Abstract
We report on the molecular and biochemical characterization of HEAT SHOCK PROTEIN 90C (HSP90C), one of the three Hsp90 chaperones encoded by the Chlamydomonas reinhardtii genome. Fractionation experiments indicate that HSP90C is a plastidic protein. In the chloroplast, HSP90C was localized to the soluble stroma fraction, but also to thylakoids and low-density membranes containing inner envelopes. HSP90C is expressed under basal conditions and is strongly induced by heat shock and moderately by light. In soluble cell extracts, HSP90C was mainly found to organize into dimers, but also into complexes of high molecular mass. Also, heterologously expressed HSP90C was mainly found in dimers, but tetramers and fewer monomers were detected, as well. HSP90C exhibits a weak ATPase activity with a Km for ATP of approximately 48 μm and a kcat of approximately 0.71 min−1. This activity was inhibited by the Hsp90-specific inhibitor radicicol. In coimmunoprecipitation experiments, we found that HSP90C interacts with several proteins, among them plastidic HSP70B. The cellular concentration of HSP70B was found to be 2.9 times higher than that of HSP90C, giving a 4.8:1 stoichiometry of HSP70B monomers to HSP90C dimers. The strong inducibility of HSP90C by heat shock implies a role of the chaperone in stress management. Furthermore, its interaction with HSP70B suggests that, similar to their relatives in cytosol and the endoplasmic reticulum, both chaperones might constitute the core of a multichaperone complex involved in the maturation of specific client proteins, e.g. components of signal transduction pathways.
Proteins of the heat shock protein 90 (Hsp90) family represent a highly conserved chaperone class. Its members are found in most bacteria (HtpG) and in the cytosol, the endoplasmic reticulum (ER; Grp94), mitochondria (TRAP1/hsp75), and chloroplasts of eukaryotic cells (Csermely et al., 1998). Most Hsp90s are abundant cellular proteins whose concentration is increased up to 10-fold in response to stress (Buchner, 1999). Hsp90s consist of three domains, an N-terminal ATP-binding domain of approximately 25 kD; a conserved, but structurally flexible, middle domain of approximately 35 kD; and a C-terminal domain of approximately 12 kD (Young et al., 2001).
The functionally active form of Hsp90 is a dimer, and dimerization is mediated by the approximately 190-amino-acid C terminus (Minami et al., 1994). The Hsp90 dimer acts as a molecular clamp that, driven by ATP-binding and hydrolysis, can open and close via transient interactions of the two N termini (Chadli et al., 2000; Prodromou et al., 2000). Hsp90 binds client proteins when it has ATP bound; ATP hydrolysis leads to an opening of the clamp and to substrate release (Young et al., 2001). Substrate binding appears to take place at the opposing inner faces of the middle segments in the closed conformation of the clamp, and apparently two client proteins can be bound simultaneously (Meyer et al., 2003). Client proteins recognized by Hsp90 are thought to be in a near-native state and thus represent late-folding intermediates (Jakob et al., 1995). However, it is not yet resolved which structural features of a substrate are recognized. In contrast to most other ATP-hydrolyzing proteins, Hsp90 binds ATP in a kinked conformation that is mimicked by the antitumor drugs geldanamycin and radicicol (Roe et al., 1999).
Cytosolic Hsp90 interacts with a large set of cohort proteins, which comprise Hsp70, Hop, p23, CHIP, Cdc37, and several immunophilins (Pratt and Toft, 2003; Wegele et al., 2004); novel cohort proteins, like Aha1 (Panaretou et al., 2002) and Tpr2 (Brychzy et al., 2003), are continuously added to this list. Whereas several cohort proteins have been characterized also for the ER resident Grp94 (Meunier et al., 2002), none have yet been identified for bacterial or organellar Hsp90s. When Hsp90 is organized with its cohort proteins into multichaperone complexes, its ATPase activity is increased severalfold compared to noncomplexed Hsp90 (Kamal et al., 2003).
A major function attributed to cytosolic Hsp90 is a role in the maturation of signal transduction proteins, like hormone receptors and kinases (Richter and Buchner, 2001; Pratt and Toft, 2003; Wegele et al., 2004). However, Hsp90 has also been implicated in the general refolding of denatured proteins (Jakob et al., 1995), and cytosolic Hsp90 also participates in the regulation of the stress response (Ali et al., 1998). The combination of these functions appears to result in a role of the chaperone as a capacitor of phenotypic variation: Decreasing Hsp90 function in Drosophila melanogaster and Arabidopsis (Arabidopsis thaliana; e.g. by the application of ansamycin drugs) results in the appearance of phenotypes that depend on the genetic background but normally are cryptic. This finding led to the following hypothesis (Rutherford and Lindquist, 1998; Queitsch et al., 2002): Mutations that have accumulated in signaling proteins are kept cryptic by the ability of Hsp90 to stabilize client proteins in their native conformation. However, upon stress, when Hsp90 is sequestered away from these client proteins while refolding stress-denatured proteins, the client proteins may change their conformation according to the accumulated mutations. This may result in a different performance of the respective clients, which, if they participate in developmental signaling pathways, may lead to phenotypic variation in a stressful environment.
Most information on chloroplast Hsp90 functions was gathered from studies with the Arabidopsis chlorate resistant 88 (cr88) mutant. This mutant, identified in a screen for plants impaired in nitrate reductase activity (Lin and Cheng, 1997), turned out to have a point mutation in the C terminus of a Hsp90 targeted to the chloroplast stroma (Cao et al., 2003). The cr88 mutant has a yellow-green appearance due to a retarded development of chloroplasts observed particularly in young leaves. The mutant exhibits reduced light-inducible expression of the nuclear NR2, CAB, and RBCS genes, and also of the plastid-encoded rbcL gene. Furthermore, the cr88 mutant showed retarded deetiolation in red light (Lin and Cheng, 1997; Cao et al., 2000). These findings suggest a role of plastidic Hsp90 in the transduction of a plastid-derived signal that is responsible for the regulation of a subset of photosynthesis-related genes. Perhaps, in analogy to its relatives in the cytosol and the ER, plastidic Hsp90 also participates in signal transduction pathways. Interestingly, phylogenetic analyses have revealed that chloroplast Hsp90s are more closely related to ER-targeted Hsp90s than to cyanobacterial HtpG (Emelyanov, 2002). Thus, it seems possible that an ER Hsp90 gene had duplicated and acquired a chloroplast transit peptide, whereas the HtpG from the cyanobacterial endosymbiont got lost.
We have previously reported on the identification of three genes in the Chlamydomonas reinhardtii genome that encode Hsp90 proteins, which were named HSP90A, HSP90B, and HSP90C (Schroda, 2004). Whereas HSP90A and HSP90B with high probability are localized to the cytosol and to the ER, respectively, HSP90C may be targeted to the chloroplast and/or to mitochondria. Here, we show that HSP90C appears to be targeted only to the chloroplast. We demonstrate that the chaperone exhibits ATPase activity, which is sensitive to radicicol. Furthermore, we show that, in cellular extracts, HSP90C exists mainly as dimers, but also forms high molecular mass complexes with several other proteins, one of which is the plastidic HSP70B chaperone.
RESULTS
Features of the HSP90C Transcript and of the Deduced Amino Acid Sequence
The amino acid sequence of HSP90C that we have reported recently (Schroda, 2004) was deduced from computer-predicted gene models, which were based on the Chlamydomonas genome sequence, and from the similarity of HSP90C with the chloroplast-targeted Hsp90 protein of Arabidopsis (Krishna and Gloor, 2001; Cao et al., 2003). Since predictions were only partially supported by cDNA sequence data, we wanted to confirm them with a full-length HSP90C cDNA sequence. When we searched the Chlamydomonas expressed sequence tag (EST) databases (Asamizu et al., 1999, 2000; Shrager et al., 2003) for ESTs with complete 5′ ends, we found four clones that had a common 5′ end 119 nucleotides upstream from the predicted AUG start codon, and three more ESTs with 5′ ends close to this position. One of the four presumably complete cDNAs (clone AV630546) was sequenced (GenBank accession no. AY705371). From this sequence, we could confirm that the start and stop codons and all exon-intron boundaries were as predicted by gene model C_160233 (nucleotides 935,091–939,742 on scaffold 16 of the 2.0 version of the Chlamydomonas genome sequence [http://genome.jgi-psf.org/chlre2/chlre2.home.html]).
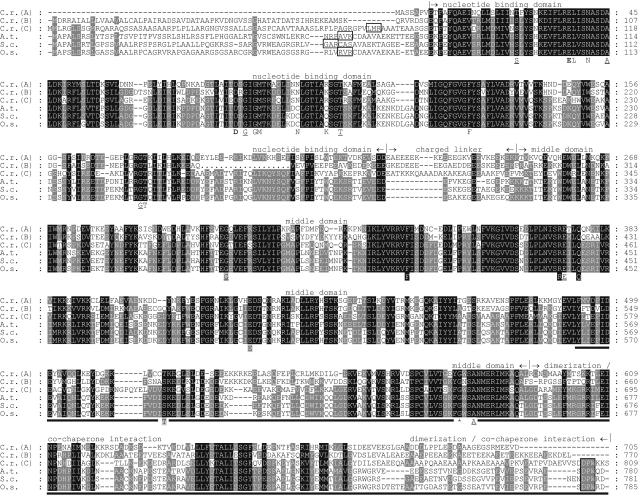
HSP90C turned out to be more closely related to plastidic Hsp90s of higher plants (approximately 50% identity, approximately 67% similarity) than to the Chlamydomonas proteins HSP90A and HSP90B, predicted to be localized to cytosol and ER, respectively (approximately 45% identity, approximately 64% similarity; Fig. 1). Several conserved amino acids that were shown to be crucial for the activity of yeast (Saccharomyces cerevisiae) cytosolic Hsp90 (Bohen and Yamamoto, 1993; Nathan and Lindquist, 1995; Prodromou et al., 1997; Panaretou et al., 1998; Meyer et al., 2003) were found at conserved positions in all three Chlamydomonas Hsp90 proteins (Fig. 1). This is true also for the plastidic Hsp90s of higher plants, except for two residues: Asn-92 and Lys-98 in yeast Hsp90 have been shown by structural analyses to participate in the binding of the Rib and of the β-phosphate of ATP, respectively (Prodromou et al., 1997). In the higher plant sequences, Asn-92 and Lys-98 have been exchanged by Cys and Gln, respectively (Fig. 1). It is not clear whether these alterations have any functional consequences. Gly-646 in Arabidopsis chloroplast Hsp90, which, upon mutation, gave rise to pleiotropic phenotypes (Cao et al., 2003), is also conserved in all Chlamydomonas and plastidic Hsp90s (Fig. 1). The charged linker that connects the nucleotide-binding and middle domains of Hsp90 is present in all Chlamydomonas and higher plant plastidic Hsp90s, although it appears to be shorter in the latter (Fig. 1; Krishna and Gloor, 2001). HSP90C and the higher plant plastidic Hsp90s contain a DPW motif at their C terminus, which is absent in mitochondrial TRAP1 (Felts et al., 2000) and in Hsp90s localized to ER and cytosol (Fig. 1). To characterize Chlamydomonas HSP90C in greater detail, we raised an antibody against its C terminus, since this region exhibited the lowest similarity to the other Chlamydomonas family members HSP90A and HSP90B (Fig. 1).
