Abstract
The physiological basis of thermoperiodic stem elongation is as yet poorly understood. Thermoperiodic control of gibberellin (GA) metabolism has been suggested as an underlying mechanism. We have investigated the influence of different day and night temperature combinations on GA levels, and diurnal steady-state expression of genes involved in GA biosynthesis (LS, LH, NA, PSGA20ox1, and PsGA3ox1) and GA deactivation (PsGA2ox1 and PsGA2ox2), and related this to diurnal stem elongation in pea (Pisum sativum L. cv Torsdag). The plants were grown under a 12-h light period with an average temperature of 17°C. A day temperature/night temperature combination of 13°C/21°C reduced stem elongation after 12 d by 30% as compared to 21°C/13°C. This was correlated with a 55% reduction of GA1. Although plant height correlated with GA1 content, there was no correlation between diurnal growth rhythms and GA1 content. NA, PsGA20ox1, and PsGA2ox2 showed diurnal rhythms of expression. PsGA2ox2 was up-regulated in 13°C/21°C (compared to 21°C/13°C), at certain time points, by up to 19-fold. Relative to PsGA2ox2, the expression of LS, LH, NA, PSGA20ox1, PsGA3ox1, and PsGA2ox1 was not or only slightly affected by the different temperature treatments. The sln mutant having a nonfunctional PsGA2ox1 gene product showed the same relative stem elongation response to temperature as the wild type. This supports the importance of PsGA2ox2 in mediating thermoperiodic stem elongation responses in pea. We present evidence for an important role of GA catabolism in thermoperiodic effect on stem elongation and conclude that PsGA2ox2 is the main mediator of this effect in pea.
The ability of plants to discriminate between temperature during the day and night in their response to flowering, fruiting, and growth is referred to as thermoperiodism (Went, 1944). Erwin et al. (1989) showed that the effects of diurnal temperature alternation on stem length in Lilium longiflorum could best be described by the mathematical difference (DIF) between day temperature (DT) and night temperature (NT). Stem elongation thus increases with an increase in DIF, from a negative to a positive value. Stem elongation in many species is affected by the relationship between DT and NT, and the DIF concept is widely used for growth control in production of ornamental plants propagated in a greenhouse (Erwin and Heins, 1995; Myster and Moe, 1995; Moe and Heins, 2000). A negative DIF treatment (low DT and high NT) is a tool to produce compact flower plants and vegetable seedlings with short internodes without a delay in production time. Also, negative DIF treatments have largely replaced the use of chemical growth retardants in a number of commercial cultures. Thus, the use of such temperature regimes has substantial practical and economic implications in addition to representing an environmentally more sustainable method than chemical growth control. However, very little is known about the mechanisms underlying the thermoperiodic responses in plants. Such knowledge will be of great importance for increasing the general understanding of the interaction between the temperature and light climate in climatic adaptation, as well as for the evaluation of the possibilities of an extended use of temperature manipulations in commercial plant culture.
GAs are involved in many aspects of plant development, particularly stem elongation. As a consequence, most studies on the basis of the effects of DIF have focused on GA. In application experiments, exogenous GA has been shown to neutralize the difference in stem elongation under negative and positive DIF (Tangerås, 1979; Zieslin and Tsujita, 1988; Moe, 1990; Ihlebekk et al., 1995; Grindal et al., 1998b). Jensen et al. (1996) reported higher levels of GA1 in Campanula isophylla grown under positive DIF than negative DIF. It has been suggested that altered stem elongation of pea (Pisum sativum) plants in response to diurnal temperature alternations may be mediated by changes in the endogenous levels of GA1 (Grindal et al., 1998a). GA1 levels were found reduced by almost 60% under a temperature regime with low DT and high NT compared to a temperature regime with high DT and low NT, with both temperature treatments having the same daily average temperature. The study of GA metabolites suggested that thermoperiodicity could affect both biosynthesis and inactivation steps of GA1 (Grindal et al., 1998a). Furthermore, based on experiments with GA biosynthesis inhibitors and applications of GA1 and GA3 (GA3 is protected from deactivation by enzymatic 2-oxidation), it has been hypothesized that reduced GA1 levels in stem tissue under negative DIF are caused by enhanced inactivation of GA1 by 2-oxidation in pea (Grindal et al., 1998b).
