitochondrial targeting sequence upon association with the newly transferred Rps10. A similar situation occurred for the transferred Rps19 gene in Arabidopsis (Sanchez et al., 1996; Adams et al., 2002). In contrast, Rps10 in maize did not acquire an N-terminal extension following transfer to the nucleus. Instead, there were base changes in the N-terminal region of the transferred Rps10 gene that allowed creation of an N-terminal noncleavable mitochondrial targeting sequence. To recreate this evolutionary process, amino acid residues in mitochondrial encoded rps10 from soybean were changed to corresponding residues in nuclear encoded Rps10 from maize until mitochondrial import into the functional location of ribosomal proteins was achieved, i.e. the mitochondrial matrix. Overall, this process mimics what has occurred in nature in order to achieve successful gene transfer, allowing all the requirements for mitochondrial import to be assessed. Some caution needs to be taken as it is possible that mitochondrial import could be achieved by altering fewer residues if they were altered in a different manner. However, given that 15 residues in total had to be changed, it is not feasible to make several thousand alterations (32,767) to investigate all these possibilities. Usually, studies investigating the requirements for protein targeting to mitochondria mutate residues to identify critical residues required for import. These studies have been informative but care must be taken when interpreting results as inserted residues may simply be inhibitory for import and the role of individual residues can be difficult to assess. This was evident with a previous study of Rps10 from rice where import could be abolished by targeting a specific region (Kubo et al., 2003), yet the overall role of the residues changed could not be assessed. Here, we demonstrated that changing these residues alone could not support import (Fig. 3B). There may be several features necessary to support mitochondrial import and abolishing import by altering one of these parameters may cause other features to be missed.
Overall, we defined four parameters that were required to be changed to achieve mitochondrial import: charge, secondary structure, hydrophobicity, and generation of a proposed TOM20 binding site. Alone, none of these parameters was sufficient to support import; in fact, mutants O and P had all but two additional hydrophobicity changes yet were not imported across the inner mitochondrial membrane. It appeared that hydrophobicity was a crucial feature in facilitating mitochondrial import and that this is difficult to assess from the physicochemical classification of amino acids alone. Analysis of the hydrophobic moment of the first 20 amino acids indicates that the hydrophobic moment is very similar for the first five amino acids, but from amino acids 7 to 12 it is greatly altered (Supplemental Table I). Comparison of the first 20 amino acids of soybean rps10 mutant Q or R to mutant L reveals that on a physicochemical basis they are identical; rather, it is that the VVM residues are required to be imported, whereas the IVI sequence is not imported (Fig. 3). This change in hydrophobicity is due to the different amino acids, not just the properties of the amino acids and thus even what would be regarded as conservative changes are important to facilitate mitochondrial import.
In the case of Rps10 from carrot and lettuce, it appears that the acquired cleavable mitochondrial targeting signals also display these features; they both contain the proposed TOM20 binding site, the potential to form an amphiphatic structure, and an overall positive charge of 3 (Supplemental Table I). In the case of carrot, these features are in the first 30 amino acids, possibly indicating that the entire region gained may not be required for import. However, for Oxalis and spinach Rps10, which appear similar to maize in that they have not gained additional sequences to support import, some of these features appear to be absent. Although the first 20 amino acids from both have the potential to form an amphiphatic structure and contain nine hydrophobic residues, both lack the proposed TOM20 binding site and also contain a negative Glu residue at position 18 as in soybean rps10 but not present in maize Rps10. Thus, the first 20 amino acids of both Oxalis and spinach Rps10 resemble soybean rps10 as much as they do maize Rps10. There are several possibilities to explain this discrepancy. First, the Oxalis and spinach Rps10 mitochondrial targeting signal may be present elsewhere in the protein. Internal and C-terminal targeting signals are well described in fungal systems (Neupert, 1997; Pfanner and Geissler, 2001). Alternatively, the targeting signal for Oxalis and spinach Rps10 may be via an N-terminal located signal but recognized by a different outer membrane surface receptor. Although TOM20 is the only receptor characterized in plants to date (Werhahn et al., 2001), an isoform of a chloroplast outer envelope protein TOC64 is present on the outer membrane of plant mitochondria (Chew et al., 2004), and a homolog of metaxin involved in protein import in animals has also been identified in plants (Armstrong et al., 1997; Lister et al., 2004). Another possibility is that in Arabidopsis, four genes exist for TOM20 and thus it is possible that each receptor may recognize different features (Werhahn et al., 2001, 2003). Plant Tom20 was first characterized by biochemical means and sequence similarity with animal and fungal TOM20 is not significant (Werhahn et al., 2001, 2003). It has been proposed that plant TOM20 represents a case of convergent evolution and thus the features important for binding to animal and fungal TOM20 may not be the same for plant TOM20 (Likic et al., 2005). An analysis of the plant TOM complex indicates that, unlike the fungal counterpart, acidic residues are much less abundant; whereas the yeast TOM complex has 26 more acidic residues than basic, the Arabidopsis TOM complex has an excess of six basic residues (Werhahn et al., 2003). Therefore, hydrophobic residues may play a more dominant role in precursor protein recognition. A common feature of the N-terminal region of all nuclear encoded Rps10 proteins is an abundance of hydrophobic residues. Therefore, it may be the degree of hydrophobicity and the ability to form an amphiphilic structure that are the essential features required to support import and the independent gene transfers achieve this in different manners.