Figure 1.
Alignment of Hsp90 protein sequences. Aligned are amino acid sequences deduced from Hsp90 genes from C. reinhardtii (C.r.; HSP90A, cytosol, gene model C_730014; HSP90B, ER, gene models C_50233/C_50234; HSP90C, chloroplast, accession AY705371); Arabidopsis (A.t.), accession AAD32922; rye (S.c.), accession Q43638; and rice (O.s.), accession XM_483065. The higher plant sequences are plastidic Hsp90s. Dots in HSP90B indicate missing sequence data. Residues highlighted in black are conserved in all sequences; those highlighted in gray are conserved in at least four of the six. Conserved amino acids were defined as N/Q, D/E, R/K, S/T, F/Y, A/G, and V/I/L/M. The chloroplast transit peptide cleavage sites as predicted by the TargetP program (Emanuelsson et al., 2000) are boxed, as is the DPW motif in chloroplast-targeted Hsp90s. The separation of Hsp90s into nucleotide-binding domain, charged linker, middle domain, and dimerization/cochaperone interaction domain was made according to Meyer et al. (2003). The amino acids given below the alignment indicate residues that in yeast Hsp90 (1) by structural analyses (nonformatted and bold letters) have been shown to bind ATP directly or via water molecules (Prodromou et al., 1997); (2) upon mutation abolish ATP binding or hydrolysis and render cells inviable (bold; Panaretou et al., 1998); (3) upon mutation result in temperature-sensitive phenotypes (underlined; Nathan and Lindquist, 1995); (4) upon mutation render the chaperone incapable of mediating glucocorticoid receptor maturation (gray shading; Bohen and Yamamoto, 1993); and (5) upon mutation decrease ATPase activity and impair growth (black shading; Meyer et al., 2003). The asterisk given below the alignment indicates a conserved Gly that, when converted to Arg in Arabidopsis chloroplast-targeted Hsp90, causes pleiotropic phenotypes (Cao et al., 2003). The solid line depicts the region of HSP90C against which an antibody was raised.
HSP90C Appears to Be Targeted to the Chloroplast
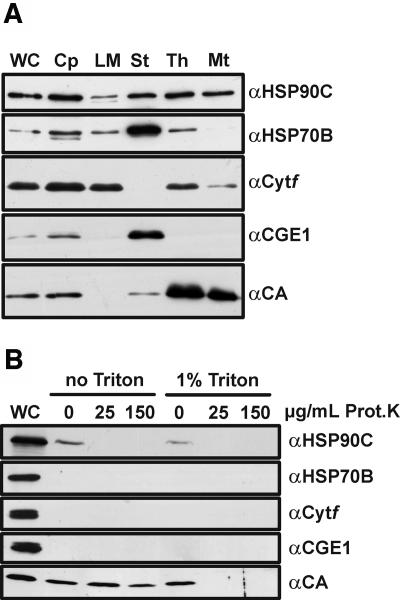
First we addressed the question of whether HSP90C is localized to chloroplasts, mitochondria, or both organelles. For this, we isolated chloroplasts and mitochondria from Chlamydomonas. Chloroplasts were subsequently subfractioned into stroma, thylakoids, and low-density membranes. Low-density membranes are considered to consist of inner envelopes and transitory membranes between inner envelope and thylakoids (Zerges and Rochaix, 1998). The purity of the fractions was tested with antibodies against mitochondrial carbonic anhydrase, chloroplast HSP70B and stromal CGE1 (the nucleotide exchange factor of HSP70B [Schroda et al., 2001]), and the integral thylakoid membrane protein cytochrome f. Chloroplasts were significantly contaminated by mitochondria and mitochondria slightly by thylakoids, as judged by the detection of carbonic anhydrase in the chloroplast fraction and cytochrome f in the mitochondrial fraction, respectively (Fig. 2A). The low-density membranes contained cytochrome f but no mitochondrial carbonic anhydrase, whereas the thylakoids were heavily contaminated with carbonic anhydrase. The stroma fraction contained no cytochrome f and, therefore, was free of thylakoidal contamination, but contained some mitochondrial carbonic anhydrase (Fig. 2A). CGE1 was detected strongly in the stroma fraction and very weakly in low-density membranes.
Figure 2.
Intracellular localization of HSP90C. A, Chlamydomonas chloroplasts (Cp) were isolated, lysed by hypo-osmotic shock, and separated into stroma (St), low-density membranes (LM), and thylakoid membranes (Th). Mitochondria (Mt) were isolated from the same strain. Whole cells (WC) and fractions (7 μg of protein each) were separated on a 7.5% to 15% SDS-polyacrylamide gel, transferred to nitrocellulose, and immunodecorated with antibodies against HSP90C, HSP70B (stroma and membrane control), thylakoidal cytochrome f (Cytf), stromal CGE1, and mitochondrial carbonic anhydrase (CA). B, Mitochondria were isolated from cells that had been disrupted with a nebulizer. Intact mitochondria and mitochondria lysed with 1% Triton X-100 were incubated with proteinase K at the concentrations indicated. Thirty micrograms of whole-cell (WC) proteins and 20 μg of mitochondrial proteins were separated on a 7.5% to 15% SDS-polyacrylamide gel, transferred to nitrocellulose, and immunodecorated with the same antibodies as in A.
Like chloroplast HSP70B, HSP90C was detected in the stroma, low-density membranes, and thylakoids, indicating that the two chaperones are both soluble and membrane-associated chloroplast proteins. In the mitochondrial fraction, however, we detected a clear signal for HSP90C but only a very weak one for HSP70B (Fig. 2A). This suggested either that HSP90C is dually targeted to chloroplasts and mitochondria or that some chloroplast-derived HSP90C is contaminating the mitochondrial fraction.
To address these possibilities, we first tried to increase the purity of our mitochondrial fractions by disrupting Chlamydomonas cells more gently with a nebulizer instead of vortexing with glass beads. As judged from the absence of chloroplast markers HSP70B, cytochrome f, and CGE1 in the mitochondrial fraction, using a nebulizer indeed yielded mitochondria of higher purity (Fig. 2B). Since HSP90C was still detected in these mitochondria, we used a proteinase K assay (Ryan et al., 2001) to test whether HSP90C is a plastidic contaminant on the mitochondrial surface or a true mitochondrial protein. Mitochondrial carbonic anhydrase was used as a mitochondrial marker protein (Eriksson et al., 1996). In three independent experiments, carbonic anhydrase was digested only when mitochondria had been lysed with Triton X-100 prior to proteinase K addition, whereas the HSP90C signal was already lost when intact mitochondria had been treated with low concentrations of proteinase K (Fig. 2B). These data indicate that the HSP90C signal detected in mitochondrial fractions originates from HSP90C located outside the mitochondria. Hence, HSP90C most likely is not a mitochondrial protein and appears to be targeted only to the chloroplast.
HSP90C Is Inducible by Heat Shock and Light
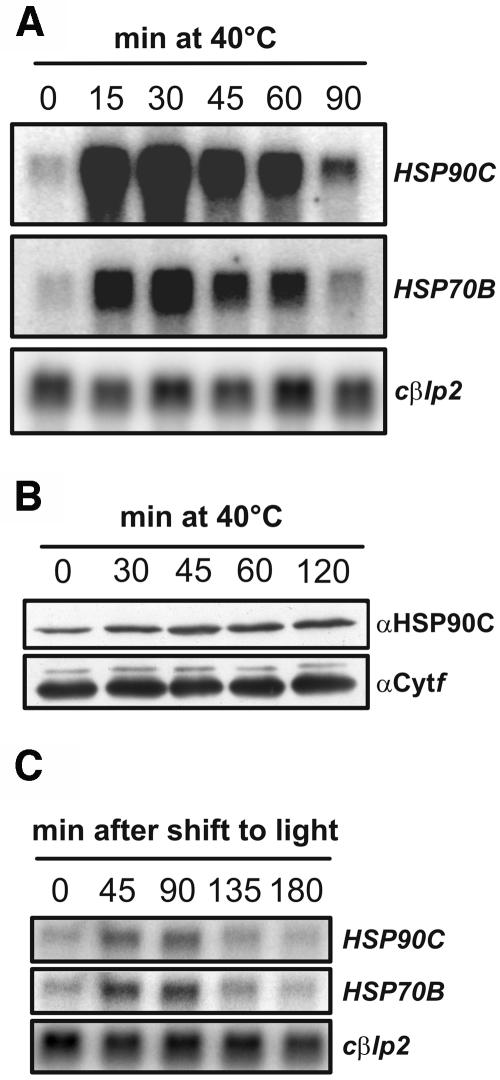
Chloroplast Hsp90 genes in rye (Secale cereale; Schmitz et al., 1996) and in Arabidopsis (Cao et al., 2003) have previously been shown to be induced by heat shock; the Arabidopsis gene was also shown to be induced by light. We found that the HSP90C transcript is present under nonstress conditions and that it accumulates strongly after heat shock (Fig. 3A). Quantification of four independent experiments revealed that heat stress induced HSP90C 37-fold (SEM ±9), whereas HSP70B was induced only 13-fold (SEM ±3). HSP90C and HSP70B mRNA accumulation peaked 30 min after shift to 40°C, then decreased again and remained at a slightly higher level 90 min after the onset of stress. Also, at the protein level, HSP90C was detectable under noninducing conditions and increased significantly after heat stress (Fig. 3B). Quantification of four independent experiments revealed that 120 min after the onset of stress, HSP90C protein levels had increased 3.9-fold (SEM ±1.5). In comparison, HSP70B protein increased at most 3-fold after heat stress (Drzymalla et al., 1996).