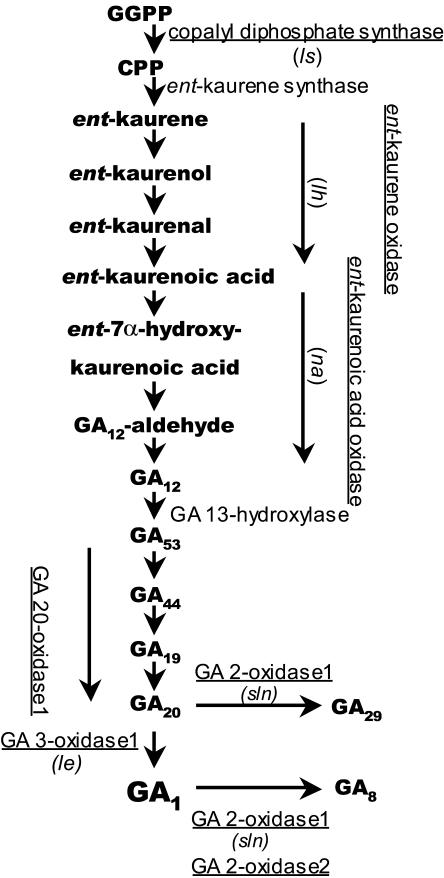
GA1 is the principal GA regulating stem length in pea (Ingram et al., 1984). Comparisons of the endogenous levels in different GA biosynthetic mutants have revealed a highly significant positive correlation between stem or internode elongation and the logarithmic increase in GA1 content in pea (Ross et al., 1989; Grindal et al., 1998a). GA metabolism (Fig. 1) is divided into three stages (Hedden and Phillips, 2000). The first stage of the pathway occurs in plastids and produces ent-kaurene from trans-geranyl geranyl diphosphate. The second-stage reactions occur on membranes outside the plastids and GA53 is produced from ent-kaurene. The last stage of GA1 production occurs in the cytoplasm, where GA53 is converted to various GA intermediates and to bioactive GA1 by a series of oxidation steps catalyzed by 2-oxoglutarate-dependent dioxygenases: GA 20-oxidases (GA20ox) and GA 3-oxidases (GA3ox). GA 2-oxidases perform deactivations of GAs by 2β-hydroxylation.
Figure 1.
Simplified 13-hydroxylation pathway of GA biosynthesis in vegetative tissue of pea. The enzymes that are cloned and characterized in pea are underlined. Corresponding mutants are given in parentheses. Diurnal steady-state expression of the characterized genes in response to diurnal temperature alternations has been investigated in this study.
In pea, most of the genes encoding enzymes involved in the GA metabolic pathway have been characterized (Fig. 1; Ait-Ali et al., 1997; Garcia-Martinez et al., 1997; Lester et al., 1997, 1999; Martin et al., 1997, 1999; Davidson et al., 2003, 2004). The GA biosynthesis genes LS, LH, NA, PsGA20ox1, PsGA3ox1 (Le, length), and the two GA deactivation 2-oxidase genes, PsGA2ox1 and PsGA2ox2, have all been shown to be expressed in expanding internodes of pea (Ait-Ali et al., 1997; Elliott et al., 2001; Ross et al., 2003; Davidson et al., 2003, 2004). The PsGA2ox1 (SLN, slender) gene product metabolizes the 2-oxidation of GA20 to GA29 to GA29 catabolite and 2-oxidation of GA1 to GA8, while the PsGA2ox2 gene product has a strong preference for GA1 rather than GA20 as a substrate (Reid et al., 1992; Lester et al., 1999). GA metabolism seems to be regulated at the transcriptional level, and the recent characterization of genes in the GA pathway has made it possible to address a possible thermoperiodic regulation of GA metabolism. The aim of this study was to investigate further the basis of thermoperiodic responses in plants by testing the hypothesis of reduced production of GA1 precursors and enhanced inactivation by GA1 under negative DIF compared to zero DIF and positive DIF (Grindal et al., 1998a, 1998b).
In this article, we present steady-state expression profiling over 2 d of seven GA metabolism genes in apical stem tissue of 18-d-old pea seedlings and present evidence of a temperature-regulated expression of PsGA2ox2. We conclude that PsGA2ox2, in contrast to PsGA2ox1, is involved in mediating thermoperiodic responses on stem elongation and that inactivation of GA1 by 2-oxidation is an important contribution to the reduced GA1 levels and reduced stem growth under negative DIF compared to zero and positive DIF in pea.