In defining the requirement for mitochondrial import, it was apparent that import could be achieved into the intermembrane space before it could be achieved into the matrix. Genes that encode proteins that do not contain mitochondrial targeting ability can acquire mitochondrial targeting signals from preexisting targeting signals or gain extensions that had no previous mitochondrial targeting ability (Kadowaki et al., 1996; Adams et al., 2000; Kubo et al., 2003). In studies assessing the potential of DNA fragments from Escherichia coli to have mitochondrial targeting ability, it was predicted that from 2% to 5% of clones tested had mitochondrial targeting activity (Baker and Schatz, 1987), or that 5% of proteins from E. coli are predicted to possess mitochondrial targeting activity (Lucattini et al., 2004). When one of these proteins from the latter study was tested for import into mitochondria, it was shown to be imported across the outer membrane only, indicating that import of a protein across the outer membrane may be an intermediate step in the natural process of gene transfer before inner membrane targeting activity is gained (Lucattini et al., 2004).
Mitochondrial encoded ribosomal proteins in plants display frequent rates of transfer to the nucleus (Adams et al., 2002). One feature thought to facilitate this transfer was that the encoded protein does not display any features that are potential barriers for import back into mitochondria, as they are generally small hydrophilic proteins. These proteins still require a typical mitochondrial targeting signal whether it is acquired from another gene or changes occur with the transferred gene itself. In the latter case, substantial changes in sequence were required to allow successful import of the maize Rps10 protein. However, no changes were required in the interior of the protein as has been previously shown for subunit 2 of cytochrome c oxidase in soybean (Daley et al., 2002). Thus, the initial site of integration and targeting signal acquisition may play an important role in determining if genes are activated upon transfer to the nucleus. As Rps10 and many other ribosomal proteins can acquire a mitochondrial targeting signal either from another gene or by changes to the transferred gene, it is puzzling why ribosomal proteins are still organelle encoded. Ribosomal proteins function in a large multisubunit complex containing proteins and RNA molecules. This machine needs to be assembled in an ordered sequential manner (Williamson, 2003; Granneman and Baserga, 2004), as has been demonstrated for several other organelle located multisubunit protein complexes (Zhang and Aro, 2002; Naithani et al., 2003; Suorsa et al., 2004). The rate of assembly can be dictated by an organelle encoded subunit as demonstrated for the assembly of complex I in mammalian cells lines where the mitochondrially encoded ND5 protein is rate limiting (Chomyn, 2001). Therefore, assembly of the functional ribosome may dictate the retention of the coding of ribosomal proteins in mitochondria. Assembly is the final step in an import pathway for a nuclear encoded organelle located protein and has been proposed previously to be a possible reason the organelle coding location of some proteins (Zerges, 2002; Daley and Whelan, 2005). Thus, even though various higher plant species may differ in organellar coding capacity for ribosomal proteins, they all have maintained some genes for ribosomal proteins, possibly due to the requirements for assembly.