Figure 3.
Accumulation of HSP90C protein after heat shock and HSP90C mRNA after heat shock and dark-to-light shift. A, mRNA from Chlamydomonas wild-type cells that were exposed to a heat shock at 40°C for the indicated times was separated on agarose gels (10 μg per lane) and transferred to nylon membranes. These were hybridized with probes generated from the coding regions of HSP90C, HSP70B (positive control), and cβlp2 (loading control). B, Whole-cell proteins (15 μg per lane) from cells that were treated as depicted in A were separated on a 10% SDS-polyacrylamide gel, transferred to nitrocellulose, and immunodetected with antibodies against HSP90C and cytochrome f (Cytf, loading control). C, mRNA from Chlamydomonas wild-type cells that were kept in the dark for 16 h (0) and then transferred to light of approximately 30 μE m−2 s−1 for the indicated times was separated on agarose gels (10 μg per lane) and transferred to nylon membranes. Membranes were hybridized with the same probes used in A.
When dark-adapted Chlamydomonas cells were shifted into the light, we found in three independent experiments that HSP90C mRNA levels increased 2.8-fold (SEM ±0.4; Fig. 3C). Transcript levels remained high between 45 and 90 min after the onset of irradiation and then decreased to basal levels. Light inducibility of HSP90C was always less pronounced than that of HSP70B, which in the same experiments exhibited 4.2-fold (SEM ±0.8) increased mRNA levels. On the protein level, no significant light induction of HSP90C was observed (data not shown). In conclusion, both heat shock and light induction kinetics of HSP90C were similar to those of HSP70B.
Native HSP90C in Soluble Cell Extracts Exists as Dimers and Higher Molecular Mass Complexes
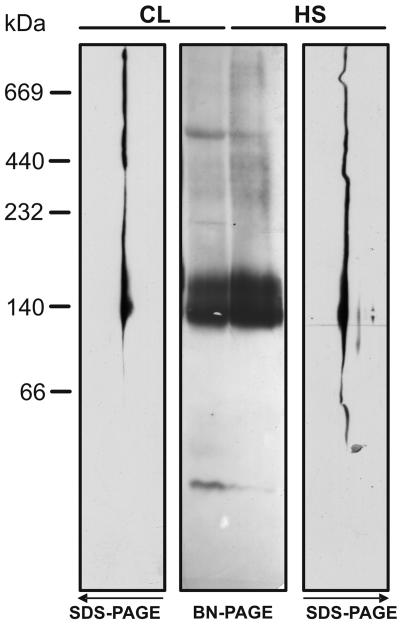
A key to understanding the roles HSP90C may play in the cell is the identification of its cochaperones and substrates, a task we intended to tackle by coimmunoprecipitation experiments. Since complexes of cytosolic Hsp90 with its substrates and cochaperones were reported to be intrinsically unstable (Stancato et al., 1993), we first wanted to test whether this holds true for HSP90C as well. In addition, we wished to determine whether the active form of HSP90C is a dimer, as is the case for its relatives in the cytosol (Minami et al., 1994) and in the ER (Wearsch and Nicchitta, 1996). To address these questions, we employed blue native (BN)-PAGE to analyze HSP90C complexes in soluble cell extracts. Since the composition of HSP90C complexes might change during stress, we prepared soluble cell extracts from nonstressed and heat-shocked cells. As shown in Figure 4, HSP90C in the first-dimension native gel migrated as a prominent double band at approximately 140 to 160 kD and as several weak, but distinct, bands in the higher molecular mass region. After separation in the second dimension, these signals could unambiguously be attributed to HSP90C (Fig. 4, SDS-PAGE images). This technique tends to overrepresent minor complexes so that the higher molecular mass bands appear more or less as a continuous smear. As judged by a slight smear of HSP90C into a region <140 kD in extracts from stressed cells, this treatment may also result in monomerization and/or degradation of the chaperone. Otherwise, the migration patterns observed in extracts from nonstressed and stressed cells were very similar.
Figure 4.
Analysis of native HSP90C complexes by BN-PAGE. Chlamydomonas total soluble proteins from cells grown in continuous light (CL) and from cells that, in the light, were exposed for 1 h to a heat shock (HS) at 40°C, were separated on a 6% to 15% polyacrylamide gradient blue native gel (BN-PAGE), and either transferred to nitrocellulose (central gel) or separated in the second dimension on 10% SDS-polyacrylamide gels (SDS-PAGE), followed by transfer to nitrocellulose (left and right gels; note that the top of the second dimension gel is shown facing the BN-PAGE gels at the center of the figure). Membranes were immunodecorated with an antibody against HSP90C.
The strong signal at approximately 140 to 160 kD suggests a dimeric state of HSP90C (expected size 165 kD). However, it is not clear why the chaperone migrates as a double band at this position. Covalent modifications (e.g. phosphorylation) of a part of the cell's HSP90C pool or fixed conformational changes (e.g. clamp formation in the ATP state) may account for this heterogeneity. The observed migration of HSP90C in the region >160 kD may be due to its organization into stable complexes or to multiple interactions of the chaperone with other proteins during electrophoresis under nondenaturing conditions.
HSP90C Interacts with the Chloroplast Chaperone HSP70B
Our finding that HSP90C seems to be at least in part organized into complexes with other proteins (presumably substrates and/or cochaperones) encouraged us to perform coimmunoprecipitations to identify interaction partners. We immunoprecipitated HSP90C from soluble cell extracts prepared from nonstressed and heat-shocked cells. As shown in Figure 5A, several proteins coprecipitated with HSP90C. The pattern of proteins coprecipitating from extracts of stressed and nonstressed cells was indistinguishable, corroborating our results from BN-PAGE that the composition of HSP90C complexes appears to be similar under both conditions (Fig. 4). No proteins were precipitated with preimmune serum. To test whether some of the proteins detected on the gel were HSP90C degradation products, we immunodecorated western blots of the coprecipitates with the HSP90C antibody. Only a minor degradation product was detected at approximately 75 kD (data not shown), suggesting that most of the proteins detected appear to be true HSP90C interaction partners.
Figure 5.
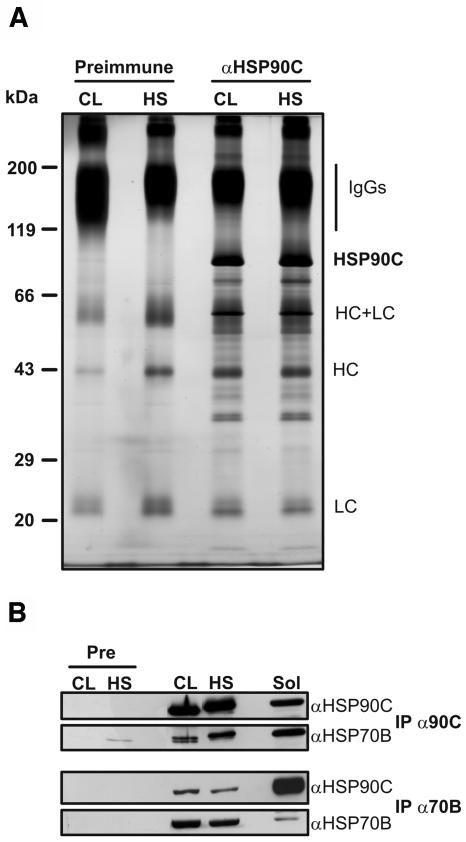
Immunoprecipitation of HSP90C. A, Chlamydomonas total soluble proteins were prepared from cells grown in continuous light (CL) and from cells that, in the light, were exposed for 1 h to a heat shock (HS) at 40°C. Next, soluble proteins were incubated with protein A Sepharose coupled to antibodies of preimmune serum (Preimmune) or to affinity-purified anti-HSP90C antibodies. Immunoadsorbed proteins were eluted in nonreducing SDS sample buffer, separated on a 7.5% to 15% SDS-polyacrylamide gel, and visualized by silver staining. The positions of precipitated HSP90C and of contaminating IgG heavy chains (HC) and light chains (LC) are indicated. B, Seven hundred microliters of soluble lysate from approximately 109 cells generated as described in A were incubated with protein A Sepharose coupled to antibodies of preimmune (Pre), anti-HSP90C (IP α90C), or anti-HSP70B (IP α70B) serum. Fifteen microliters of the lysate (Sol) and one-third of each immunoprecipitation were separated on a 7.5% to 15% SDS-polyacrylamide gel, transferred to nitrocellulose, and immunodecorated with antibodies against HSP90C and HSP70B.
The analysis of these HSP90C coprecipitating proteins by mass spectrometry led to the identification of some of them; however, peptide coverage was low, since coprecipitating proteins were only of low abundance. Thus, the identity of these putative HSP90C client proteins needs first to be verified by other approaches before publication. We assume that complexes of HSP90C with its client proteins have largely fallen apart during the washing steps, as was observed for cytosolic Hsp90 complexes (Stancato et al., 1993). We have tried to stabilize HSP90C complexes by the addition of 20 mm sodium molybdate to cell extracts and washing buffers as described by Hutchison et al. (1992), but with no improvement (data not shown). Note that molybdate also had no stabilizing effect on hormone receptor-plant Hsp90 complexes (Stancato et al., 1996).
Hsp90s in the eukaryotic cytosol and in the ER have been reported to interact with members of the Hsp70 family (Csermely et al., 1998; Wegele et al., 2004). We therefore tested whether plastidic HSP70B (Schroda et al., 1999, 2001) coimmunoprecipitated with HSP90C. As shown in Figure 5B, HSP70B could indeed be detected in coprecipitates of HSP90C. In contrast, proteins against which we had antibodies available, i.e. rbcL, mitochondrial carbonic anhydrase, ClpB, CF1β, OEE2, and VIPP1, could not be detected in HSP90C coprecipitates (data not shown), suggesting that the interaction of HSP90C with HSP70B is specific. We found that HSP90C preimmune serum immunoprecipitated traces of HSP70B from extracts of heat-shocked cells (Fig. 5B); note, however, that precipitation of some HSP70 with preimmune serum has been reported before and was attributed to an interaction of HSP70 with IgGs and protein A (Stancato et al., 1993).
We also performed the reciprocal experiment and tested whether HSP90C coprecipitated with HSP70B. As shown in Figure 5B, this also was the case. The amounts of HSP70B and HSP90C that coprecipitated with HSP90C and HSP70B, respectively, were about the same, regardless of whether extracts from heat-shocked or nonstressed cells were used (Fig. 5B). However, when comparing the signals obtained for coprecipitated HSP90C and HSP70B with those from whole-cell extracts (Fig. 5B), it appears that more HSP70B coprecipitated with HSP90C than vice versa. This could be due to higher concentrations of HSP70B than of HSP90C in the chloroplast, resulting in a smaller fraction of HSP70B that is in complex with HSP90C. Alternatively, binding of the αHSP90C antibody to HSP90C might lock the chaperone in a conformation that leads to stabilization of the complex with HSP70B. To test the former possibility, we quantified the cellular concentrations of the two chaperones. For this, we detected HSP90C and HSP70B immunologically in dilution series of whole-cell proteins and of heterologously expressed and purified chaperones (Fig. 6). In two independent experiments, we found that HSP90C represented 0.055% and 0.056% of total cellular proteins and that these contained 0.148% and 0.115% HSP70B. On a mass basis, this means that, on average, HSP70B is approximately 2.4 times more abundant than HSP90C. Taking into account the different molecular masses of the two chaperones (82.4 kD for HSP90C and 67.9 kD for HSP70B [Schroda, 2004]), cells appear to contain HSP70B and HSP90C chaperones in a ratio of approximately 2.9:1, i.e. 5.8 HSP70B monomers per HSP90C dimer. However, these slightly higher concentrations of HSP70B compared to HSP90C cannot explain why so much more HSP70B coprecipitates with HSP90C than vice versa (Fig. 5B). Hence, the possibility that binding of the αHSP90C antibody to HSP90C stabilizes complex formation with HSP70B appears to be the most plausible.
Figure 6.
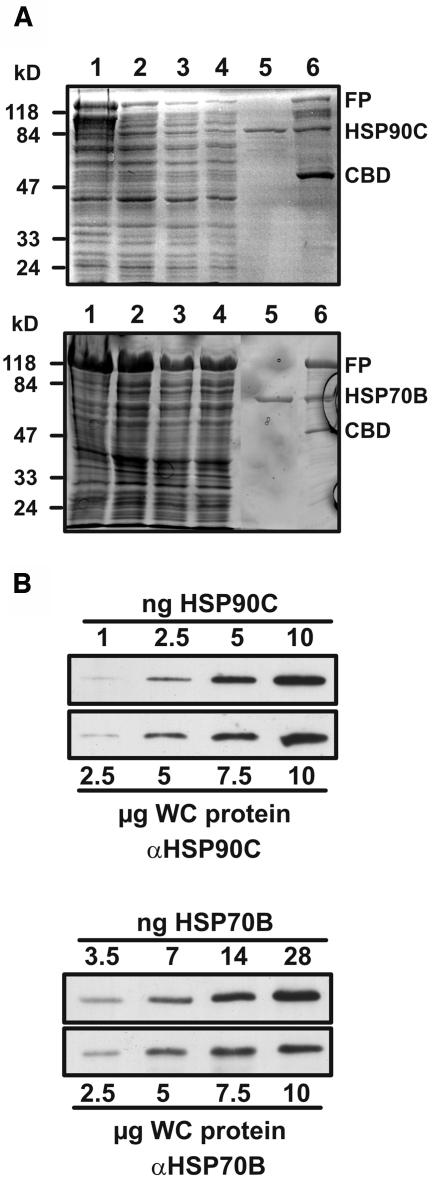
Heterologous expression of HSP90C and HSP70B and quantification of cellular HSP90C and HSP70B concentrations. A, HSP90C and HSP70B were expressed as C-terminal fusions to the Sce VMA intein containing a chitin-binding domain, purified by chitin affinity chromatography, and eluted after thiol-induced cleavage of the intein. Aliquots obtained during the purification steps were separated on 10% SDS-polyacrylamide gels and stained with Coomassie Brilliant blue. Lane 1, Lysate of E. coli ER2566 host cells after induction with IPTG. Lane 2, Supernatant of cell lysates after a 30-min centrifugation at 20,000g. Lanes 3 and 4, First and second flow-through, respectively. Lane 5, Eluate after thiol-induced cleavage. Lane 6, Proteins remaining on the chitin column after elution. The positions of the fusion proteins (FP), cleaved HSP90C/HSP70B, and the Sce VMA intein/chitin-binding domain (CBD) are indicated. B, HSP90C/HSP70B purified as shown in A and Chlamydomonas whole-cell (WC) proteins were diluted and the protein amounts indicated were separated on a 7.5% to 15% SDS-polyacrylamide gel, transferred to nitrocellulose, and immunodecorated with antibodies against HSP90C and HSP70B, respectively.
In general, Hsp90 and Hsp70 form stable complexes only in the presence of specific cochaperones like Hop (Smith et al., 1993) or Tpr2 (Brychzy et al., 2003). In Neurospora, however, cytosolic Hsp90 and Hsp70 have been shown to interact directly (Freitag et al., 1997). To test whether HSP90C and HSP70B may also interact directly, we mixed heterologously expressed HSP90C (0.012 μm) and HSP70B (0.015 μm) in the presence and absence of Mg-ATP in vitro. When we immunoprecipitated HSP90C from the mixture, we could not detect any coprecipitating HSP70B, and, vice versa, when HSP70B was immunoprecipitated, no HSP90C was found to coprecipitate (data not shown). Thus, HSP90C and HSP70B in the chloroplast may not interact directly, but via other proteins within a common complex. Alternatively, a specialized cochaperone might be required to mediate a stable interaction of the two chaperones. Finally, the HSP90C-HSP70B interaction might take place only at very high protein concentrations as they are encountered in the stroma of the chloroplast.
We conclude that HSP90C is in complex with several proteins, although these complexes appear to be unstable, that the pattern of interacting proteins is similar in stressed and unstressed cells, that chloroplast HSP70B is among the HSP90C-interacting proteins, and that HSP70B monomers are approximately 5.8 times more abundant than HSP90C dimers.
Heterologously Expressed HSP90C Forms Dimers and Tetramers
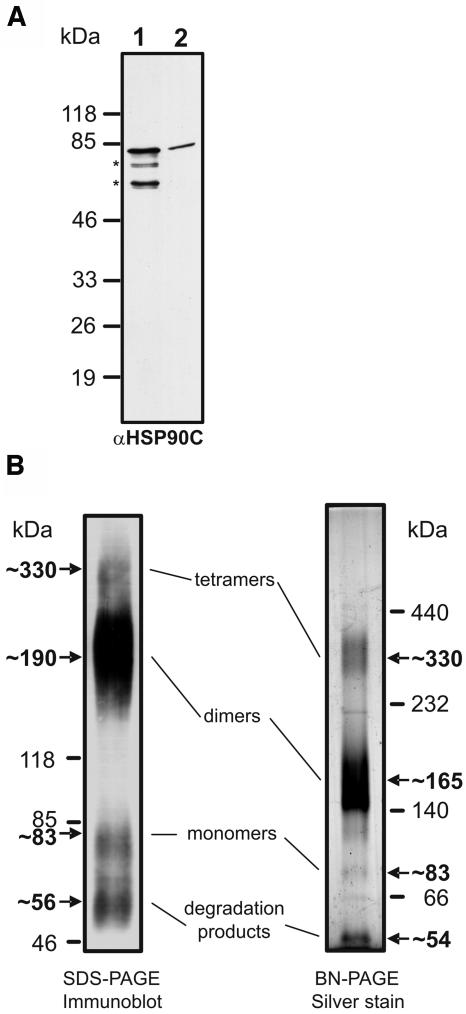
To further characterize HSP90C biochemically, we wished to determine the N-terminal amino acid sequence of mature HSP90C in order to deduce the cleavage site of its transit peptide. We immunoprecipitated HSP90C from Chlamydomonas cell extracts and subjected the precipitated protein to Edman degradation. Unfortunately, HSP90C was N-terminally blocked. As an alternative, we separated heterologously expressed and purified HSP90C that lacks the putative 71-amino-acid N-terminal transit peptide (Fig. 1) next to whole-cell extracts on a high-resolution SDS-polyacrylamide gel and detected HSP90C by immunoblotting. Note that, although we used an efficient protease inhibitor cocktail during purification from the overexpressing Escherichia coli strain, our HSP90C preparations always contained some degradation products at approximately 75, 63, and 33 kD (Fig. 6A); those at approximately 75 and 63 kD were also recognized by the antibody against the HSP90C C terminus (Fig. 7A, lane 1). Since in high-resolution gels purified HSP90C (lane 1) migrated at the same position as mature Chlamydomonas HSP90C (lane 2), the mature protein appears to be truncated by a sequence stretch that could well be of the dimension of the predicted 71-amino-acid transit peptide. Thus, the LMR/A transit peptide cleavage site predicted by the TargetP program (Emanuelsson et al., 2000) might indeed be used in vivo (Fig. 1).
Figure 7.
Analysis of migration and oligomerization properties of heterologously expressed HSP90C. A, Fifty nanograms of heterologously expressed HSP90C lacking the putative 71-amino-acid transit peptide (lane 1) were separated on a 20-cm-long 7.5% to 15% SDS-polyacrylamide gel next to 50 μg of whole-cell Chlamydomonas proteins (lane 2). Proteins were transferred to nitrocellulose and immunodecorated with the HSP90C antibody. B, Four micrograms of heterologously expressed HSP90C were cross-linked with 0.1% glutaraldehyde for 10 min at 30°C, separated on a 4% to 18% SDS-polyacrylamide gel, transferred to nitrocellulose, and detected with the αHSP90C antibody (left gel). Alternatively, 2 μg of heterologously expressed HSP90C were separated on a 6% to 15% BN gel and visualized by silver staining (right gel).
Next, we used two methods to monitor the oligomerization properties of heterologously expressed HSP90C: cross-linking and BN-PAGE. After cross-linking with glutaraldehyde and electrophoresis in a denaturing SDS gel, we found the majority of HSP90C to migrate at approximately 190 kD, and much smaller amounts to migrate at approximately 330, 83, and 56 kD (Fig. 7B, left gel). In BN-PAGE, purified HSP90C migrated at approximately 330, 165, 83, and 54 kD; most of the protein was found in the approximately 165-kD band (Fig. 7B, right gel). Whereas the signals at approximately 330, 190/165, and 83 kD most likely correspond to HSP90C tetramers, dimers, and monomers, respectively, those at approximately 56/54 kD may represent degradation products.