MATERIALS AND METHODS
Precursor Proteins, Potato Mitochondrial Isolation, and Outer Membrane Ruptured Mitochondria
The cDNAs encoding the Rps10 proteins have been described previously (Adams et al., 2000). Constructs of carrot (Daucus carota) Rps10 and lettuce Rps10 lacking the N-terminal extensions (see Fig. 2) were created by PCR amplification of cDNA and cloning into pBluescript. The coding regions of soybean AOX and FAd with the targeting signals removed (Tanudji et al., 2001) were placed downstream in frame of the coding region of various Rps10 proteins using standard cloning techniques, inserting compatible restriction site using site directed mutagenesis with the Quikchange site directed mutagenesis kit according to manufacturer's instructions (Stratagene, Sydney). Precursor proteins were produced in a coupled transcription-translation system according to manufacturer's instructions (Promega, Melbourne).
Percoll density gradient potato (Solanum tuberosum) cv Desiree tuber mitochondria were isolated according to Millar et al. (2001). To prepare outer membrane ruptured mitochondria for import assays, 200 μg of pelleted mitochondria were resuspended in 10 μL SEH buffer (250 mm Suc, 1 mm EDTA, 10 mm HEPES, pH 7.4). One hundred and fifty-five μL of 20 mm HEPES, pH 7.4, was added and incubated on ice 20 min; 25 μL of 2 m Suc and 10 μL of 3 m KCl were added and mixed. These mitochondria are referred to as outer membrane ruptured mitochondria. This procedure was carried out to test if precursor proteins could be imported across the inner membrane as the outer membrane is ruptured, and thus upon protease treatment after the import assay only proteins imported across the inner membrane will be protected from digestion.
Protein Import into Mitochondria and Outer Membrane Ruptured Mitochondria
Import assays into potato mitochondria were carried out at as outlined previously (Lister et al., 2002; Murcha et al., 2004). A total of 200 μg of mitochondria was used in each import assay. Import assays were carried out at 23°C for 20 min. Import assays were carried out in 15-mL polypropylene tubes in a volume of 200 μL in 0.3 m Suc, 50 mm KCl, 10 mm MOPS, pH 7.2, 5 mm KH2PO4, 1% (w/v) bovine serum albumin, 1 mm MgCl2, 1 mm Met, 0.2 mm ADP, 0.75 mm ATP, 5 mm succinate, and 5 mm dithiothreitol. Assays were stopped by placing on ice, divided into two aliquots and addition of Proteinase K to one aliquot to a final concentration of 50 μg/mL. Proteinase K digestion was stopped by the addition of phenylmethylsulfonyl fluoride to 2 mm after 15 min. Mitochondria were diluted into 1 mL of ice cold import buffer and pelleted by centrifugation for 3 min in a microfuge. Mitochondrial pellets were resuspended in SDS-PAGE gel sample buffer, subjected to SDS-PAGE, gels dried, and products visualized by exposing to a BAS TR2040 plate for 24 h and reading in a BAS 2500 Bio imaging analyzer (Fuji, Tokyo). Import assays into outer membrane ruptured mitochondria were carried out as for mitochondria except that 200 μg of outer membrane ruptured mitochondria were added in place of mitochondria.
Analysis of the Mitochondrial Targeting Signal of Rps10
The predicted secondary structure characteristics of the first 20 amino acids of maize Rps10 and soybean rps10 were analyzed using the Expasy suite of analysis tools (http://au.expasy.org/tools/). The tools helical wheel, helical draw, and hydrophobic moment were used on default settings.
Supplementary Material
This work was supported by the Australian Research Council (to J.W.).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.062745.