We conclude that HSP90C in vivo is processed to a mature form of approximately 83 kD, possibly at the predicted LMR/A cleavage site. Heterologously expressed HSP90C appears to be organized mainly into dimers, but in small amounts it is also found in monomers and tetramers.
HSP90C Exhibits Weak ATPase Activity That Is Inhibited by Radicicol
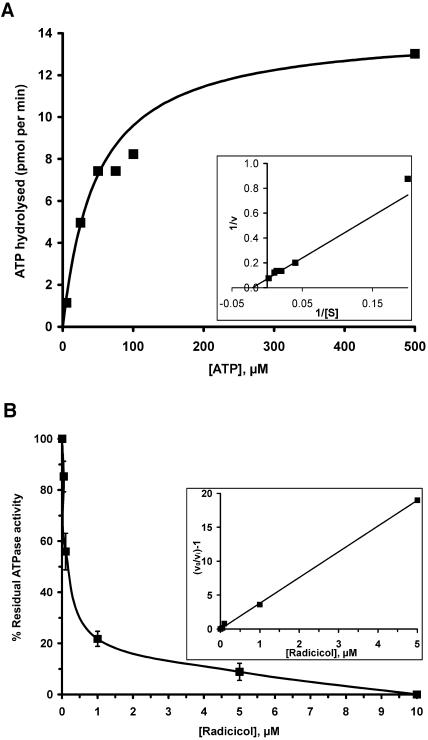
Some members of the Hsp90 family characterized so far have been shown to possess a weak, but essential, ATPase activity, which is inhibited by the antitumor agents geldanamycin and radicicol (Panaretou et al., 1998; Felts et al., 2000). To test whether HSP90C can also hydrolyze ATP, we incubated heterologously expressed HSP90C with [γ-32P]ATP and found that the latter was indeed converted to 32Pi and ADP (Fig. 8A). When we determined the initial rate of ATP hydrolysis by HSP90C at increasing ATP concentrations, we obtained values that follow Michaelis-Menten kinetics (Fig. 8A). To determine values for Vmax and Km, we used a direct linear plot according to Eisenthal and Cornish-Bowden (1974) and obtained values of approximately 48 μm for Km and of approximately 14.2 pmol min−1 for Vmax, giving a kcat value of approximately 0.71 min−1. To ensure that the ATPase activity measured was not influenced by contamination of ATP-hydrolyzing enzymes in our HSP90C preparation, we took advantage of the finding that radicol specifically inhibits Hsp90's ATPase activity by occupying its ATP-binding site (Roe et al., 1999). As shown in Figure 8B, radicicol indeed inhibited HSP90C's ATPase activity with a Ki of approximately 0.13 μm at 50 μm ATP. As judged from the complete loss of ATPase activity at radicicol concentrations higher than 10 μm, our HSP90C preparation appeared to be free of contaminating ATP-hydrolyzing enzymes.
Figure 8.
ATPase activity of purified HSP90C and its inhibition by radicicol. A, Initial rates of ATP hydrolysis (i.e. the conversion of [γ-32P]ATP to 32Pi and ADP) were measured as a function of ATP concentration using one micromolar heterologously expressed HSP90C. Values for Km and Vmax (48 μm and 14.2 pmol min−1, respectively) were determined using a direct linear plot according to Eisenthal and Cornish-Bowden (1974). Km and Vmax values were inserted into the Michaelis-Menten equation v = Vmax[S]/(Km + [S]) to draw the curve shown. Inset, The data are replotted double reciprocally in a Lineweaver-Burk plot. Km and Vmax values were inserted into equation 1/v = {(Km/Vmax)1/[S]} + 1/Vmax to draw the straight line shown. B, Inhibition of HSP90C's ATPase activity by increasing concentrations of radicicol. One micromolar HSP90C was used at a fixed ATP concentration of 50 μm. Each data point represents the mean of three independent experiments; error bars indicate SEM. Inset, The data are replotted according to equation v0/vi − 1 = [I]1/Ki(app). The value obtained for Ki(app) is approximately 0.26 μm, giving a Ki for radicicol of approximately 0.13 μm at 50 μm ATP.
DISCUSSION
We report on the molecular and biochemical characterization of HSP90C, one of three Hsp90 chaperones encoded by the Chlamydomonas genome. We provide the following evidence that HSP90C indeed is a bona fide chaperone of the Hsp90 family. (1) HSP90C is homologous to other Hsp90s; all residues that were found to be essential for Hsp90 function are conserved in HSP90C (Fig. 1). (2) HSP90C exhibits an ATPase activity with a Km of approximately 48 μm and a kcat of approximately 0.71 min−1 at 30°C (Fig. 8A). These values are similar to those of other members of the Hsp90 family, e.g. yeast cytosolic Hsp90 (kcat ≅ 0.1 min−1 and Km ≅ 100 μm at 30°C [Panaretou et al., 1998]), human mitochondrial TRAP1 (kcat ≅ 0.1 min−1 and Km ≅ 33 μm at 30°C [Felts et al., 2000]), and E. coli HtpG (kcat ≅ 0.47 min−1 at 37°C [Panaretou et al., 1998]). (3) HSP90C's ATPase activity is inhibited by radicicol, a drug that was reported to act specifically on Hsp90s (Schulte et al., 1998; Sharma et al., 1998; Fig. 8B). The kinetics of inhibition of HSP90C's ATPase by increasing radicicol concentrations were similar to those of yeast Hsp90 (Roe et al., 1999) and human TRAP1 (Felts et al., 2000). (4) HSP90C is heat shock inducible, a feature shared by many Hsp90s characterized to date (e.g. Welch and Feramisco, 1982; Mason et al., 1999; Cao et al., 2003). (5) Bulk HSP90C in vivo and in vitro is constitutively organized into dimers (Figs. 4 and 7B) and dimerization has been demonstrated to be required for Hsp90 function in vivo (Minami et al., 1994). (6) HSP90C interacts (directly or indirectly) with HSP70B (Fig. 5B); in the cytosol, complex formation of Hsp90 with HSP70 into the foldosome was observed to be essential for the maturation of hormone receptors and signaling kinases (Pratt and Toft, 2003).
In fractionation experiments, we reproducibly detected HSP90C in chloroplast and mitochondrial fractions of Chlamydomonas, which suggested a dual targeting of the chaperone to both organelles (Fig. 2A). In favor of a localization of HSP90C in the chloroplast, we found that it is in complex with the plastidic HSP70B protein (Fig. 5B), and that HSP90C contains a DPW motif at its C terminus. The DPW motif is present in all plastidic Hsp90s identified to date (Schmitz et al., 1996; Cao et al., 2003), but not in cytosolic, mitochondrial, or ER lumenal Hsp90s (Fig. 1). To test whether HSP90C is also localized to mitochondria, we treated our mitochondrial fraction with proteinase K, a procedure that is routinely used in yeast to test the mitochondrial localization of a given protein (Ryan et al., 2001). Since HSP90C became undetectable in intact mitochondria already at the lowest proteinase K concentration used (Fig. 2B), we have to assume that HSP90C is not a true mitochondrial protein. Generally, Hsp90s are hydrophobic proteins that tend to stick to membranes (Csermely et al., 1998). In fact, the high affinity of Grp94 for membranes of the ER and Golgi apparatus had led to the early assumption that it was a transmembrane protein (Mazzarella and Green, 1987). Thus, it seems likely that HSP90C was released from chloroplasts that had been disrupted during fractionation and that some of the liberated chaperones then adhered to mitochondrial outer membranes. Hence, our localization of HSP90C to low-density membranes and thylakoids (Fig. 2A) might also be due to the hydrophobicity of the chaperone and may not be of functional significance.
In the Chlamydomonas genome, only three HSP90 genes were identified that encode cytosolic, ER lumenal, and plastidic chaperones (HSP90A-C; Schroda, 2004). The lack of a fourth, mitochondrial HSP90 gene is corroborated by the finding that all 52 ESTs, which we found among the approximately 200,000 Chlamydomonas ESTs to contain Hsp90 coding regions, could be assigned to one of the three HSP90 genes: 20 to HSP90A, 12 to HSP90B, and 20 to HSP90C (data not shown). Phylogenetic analyses indicate that organellar Hsp90 genes of modern eukaryotes originate from duplications of ancient Hsp90 genes, which acquired sequences encoding organellar targeting peptides in a secondary event, whereas the endosymbionts' htpG genes were lost (Emelyanov, 2002; Stechmann and Cavalier-Smith, 2004). Apparently, gene duplications giving rise to mitochondrial Hsp90 genes were manifested only in some lineages, hence explaining why genes encoding mitochondrial Hsp90 proteins exist in some organisms, like Dictyostelium, Drosophila, Arabidopsis, and humans (Felts et al., 2000; Krishna and Gloor, 2001; Morita et al., 2005), but appear to be absent in others, like Chlamydomonas and yeast. The question arises as to which evolutionary pressure may have triggered the reinvention of mitochondrial Hsp90s in some lineages, and how other lineages could otherwise address this pressure.
The expression pattern of HSP90C is very similar to that reported for plastidic Hsp90s in Arabidopsis and rye (Schmitz et al., 1996; Cao et al., 2003) and also resembles that of plastidic Hsp70 and its nucleotide exchange factor CGE1 (Drzymalla et al., 1996; Lin et al., 2001; Schroda et al., 2001): significant basal expression and inducibility by heat shock and light (Fig. 3). This pattern suggests a constitutive function of plastidic Hsp90s in the maturation of plastidic proteins, whose increased synthesis after the onset of light appears to require higher expression levels of the chaperone. The strong inducibility of plastidic Hsp90s by heat shock suggests that they also play a role in the refolding of denatured plastid proteins. The similar pattern of proteins that interact with HSP90C under basal and heat stress conditions (Fig. 5A) indicates that the chaperone constitutively sticks to a defined set of interaction partners and that the interaction with denatured plastid proteins appears to be transient.
Most interesting is our finding that HSP90C and plastidic HSP70B, which in the cell are present at a molar ratio of 1:2.9 (Fig. 6B), appear to form common complexes (Fig. 5B). The organization of cytosolic Hsp90 and Hsp70 and the ER lumenal family members Grp94 and BiP into common complexes has been well documented (Csermely et al., 1998; Pratt and Toft, 2003; Wegele et al., 2004), but is novel for plastidic Hsp70 and Hsp90 and for organellar family members in general. In the eukaryotic cytosol, Hsp90 and Hsp70 represent the essential core of dynamic complexes that catalyze the maturation of a number of client proteins (Pratt and Toft, 2003). Specificity is mediated by the set of cohort proteins: With Hsp40, Hop, and p23, Hsp70-Hsp90 catalyzes the folding of many receptors into a conformation competent for ligand binding (Morishima et al., 2000). Together with Cdc37, the Hsp70-Hsp90 complex mediates the maturation of several signaling kinases (Stancato et al., 1993; Stewart et al., 1999). Also in the ER, Grp94 and BiP form the core of a multichaperone complex that, among others, contains ERdj3 (an Hsp40 homolog), cyclophilin B (an immunophilin), and several protein disulfide isomerases as cohort proteins (Meunier et al., 2002). This multichaperone complex is required for the maturation of proteins traversing the ER lumen, like IgGs (Melnick et al., 1992), but also of some kinases (Csermely et al., 1998). It is assumed that Hsp70 mediates the initial folding steps and then hands over late-folding intermediates to Hsp90 (Pratt and Toft, 2003).
Therefore, it appears likely that a multichaperone complex with an Hsp70-Hsp90 core exists also in organelles. The set of cohort proteins of such an organellar multichaperone complex is likely to differ from that in the cytosol, since human mitochondrial TRAP1 did not interact with human cytosolic Hop or p23 (Felts et al., 2000). One potential candidate might, however, be an Hsp40-like protein for which homologs have been identified in mitochondria (Rowley et al., 1994) and chloroplasts (Schlicher and Soll, 1997; Liu et al., 2005). A mutation in the C terminus of plastidic Hsp90 impaired plastid-to-nucleus signal transduction in Arabidopsis (Lin and Cheng, 1997; Cao et al., 2000, 2003). Thus, it appears plausible that a plastidic Hsp70-Hsp90-based multichaperone complex might be required for the maturation of components of plastid-to-nucleus signal transduction pathways.
MATERIALS AND METHODS
Strains and Culture Conditions
Chlamydomonas reinhardtii strains were grown mixotrophically in Tris-acetate phosphate (TAP) medium (Harris, 1989) on a rotatory shaker at 25°C and an illumination of approximately 30 μE m−2 s−1. For chloroplast isolation, cells were grown in TAP medium supplemented with 0.5% (w/v) peptone.
Heat Shock and Dark-to-Light Shift Kinetics, RNA and Protein Extractions, RNA Gels, and Hybridizations
Heat shock and dark-to-light shift kinetics, isolation of protein and RNA, and preparation of RNA blots were carried out as described in Liu et al. (2005). Membranes were hybridized with DNA probes prepared by the random-priming technique (Feinberg and Vogelstein, 1983) using [α-32P]dCTP (Amersham, Freiburg, Germany). Hybridization was done as described previously (Schroda et al., 1999). The probes used were a 2-kb NheI-AatII fragment containing the HSP70B coding region, a 736-bp PCR product encoding the HSP90C C terminus, and a 1-kb cDNA of cβlp2 (Von Kampen et al., 1994). Radioactive signals were detected with BAS-IP MS 2040 phosphorimager plates (Raytest, Straubenhardt, Germany), scanned with a molecular imager FX phosphorimager (Bio-Rad, Munich), and quantified using the Quantity One-4.5.1 program (Bio-Rad). HSP90C and HSP70B signal intensities were corrected for unequal loading by using the cβlp2 signals obtained for the respective lanes.
SDS-PAGE and Gel-Blot Analyses
SDS-PAGE was performed as described earlier (Laemmli, 1970). For heat shock kinetics, proteins were loaded on the basis of equal chlorophyll concentrations. For fractionation experiments, 1 volume of 2× Laemmli sample buffer (125 mm Tris-HCl, pH 6.8, 20% glycerol, 4% SDS, 10% β-mercaptoethanol, 0.005% bromphenol blue) was added to the samples and protein concentrations were determined by amido black (Popov et al., 1975). BN-PAGE for soluble proteins was carried out according to a published protocol (Schägger et al., 1994). Soluble proteins were prepared as described previously (Schroda et al., 2001). The native high molecular mass marker (66–669 kD) was purchased from Amersham.
Proteins in gels were stained with silver nitrate or transferred to nitrocellulose membranes (Hybond-ECL, Amersham) by semidry blotting using a discontinuous transfer system. Blocking and immunodecorations were performed in phosphate-buffered saline (PBS; Sambrook et al., 1989) containing 3% nonfat dry milk, and immunodetection was done by enhanced chemiluminescence (ECL; Amersham). Antisera were against HSP70B (Schroda et al., 1999), CGE1 (Schroda et al., 2001), mitochondrial carbonic anhydrase (Eriksson et al., 1996), and cytochrome f (Pierre and Popot, 1993). ECL signals were detected with Hyperfilm-ECL (Amersham). Films were scanned using a GS-710 calibrated imaging densitometer (Bio-Rad) and signals were quantified using the Quantity One-4.5.1 program (Bio-Rad). HSP90C signal intensities were corrected for unequal loading by using the cytochrome f signals obtained for the respective lanes. Relative cellular concentrations of HSP90C and HSP70B (Fig. 6) were determined from two independent experiments as follows. Signal intensities obtained from four different concentrations of the purified chaperones were plotted against the protein concentrations used and a trendline was drawn. Signal intensities obtained for the chaperones from four different concentrations of whole-cell protein were inserted into the trendline equation derived from the purified proteins and the mean values of the resulting protein concentrations were calculated.
Cell Fractionations
Isolation of chloroplasts and fractionation into stroma, thylakoids, and low-density membranes was done as described previously (Zerges and Rochaix, 1998). Mitochondria were isolated according to Eriksson et al. (1995). For the proteinase K assay, mitochondria were isolated from cells that have been disrupted with a BioNebulizer (Glas-Col, Terre Haute, IN) instead of vortexing with glass beads. Mitochondria were resuspended in SEM (250 mm Suc, 1 mm EDTA, 10 mm MOPS-KOH, pH 7.2) buffer. If indicated, mitochondria were solubilized by adding Triton X-100 (25% stock) to a final concentration of 1%, followed by pipetting up and down several times and incubating on ice for 5 min. For each assay, proteinase K (1 mg/mL stock) was added to mitochondria equivalent to 20 μg of protein to give a final proteinase K concentration of 25 or 150 μg/mL. Digestion was carried out for 15 min on ice and stopped by adding phenylmethylsulfonyl fluoride (0.1 m stock) to a final concentration of 2 mm.
Immunoprecipitations
Chlamydomonas CF185 cells (Schroda et al., 1999) were grown to a density of approximately 8 × 106 cells/mL, harvested in two equal fractions by centrifugation, and either resuspended in 50 mL TAP medium prewarmed to 40°C for heat shock or in 50 mL TAP medium at 25°C for control. Heat shock and control treatments were performed for 1 h under shaking, then cells were harvested and resuspended in lysis buffer (20 mm HEPES, pH 7.2, 10 mm KCl, 1 mm MgCl2, 154 mm NaCl, 0.25× protease inhibitor cocktail [Roche, Mannheim, Germany]). Cells were lysed by sonication on ice. Lysates were loaded onto Suc cushions (20 mm HEPES-KOH, pH 7.2, 0.6 m Suc) and centrifuged in a TI50 rotor for 30 min at 152,000g and 4°C. The supernatants were supplied with Triton X-100 to a final concentration of 0.5% and incubated for 5 min. Protein A Sepharose beads with coupled antibodies were equilibrated in lysis buffer and incubated with the cell lysates under agitation for 1 h at 10°C. Beads were washed four times with lysis buffer (containing 0.1% Triton X-100) and twice with 10 mm Tris-HCl, pH 7.5, and proteins were eluted by boiling 45 s in 2× Laemmli sample buffer (Fig. 5B) or by shaking 30 min at 25°C with 2× Laemmli sample buffer lacking β-mercaptoethanol (Fig. 5A).
Cloning, Expression, and Purification of HSP90C and HSP70B
DNA encoding the C-terminal 238 amino acids of HSP90C was amplified by PCR from cDNA clone AV630546 with primers 5′-AAGGATCCATGTACCTGACCGAGCCCATTGA-3′ and 5′-CCTGGGTAACCCGCGGACTTCTTCCAGGGGT-3′. The 736-bp PCR product was digested with BamHI and cloned into BamHI-EcoRV-digested pBluescript (Stratagene, La Jolla, CA), giving pMS313. pMS313 was digested partially with BstEII and completely with BamHI, and the resulting 720-bp fragment was cloned into BamHI-BstEII-digested pCB785 (a pQE-9 derivative encoding N- and C-terminal hexahistidine tags containing the HSP70B coding region cloned into BamHI and BstEII restriction sites), giving pMS317. pMS317 was expressed in Escherichia coli M15 (Qiagen, Hilden, Germany) and purified by nickel-nitrilotriacetic acid agarose (Ni-NTA) according to the manufacturer's instructions (Qiagen), with the following alterations. Cells expressing pMS317 were lysed in lysis buffer (6 m guanidine-HCl, 0.5 m NaCl, 20 mm Tris-HCl, pH 8.0, 5 mm imidazole), and lysates were applied to a column containing 2 mL Ni-NTA agarose. The column was washed with lysis buffer containing 6 m urea instead of guanidine-HCl and with urea buffer containing 30 mm imidazole instead of 5 mm. The HSP90C C terminus containing two flanking hexahistidine tags was eluted with 200 mm imidazole in urea buffer and used for the immunization of rabbits.
For the heterologous expression of the entire HSP90C protein, we first amplified the coding region of the N-terminal part of mature HSP90C (lacking its putative N-terminal 71-amino-acid transit peptide) by PCR from cDNA clone AV630546 with primers 5′-GGACTAGTGCTCTTCGAACGCGAACCTGTTCTCTCGC-3′ and 5′-ACGTCCTCGAGGCTGCGCGTCCA-3′. Next, the 867-bp PCR product was digested with SpeI and NruI, and the resulting 291-bp SpeI-NruI fragment was ligated into SpeI-NruI-digested cDNA clone AV630546, giving pMS334. Finally, pMS334 was digested with SapI and XhoI, and the 2,635-bp SapI-XhoI fragment was ligated into SapI-XhoI-digested pTYB11 (NEB, Frankfurt), giving pMS335.