References
- Abe Y, Shodai T, Muto T, Mihara K, Torii H, Nishikawa S, Endo T, Kohda D (2000) Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell 100: 551–560 [DOI] [PubMed] [Google Scholar]
- Adams KL, Daley DO, Qiu YL, Whelan J, Palmer JD (2000) Repeated, recent and diverse transfers of a mitochondrial gene to the nucleus in flowering plants. Nature 408: 354–357 [DOI] [PubMed] [Google Scholar]
- Adams KL, Palmer JD (2003) Evolution of mitochondrial gene content: gene loss and transfer to the nucleus. Mol Phylogenet Evol 29: 380–395 [DOI] [PubMed] [Google Scholar]
- Adams KL, Qiu YL, Stoutemyer M, Palmer JD (2002) Punctuated evolution of mitochondrial gene content: high and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. Proc Natl Acad Sci USA 99: 9905–9912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambard-Bretteville F, Small I, Grandjean O, Colas des Francs-Small C (2003) Discrete mutations in the presequence of potato formate dehydrogenase inhibit the in vivo targeting of GFP fusions into mitochondria. Biochem Biophys Res Commun 311: 966–971 [DOI] [PubMed] [Google Scholar]
- Andersson SG, Karlberg O, Canback B, Kurland CG (2003) On the origin of mitochondria: a genomics perspective. Philos Trans R Soc Lond B Biol Sci 358: 165–177; discussion 177–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong LC, Komiya T, Bergman BE, Mihara K, Bornstein P (1997) Metaxin is a component of a preprotein import complex in the outer membrane of the mammalian mitochondrion. J Biol Chem 272: 6510–6518 [DOI] [PubMed] [Google Scholar]
- Baker A, Schatz G (1987) Sequences from a prokaryotic genome or the mouse dihydrofolate reductase gene can restore the import of a truncated precursor protein into yeast mitochondria. Proc Natl Acad Sci USA 84: 3117–3121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger G, Gray MW, Lang BF (2003) Mitochondrial genomes: anything goes. Trends Genet 19: 709–716 [DOI] [PubMed] [Google Scholar]
- Chew O, Lister R, Qbadou S, Heazlewood JL, Soll J, Schleiff E, Millar AH, Whelan J (2004) A plant outer mitochondrial membrane protein with high amino acid sequence identity to a chloroplast protein import receptor. FEBS Lett 557: 109–114 [DOI] [PubMed] [Google Scholar]
- Chomyn A (2001) Mitochondrial genetic control of assembly and function of complex I in mammalian cells. J Bioenerg Biomembr 33: 251–257 [DOI] [PubMed] [Google Scholar]
- Daley DO, Clifton R, Whelan J (2002) Intracellular gene transfer: reduced hydrophobicity facilitates gene transfer for subunit 2 of cytochrome c oxidase. Proc Natl Acad Sci USA 99: 10510–10515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley DO, Whelan J (2005) All wired up or too hot to handle: Why do genes persist in organelle genomes? Genome Biol 6: 110.1–110.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duby G, Degand H, Boutry M (2001. a) Structure requirement and identification of a cryptic cleavage site in the mitochondrial processing of a plant F1-ATPase beta-subunit presequence. FEBS Lett 505: 409–413 [DOI] [PubMed] [Google Scholar]
- Duby G, Oufattole M, Boutry M (2001. b) Hydrophobic residues within the predicted N-terminal amphiphilic alpha-helix of a plant mitochondrial targeting presequence play a major role in in vivo import. Plant J 27: 539–549 [DOI] [PubMed] [Google Scholar]
- Glaser E, Sjoling S, Tanudji M, Whelan J (1998) Mitochondrial protein import in plants. Signals, sorting, targeting, processing and regulation. Plant Mol Biol 38: 311–338 [DOI] [PubMed] [Google Scholar]
- Granneman S, Baserga SJ (2004) Ribosome biogenesis: of knobs and RNA processing. Exp Cell Res 296: 43–50 [DOI] [PubMed] [Google Scholar]
- Gray MW, Burger G, Lang BF (1999) Mitochondrial evolution. Science 283: 1476–1481 [DOI] [PubMed] [Google Scholar]
- Hammen PK, Gorenstein DG, Weiner H (1996. a) Amphiphilicity determines binding properties of three mitochondrial presequences to lipid surfaces. Biochemistry 35: 3772–3781 [DOI] [PubMed] [Google Scholar]
- Hammen PK, Waltner M, Hahnemann B, Heard TS, Weiner H (1996. b) The role of positive charges and structural segments in the presequence of rat liver aldehyde dehydrogenase in import into mitochondria. J Biol Chem 271: 21041–21048 [PubMed] [Google Scholar]