For the heterologous expression of the entire HSP70B protein, we first digested a vector containing the HSP70B cDNA with MscI and HpaI and religated the vector to give pMS303. The latter was used as a template for PCR to amplify a 671-bp fragment using primers 5′-GTCAGCTCTTCTAACGAGAAGGTCGTGGGT-3′ and T7. The PCR product was then digested with ApaI and ligated into SmaI-ApaI-digested pBluescript, yielding pMS304. Next, pMS304 was digested with SpeI-XhoI, and the resulting 638-bp fragment was ligated into pTYB11 that had been cleaved with SpeI and XhoI to give pMS305. The latter was digested with SapI and, after removal of a 39-bp fragment, was religated to yield pMS306. Finally, pCB644, which contains the HSP70B cDNA into which the coding region for a hexahistidine tag had been engineered immediately upstream of the stop codon, was digested with NotI-XhoI, and the resulting 1,871-bp fragment was ligated into NotI-XhoI-digested pMS306 to give pMS307.
pMS335 and pMS307 were expressed in E. coli ER2566 and purified by chitin affinity chromatography according to the manufacturer's instructions (NEB). The chaperones were dialyzed extensively at 4°C against KMH (20 mm HEPES-KOH, pH 7.2, 80 mm KCl, 2.5 mm MgCl2) buffer, supplemented with 10% glycerol, and stored at −80°C. Yields were around 300 μg pure chaperones per liter E. coli culture. The calculated masses of mature HSP90C (82,427.6 D) and hexahistidine-tagged HSP70B (68,936) matched the masses determined by mass spectrometry (Finnigan TSQ 700) for heterologously expressed HSP90C (82,431 D) and HSP70B (68,942), with a deviation below 0.01%.
Affinity Purification of HSP90C Antibodies
A 5- × 10-cm nitrocellulose membrane (Hybond-ECL, Amersham) was incubated for 1 h with 1 mg of purified HSP90C C termini in 6 m urea, 0.5 m NaCl, and 20 mm Tris-HCl, pH 8.0. The membrane was blocked with PBS-T (PBS plus Tween 20 at a final concentration of 0.1%) containing 3% nonfat dry milk, washed with PBS-T, and incubated for 2 h at 4°C with 2 mL antiserum in PBS-T supplemented with bovine serum albumin at a final concentration of 10 mg/mL. After thorough washing, bound antibodies were eluted two times for 30 s with pH shock buffer (100 mm Gly-HCl, pH 2.5, 137 mm NaCl, 0.1% Tween 20), and the pH was raised to 7.0 with 1 m Tris-HCl, pH 8.0. Affinity-purified antibodies were stored at −80°C after addition of 10% glycerol and 0.02% sodium azide.
ATPase Assays
One micromolar heterologously expressed HSP90C protein was incubated in KMH buffer containing 3 mg/mL bovine serum albumin with 0.4 μCi of [γ-32P]ATP (3,000 Ci/mmol; Amersham) at 30°C for 0 to 240 min. The ATP concentrations tested were adjusted accordingly by adding unlabeled ATP. The inhibition of HSP90C's ATPase activity by radicicol was measured with 1 μm HSP90C, 50 μm ATP, 0.4 μCi [γ-32P]ATP, and the following inhibitor concentrations (diluted from a 5 mm ethanol stock): 0, 0.05, 0.1, 1, 5, and 10 μm. The same volume of ethanol was given to the control reactions. Reactions were incubated for 1 h at 30°C. For each inhibitor concentration, three independent experiments were performed. Each reaction was spotted directly onto a thin-layer chromatography plate (polyethyleneimine cellulose F; Merck, Darmstadt Germany) and resolved at room temperature by ascending thin-layer chromatography in 0.4 m LiCl and 3.6% acetic acid buffer for about 15 min. The plates were air dried and the radioactive spots were detected and quantified as described above.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requester.
Sequence data from this article have been deposited to the GenBank data library under the accession number AY705371.
Acknowledgments
We would like to thank the Kazusa DNA Research Institute for providing cDNA clone AV630546. We would also like to thank Francis-André Wollman for the antibody against cytochrome f, Mats Eriksson for the antibody against mitochondrial carbonic anhydrase, and Adriane Atteia for suggesting the use of a nebulizer for mitochondrial preparations. We are grateful to Christoph Beck and Olivier Vallon for critically reading the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft (Schr 617/2–1).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.063578.
References
- Ali A, Bharadwaj S, O'Carrol R, Ovsenek N (1998) Hsp90 interacts with and regulates the activity of heat shock factor 1 in Xenopus oocytes. Mol Cell Biol 18: 4949–4960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asamizu E, Miura K, Kucho K, Inoue Y, Fukuzawa H, Ohyama K, Nakamura Y, Tabata S (2000) Generation of expressed sequence tags from low-CO2 and high-CO2 adapted cells of Chlamydomonas reinhardtii. DNA Res 7: 305–307 [DOI] [PubMed] [Google Scholar]
- Asamizu E, Nakamura Y, Sato S, Fukuzawa H, Tabata S (1999) A large scale structural analysis of cDNAs in a unicellular green alga, Chlamydomonas reinhardtii. Generation of 3433 non-redundant expressed sequence tags. DNA Res 6: 369–373 [DOI] [PubMed] [Google Scholar]
- Bohen SP, Yamamoto KR (1993) Isolation of Hsp90 mutants by screening for decreased steroid receptor function. Proc Natl Acad Sci USA 90: 11424–11428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brychzy A, Rein T, Winklhofer KF, Hartl FU, Young JC, Obermann WMJ (2003) Cofactor Tpr2 combines two TPR domains and a J domain to regulate the Hsp70/Hsp90 chaperone system. EMBO J 22: 3613–3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchner J (1999) HSP90 & Co.—a holding for folding. Trends Biochem Sci 24: 136–141 [DOI] [PubMed] [Google Scholar]
- Cao D, Froehlich JE, Zhang H, Cheng C-L (2003) The chlorate-resistant and photomorphogenesis-defective mutant cr88 encodes a chloroplast-targeted HSP90. Plant J 33: 107–118 [DOI] [PubMed] [Google Scholar]
- Cao D, Lin Y, Cheng C-L (2000) Genetic interactions between the chlorate-resistant mutant cr88 and the photomorphogenic mutants cop1 and hy5. Plant Cell 12: 199–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadli A, Bouhouche I, Sullivan W, Stensgard B, McMahon N, Catelli MG, Toft DO (2000) Dimerization and N-terminal domain proximity underlie the function of the molecular chaperone heat shock protein 90. Proc Natl Acad Sci USA 97: 12524–12529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csermely P, Schnaider T, Söti C, Prohászka Z, Nardai G (1998) The 90-kDa molecular chaperone family: structure, function, and clinical applications. A comprehensive review. Pharmacol Ther 79: 129–168 [DOI] [PubMed] [Google Scholar]
- Drzymalla C, Schroda M, Beck CF (1996) Light inducible gene HSP70B encodes a chloroplast-localized heat shock protein in Chlamydomonas reinhardtii. Plant Mol Biol 31: 1185–1194 [DOI] [PubMed] [Google Scholar]
- Eisenthal R, Cornish-Bowden A (1974) The direct linear plot. Biochem J 139: 715–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Emelyanov VV (2002) Phylogenetic relationship of organellar Hsp90 homologs reveal fundamental differences to organellar Hsp70 and Hsp60 evolution. Gene 299: 125–133 [DOI] [PubMed] [Google Scholar]
- Eriksson M, Gardeström P, Samuelsson G (1995) Isolation, purification, and characterization of mitochondria from Chlamydomonas reinhardtii. Plant Physiol 107: 479–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson M, Karlsson J, Ramazanov Z, Gardestrom P, Samuelsson G (1996) Discovery of an algal mitochondrial carbonic anhydrase: molecular cloning and characterization of a low-CO2-induced polypeptide in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 93: 12031–12034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B (1983) A technique for radiolabelling DNA restriction endonuclease fragments to high activity. Anal Biochem 132: 6–13 [DOI] [PubMed] [Google Scholar]
- Felts SJ, Owen BAL, Nguyen P, Trepel J, Donner DB, Toft DO (2000) The hsp90-related protein TRAP1 is a mitochondrial protein with distinct functional properties. J Biol Chem 275: 3305–3312 [DOI] [PubMed] [Google Scholar]
- Freitag DG, Ouimet PM, Girvitz TL, Kapoor M (1997) Heat shock protein 80 of Neurospora crassa, a cytosolic molecular chaperone of the eukaryotic stress 90 family, interacts directly with heat shock protein 70. Biochemistry 36: 10221–10229 [DOI] [PubMed] [Google Scholar]
- Harris EH (1989) The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. Academic Press, San Diego [DOI] [PubMed]
- Hutchison KA, Brott BK, De Leon JH, Perdew GH, Jove R, Pratt WB (1992) Reconstitution of the multiprotein complex of pp60src, hsp90, and p50 in a cell-free system. J Biol Chem 267: 2902–2908 [PubMed] [Google Scholar]
- Jakob U, Lilie H, Meyer I, Buchner J (1995) Transient interactions of Hsp90 with early unfolding intermediates of citrate synthase. Implications for heat shock in vivo. J Biol Chem 270: 7288–7294 [DOI] [PubMed] [Google Scholar]
- Kamal A, Thao L, Sensintaffar J, Zhang L, Boehm MF, Fritz LC, Burrows FJ (2003) A high-affinity conformation of Hsp90 confers tumour selectivity on Hsp90 inhibitors. Nature 425: 407–410 [DOI] [PubMed] [Google Scholar]