- Heazlewood JL, Tonti-Filippini JS, Gout AM, Day DA, Whelan J, Millar AH (2004) Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. Plant Cell 16: 241–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogenraad NJ, Ward LA, Ryan MT (2002) Import and assembly of proteins into mitochondria of mammalian cells. Biochim Biophys Acta 1592: 97–105 [DOI] [PubMed] [Google Scholar]
- Humphries AD, Streimann IC, Stojanovski D, Johnston AJ, Yano M, Hoogenraad NJ, Ryan MT (2005) Dissection of the mitochondrial import and assembly pathway for human TOM40. J Biol Chem 280: 11535–11543 [DOI] [PubMed] [Google Scholar]
- Kadowaki K, Kubo N, Ozawa K, Hirai A (1996) Targeting presequence acquisition after mitochondrial gene transfer to the nucleus occurs by duplication of existing targeting signals. EMBO J 15: 6652–6661 [PMC free article] [PubMed] [Google Scholar]
- Knoop V, Ehrhardt T, Lattig K, Brennicke A (1995) The gene for ribosomal protein S10 is present in mitochondria of pea and potato but absent from those of Arabidopsis and Oenothera. Curr Genet 27: 559–564 [DOI] [PubMed] [Google Scholar]
- Kubo N, Arimura S, Tsutsumi N, Hirai A, Kadowaki K (2003) Involvement of N-terminal region in mitochondrial targeting of rice RPS10 and RPS14 proteins. Plant Sci 164: 1047–1055 [Google Scholar]
- Kubo N, Jordana X, Ozawa K, Zanlungo S, Harada K, Sasaki T, Kadowaki K (2000) Transfer of the mitochondrial rps10 gene to the nucleus in rice: acquisition of the 5′ untranslated region followed by gene duplication. Mol Gen Genet 263: 733–739 [DOI] [PubMed] [Google Scholar]
- Likic VA, Perry A, Hulett J, Derby M, Traven A, Waller RF, Keeling PJ, Koehler CM, Curran SP, Gooley PR, et al (2005) Patterns that define the four domains conserved in known and novel isoforms of the protein import receptor Tom20. J Mol Biol 347: 81–93 [DOI] [PubMed] [Google Scholar]
- Lister R, Chew O, Lee MN, Heazlewood JL, Clifton R, Parker KL, Millar AH, Whelan J (2004) A transcriptomic and proteomic characterization of the Arabidopsis mitochondrial protein import apparatus and its response to mitochondrial dysfunction. Plant Physiol 134: 777–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Mowday B, Whelan J, Millar AH (2002) Zinc-dependent intermembrane space proteins stimulate import of carrier proteins into plant mitochondria. Plant J 30: 555–566 [DOI] [PubMed] [Google Scholar]
- Lucattini R, Likic VA, Lithgow T (2004) Bacterial proteins predisposed for targeting to mitochondria. Mol Biol Evol 21: 652–658 [DOI] [PubMed] [Google Scholar]
- Macasev D, Whelan J, Newbigin E, Silva-Filho MC, Mulhern TD, Lithgow T (2004) Tom22', an 8-kDa trans-site receptor in plants and protozoans, is a conserved feature of the TOM Complex that appeared early in the evolution of eukaryotes. Mol Biol Evol 21: 1557–1564 [DOI] [PubMed] [Google Scholar]
- Millar AH, Heazlewood JL, Kristensen BK, Braun HP, Moller IM (2005) The plant mitochondrial proteome. Trends Plant Sci 10: 36–43 [DOI] [PubMed] [Google Scholar]
- Millar AH, Liddell A, Leaver CJ (2001) Isolation and sub-fractionation of mitochondria from plants. In E Schon, ed, Methods in Cell Biology on Mitochondria. Academic Press, San Diego, pp 53–74 [DOI] [PubMed]
- Murcha MW, Elhafez D, Millar AH, Whelan J (2004) The N-terminal extension of plant mitochondrial carrier proteins is removed by two-step processing: the first cleavage is by the mitochondrial processing peptidase. J Mol Biol 344: 443–454 [DOI] [PubMed] [Google Scholar]
- Muto T, Obita T, Abe Y, Shodai T, Endo T, Kohda D (2001) NMR identification of the Tom20 binding segment in mitochondrial presequences. J Mol Biol 306: 137–143 [DOI] [PubMed] [Google Scholar]
- Naithani S, Saracco SA, Butler CA, Fox TD (2003) Interactions among COX1, COX2, and COX3 mRNA-specific translational activator proteins on the inner surface of the mitochondrial inner membrane of Saccharomyces cerevisiae. Mol Biol Cell 14: 324–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert W (1997) Protein import into mitochondria. Annu Rev Biochem 66: 863–917 [DOI] [PubMed] [Google Scholar]
- Pfanner N, Chacinska A (2002) The mitochondrial import machinery: preprotein-conducting channels with binding sites for presequences. Biochim Biophys Acta 1592: 15–24 [DOI] [PubMed] [Google Scholar]
- Pfanner N, Geissler A (2001) Versatility of the mitochondrial protein import machinery. Nat Rev Mol Cell Biol 2: 339–349 [DOI] [PubMed] [Google Scholar]
- Prokisch H, Scharfe C, Camp DG II, Xiao W, David L, Andreoli C, Monroe ME, Moore RJ, Gritsenko MA, Kozany C, et al (2004) Integrative analysis of the mitochondrial proteome in yeast. PLoS Biol 2: e160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roise D, Schatz G (1988) Mitochondrial presequences. J Biol Chem 263: 4509–4511 [PubMed] [Google Scholar]
- Sanchez H, Fester T, Kloska S, Schroder W, Schuster W (1996) Transfer of rps19 to the nucleus involves the gain of an RNP-binding motif which may functionally replace RPS13 in Arabidopsis mitochondria. EMBO J 15: 2138–2149 [PMC free article] [PubMed] [Google Scholar]
- Schatz G, Dobberstein B (1996) Common principles of protein translocation across membranes. Science 271: 1519–1526 [DOI] [PubMed] [Google Scholar]
- Sickmann A, Reinders J, Wagner Y, Joppich C, Zahedi R, Meyer HE, Schonfisch B, Perschil I, Chacinska A, Guiard B, et al (2003) The proteome of Saccharomyces cerevisiae mitochondria. Proc Natl Acad Sci USA 100: 13207–13212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjoling S, Glaser E (1998) Mitochondrial targeting peptides in plants. Trends Plant Sci 3: 136–140 [Google Scholar]
- Suorsa M, Regel RE, Paakkarinen V, Battchikova N, Herrmann RG, Aro EM (2004) Protein assembly of photosystem II and accumulation of subcomplexes in the absence of low molecular mass subunits PsbL and PsbJ. Eur J Biochem 271: 96–107 [DOI] [PubMed] [Google Scholar]
- Tanudji M, Dessi P, Murcha M, Whelan J (2001) Protein import into plant mitochondria: precursor proteins differ in ATP and membrane potential requirements. Plant Mol Biol 45: 317–325 [DOI] [PubMed] [Google Scholar]
- Tanudji M, Sjoling S, Glaser E, Whelan J (1999) Signals required for the import and processing of the alternative oxidase into mitochondria. J Biol Chem 274: 1286–1293 [DOI] [PubMed] [Google Scholar]
- Taylor SW, Fahy E, Zhang B, Glenn GM, Warnock DE, Wiley S, Murphy AN, Gaucher SP, Capaldi RA, Gibson BW, et al (2003) Characterization of the human heart mitochondrial proteome. Nat Biotechnol 21: 281–286 [DOI] [PubMed] [Google Scholar]
- von Heijne G, Steppuhn J, Herrmann RG (1989) Domain structure of chondrial and chloroplast targeting peptides. Eur J Biochem 180: 535–545 [DOI] [PubMed] [Google Scholar]
- Werhahn W, Jansch L, Braun H-P (2003) Identification of novel subunits of the TOM complex from Arabidopsis thaliana. Plant Physiol Biochem 41: 407–416 [Google Scholar]
- Werhahn W, Niemeyer A, Jansch L, Kruft V, Schmitz UK, Braun H (2001) Purification and characterization of the preprotein translocase of the outer mitochondrial membrane from Arabidopsis. Identification of multiple forms of TOM20. Plant Physiol 125: 943–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson JR (2003) After the ribosome structures: how are the subunits assembled? RNA 9: 165–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wischmann C, Schuster W (1995) Transfer of rps10 from the mitochondrion to the nucleus in Arabidopsis thaliana: evidence for RNA-mediated transfer and exon shuffling at the integration site. FEBS Lett 374: 152–156 [DOI] [PubMed] [Google Scholar]
- Zerges W (2002) Does complexity constrain organelle evolution? Trends Plant Sci 7: 175–182 [DOI] [PubMed] [Google Scholar]
- Zhang L, Aro EM (2002) Synthesis, membrane insertion and assembly of the chloroplast-encoded D1 protein into photosystem II. FEBS Lett 512: 13–18 [DOI] [PubMed] [Google Scholar]
- Zhang XP, Sjoling S, Tanudji M, Somogyi L, Andreu D, Eriksson LE, Graslund A, Whelan J, Glaser E (2001) Mutagenesis and computer modelling approach to study determinants for recognition of signal peptides by the mitochondrial processing peptidase. Plant J 27: 427–438 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.