- Krishna P, Gloor G (2001) The Hsp90 family of proteins in Arabidopsis thaliana. Cell Stress Chaperones 6: 238–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lin B-L, Wang J-S, Liu H-C, Chen R-W, Meyer Y, Barakat A, Delseny M (2001) Genomic analysis of the Hsp70 superfamily in Arabidopsis thaliana. Cell Stress Chaperones 6: 201–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Cheng CL (1997) A chlorate-resistant mutant defective in the regulation of nitrate reductase gene expression in Arabidopsis defines a new HY locus. Plant Cell 9: 21–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Willmund F, Whitelegge JP, Hawat S, Knapp B, Lodha M, Schroda M (2005) J-domain protein CDJ2 and HSP70B are a plastidic chaperone pair that interacts with vesicle inducing protein in plastids 1 (VIPP1). Mol Biol Cell 16: 1165–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason CA, Dunner J, Indra P, Colangelo T (1999) Heat-induced expression and chemically induced expression of the Escherichia coli stress protein HtpG are affected by the growth environment. Appl Environ Microbiol 65: 3433–3440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzarella RA, Green M (1987) ERp99, an abundant, conserved glycoprotein of the endoplasmic reticulum, is homologous to the 90-kDa heat shock protein (hsp90) and the 94-kDa glucose regulated protein (GRP94). J Biol Chem 262: 8875–8883 [PubMed] [Google Scholar]
- Melnick J, Aviel S, Argon Y (1992) The endoplasmic reticulum stress protein GRP94, in addition to BiP, associates with unassembled immunoglobulin chains. J Biol Chem 267: 21303–21306 [PubMed] [Google Scholar]
- Meunier L, Usherwood YK, Chung KT, Hendershot LM (2002) A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol Biol Cell 13: 4456–4469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer P, Prodromou C, Hu B, Vaughan C, Roe SM, Panaretou B, Piper PW, Pearl LH (2003) Structural and functional analysis of the middle segment of hsp90: implications for ATP hydrolysis and client protein and cochaperone interactions. Mol Cell 11: 647–658 [DOI] [PubMed] [Google Scholar]
- Minami Y, Kimura Y, Kawasaki H, Suzuki K, Yahara I (1994) The carboxy-terminal region of mammalian HSP90 is required for its dimerization and function in vivo. Mol Cell Biol 14: 1459–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishima Y, Kanelakis KC, Silverstein AM, Dittmar KD, Estrada L, Pratt WB (2000) The Hsp organizer protein hop enhances the rate of but is not essential for glucocorticoid receptor folding by the multiprotein Hsp90-based chaperone system. J Biol Chem 275: 6894–6900 [DOI] [PubMed] [Google Scholar]
- Morita T, Yamaguchi H, Amagai A, Maeda Y (2005) Involvement of the TRAP-1 homologue, Dd-TRAP1, in spore differentiation during Dictyostelium development. Exp Cell Res 303: 425–431 [DOI] [PubMed] [Google Scholar]
- Nathan DF, Lindquist S (1995) Mutational analysis of Hsp90 function: interactions with a steroid receptor and a protein kinase. Mol Cell Biol 15: 3917–3925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaretou B, Prodromou C, Roe SM, O'Brien R, Ladbury JE, Piper PW, Pearl LH (1998) ATP binding and hydrolysis are essential to the function of the Hsp90 molecular chaperone in vivo. EMBO J 17: 4829–4836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaretou B, Siligardi G, Meyer P, Maloney A, Sullivan JK, Singh S, Millson SH, Clarke PA, Naaby-Hansen S, Stein R, et al (2002) Activation of the ATPase activity of Hsp90 by the stress-regulated cochaperone Aha1. Mol Cell 10: 1307–1318 [DOI] [PubMed] [Google Scholar]
- Pierre Y, Popot J-L (1993) Identification of two 4 kDa mini-proteins in the cytochrome b6f complex from Chlamydomonas reinhardtii. C R Acad Sci Ser III Sci Vie 316: 1404–1409 [PubMed] [Google Scholar]
- Popov N, Schmitt S, Matthies H (1975) Eine störungsfreie Mikromethode zur Bestimmung des Proteingehalts in Gewebshomogenaten. Acta Biol Germ 34: 1441–1446 [PubMed] [Google Scholar]
- Pratt WB, Toft DO (2003) Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp Biol Med 228: 111–133 [DOI] [PubMed] [Google Scholar]
- Prodromou C, Panaretou B, Chohan S, Siligardi G, O'Brian R, Ladburry JE, Roe SM, Piper PW, Pearl LH (2000) The ATPase cycle of Hsp90 drives a molecular “clamp” via transient dimerisation of the N-terminal domains. EMBO J 19: 4383–4392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prodromou C, Roe SM, O'Brien O, Ladbury JE, Piper PW, Pearl LH (1997) Identification and structural characterization of the ATP/ADP-binding site in the Hsp90 molecular chaperone. Cell 90: 65–75 [DOI] [PubMed] [Google Scholar]
- Queitsch C, Sangster TA, Lindquist S (2002) Hsp90 as a capacitor of phenotypic variation. Nature 417: 618–624 [DOI] [PubMed] [Google Scholar]
- Richter K, Buchner J (2001) Hsp90: chaperoning signal transduction. J Cell Physiol 188: 281–290 [DOI] [PubMed] [Google Scholar]
- Roe SM, Prodromou C, O'Brien R, Ladbury JE, Piper PW, Pearl LH (1999) Structural basis for inhibition of the Hsp90 molecular chaperone by the antitumor antibiotics radicicol and geldanamycin. J Med Chem 42: 260–266 [DOI] [PubMed] [Google Scholar]
- Rowley N, Prip-Buus C, Westermann B, Brown C, Schwarz E, Barrell B, Neupert W (1994) Mdj1p, a novel chaperone of the DnaJ family, is involved in mitochondrial biogenesis and protein folding. Cell 77: 249–259 [DOI] [PubMed] [Google Scholar]
- Rutherford SL, Lindquist S (1998) Hsp90 as a capacitor for morphological evolution. Nature 396: 336–342 [DOI] [PubMed] [Google Scholar]
- Ryan MT, Voos W, Pfanner N (2001) Assaying protein import into mitochondria. Methods Cell Biol 65: 189–215 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schägger H, Cramer WA, von Jagow G (1994) Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal Biochem 217: 220–230 [DOI] [PubMed] [Google Scholar]
- Schlicher T, Soll J (1997) Chloroplastic isoforms of DnaJ and GrpE in pea. Plant Mol Biol 33: 181–185 [DOI] [PubMed] [Google Scholar]
- Schmitz G, Schmidt M, Feierabend J (1996) Characterization of a plastid-specific HSP90 homologue: identification of a cDNA sequence, phylogenetic descendence and analysis of its mRNA and protein expression. Plant Mol Biol 30: 479–492 [DOI] [PubMed] [Google Scholar]
- Schroda M (2004) The Chlamydomonas genome reveals its secrets: chaperone genes and the potential roles of their gene products in the chloroplast. Photosynth Res 82: 221–240 [DOI] [PubMed] [Google Scholar]
- Schroda M, Vallon O, Whitelegge JP, Beck CF, Wollman F-A (2001) The chloroplastic GrpE homolog of Chlamydomonas: two isoforms generated by differential splicing. Plant Cell 13: 2823–2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroda M, Vallon O, Wollman F-A, Beck CF (1999) A chloroplast-targeted heat shock protein 70 (HSP70) contributes to the photoprotection and repair of photosystem II during and after photoinhibition. Plant Cell 11: 1165–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte TW, Akinaga S, Soga S, Sullivan W, Stensgard B, Toft D, Neckers LM (1998) Antibiotic radicicol binds to the N-terminal domain of Hsp90 and shares important biologic activities with geldanamycin. Cell Stress Chaperones 3: 100–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SV, Agatsuma T, Nakano H (1998) Targeting of the protein chaperone, HSP90, by the transformation suppressing agent, radicicol. Oncogene 16: 2639–2645 [DOI] [PubMed] [Google Scholar]
- Shrager J, Hauser C, Chang CW, Harris EH, Davies J, McDermott J, Tamse R, Zhang Z, Grossman AR (2003) The Chlamydomonas reinhardtii genome project. A guide to the generation and use of the cDNA information. Plant Physiol 131: 401–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DF, Sullivan WP, Marion TN, Zaitsu K, Madden B, McCormick DJ, Toft DO (1993) Identification of a 60-kilodalton stress-related protein, p60, which interacts with hsp90 and hsp70. Mol Cell Biol 13: 869–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stancato LF, Chow YH, Hutchison KA, Perdew GH, Jove R, Pratt WB (1993) Raf exists in a native heterocomplex with hsp90 and p50 that can be reconstituted in a cell-free system. J Biol Chem 268: 21711–21716 [PubMed] [Google Scholar]
- Stancato LF, Hutchison KA, Krishna P, Pratt WB (1996) Animal and plant cell lysates share a conserved chaperone system that assembles the glucocorticoid receptor into a functional heterocomplex with hsp90. Biochemistry 35: 554–561 [DOI] [PubMed] [Google Scholar]
- Stechmann A, Cavalier-Smith T (2004) Evolutionary origins of Hsp90 chaperones and a deep paralogy in their bacterial ancestors. J Eukaryot Microbiol 51: 364–373 [DOI] [PubMed] [Google Scholar]
- Stewart S, Sundaram M, Zhang Y, Lee J, Han M, Guan KL (1999) Kinase suppressor of Ras forms a multiprotein signaling complex and modulates MEK localization. Mol Cell Biol 19: 5523–5534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Kampen J, Nieländer U, von Wettern M (1994) Stress-dependent transcription of a gene encoding a Gb-like polypeptide from Chlamydomonas reinhardtii. J Plant Physiol 143: 756–758 [Google Scholar]
- Wearsch PA, Nicchitta CV (1996) Purification and partial molecular characterization of GRP94, an ER resident chaperone. Protein Expr Purif 7: 114–121 [DOI] [PubMed] [Google Scholar]
- Wegele H, Müller L, Buchner J (2004) Hsp70 and Hsp90-a relay team for protein folding. Rev Physiol Biochem Pharmacol 151: 1–44 [DOI] [PubMed] [Google Scholar]
- Welch WJ, Feramisco JR (1982) Purification of the major mammalian heat shock proteins. J Biol Chem 257: 14949–14959 [PubMed] [Google Scholar]
- Young JC, Moarefi I, Hartl FU (2001) Hsp90: a specialized but essential protein-folding tool. J Cell Biol 154: 267–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerges W, Rochaix J-D (1998) Low density membranes are associated with RNA-binding proteins and thylakoids in the chloroplast of Chlamydomonas reinhardtii. J Cell Biol 140: 101–110 [DOI] [PMC free article] [PubMed] [Google Scholar]