RESULTS
Negative DIF Inhibits Stem Elongation during the Daytime
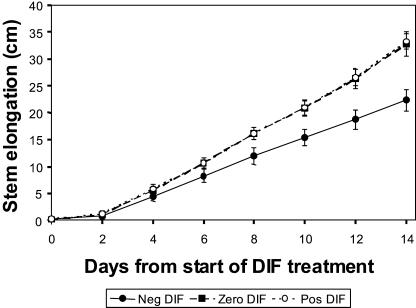
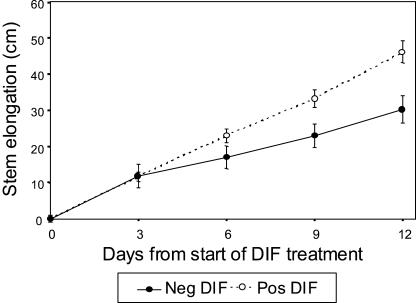
To examine the effect of negative DIF on stem elongation, plants subjected to negative DIF (DT 13°C/NT 21°C), zero DIF (DT 17°C/NT 17°C), and positive DIF (DT 21°C/NT 13°C) were compared. After 14 d of treatment, a negative DIF treatment resulted in approximately 30% shorter plants than those grown at zero DIF and positive DIF (Fig. 2). Plants grown at zero DIF were similar in height to those grown at positive DIF. The leaf number was not affected by negative DIF compared to zero and positive DIF (data not shown; Grindal et al., 1998a). These results confirm the effect of negative DIF on the reduction of internode length, and that reduction in internode length occurs without any delay in plant development.
Figure 2.
Effect of different DT/NT combinations on the stem elongation rate of pea. Seedlings were grown for 6 d at constant temperature of 17°C prior to start of DIF treatments. The temperature regimes were negative DIF, 13°C/21°C; zero DIF, 17°C/17°C; and positive DIF, 21°C/13°C. Results are average of 18 plants ±sd.
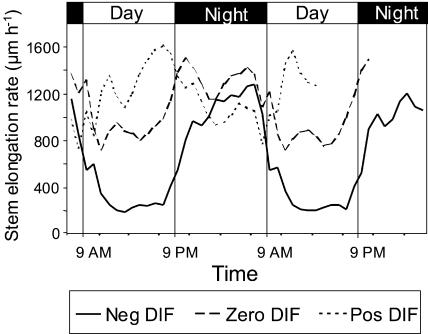
To assess the effects of the DIF treatments on diurnal stem elongation rhythms, stem elongation rate was measured in a triangular displacement transducer. Under zero DIF, the stem elongation rate in the light period (on average 1,000 μm h−1) was lower than in the dark period (on average 1,250 μm h−1; Fig. 3), showing that stem elongation was inhibited in the light period as compared to the dark period. Under negative DIF, this effect was more pronounced, as a strong inhibition of the rate of stem elongation in the light period was observed (about 250 μm h−1). However, in the dark period, the stem elongation rate increased 5-fold up to levels similar to or somewhat lower than those observed when the plants were grown under positive and zero DIF (1,000 μm h−1). Positive DIF stimulated the rate of stem elongation in the light period (on average 1,250 μm h−1) compared to the zero DIF (1,000 μm h−1), but reduced the elongation rate in the dark period (on average 1,000 μm h−1) compared to zero DIF (1,250 μm h−1). However, the average daily stem elongation rate of the positive and zero DIF treatment was approximately the same (as can be seen in Fig. 2). The positive DIF treatment showed that the inhibition of stem elongation observed in the light period at zero DIF disappeared when the DT was higher than the NT.
Figure 3.
Diurnal growth rhythms in pea as affected by different DT/NT combinations. Seedlings were grown for 6 d at constant temperature of 17°C prior to start of DIF treatments. The temperature regimes were negative DIF, 13°C/21°C; zero DIF, 17°C/17°C; and positive DIF, 21°C/13°C. Results are average of six individual plants and represent growth rhythms at day 12 to day 14 after start of DIF treatments.
Thermoperiodic Effects on Transcriptional Regulation of GA Metabolism Genes
A negative DIF treatment has been shown to reduce GA1 levels compared to zero and positive DIF (Grindal et al., 1998a). To investigate whether the transcription of genes encoding enzymes involved in GA metabolism (Fig. 1) is affected by DT/NT alternations, steady-state mRNA levels of the corresponding genes were analyzed using real-time reverse transcription (RT)-PCR. As internal controls in this experiment, both α-tubulin and actin were tested. Both reference genes gave similar reproducible patterns of mRNA levels. In Figures 4 and 5, relative transcript levels of genes encoding enzymes involved in GA metabolism in apical stem tissues of pea are shown. The results are normalized to actin and to the lowest mRNA level within each plate. The values presented are an average from two independent experiments. All tendencies discussed in this article were clear both days in both replicate experiments.
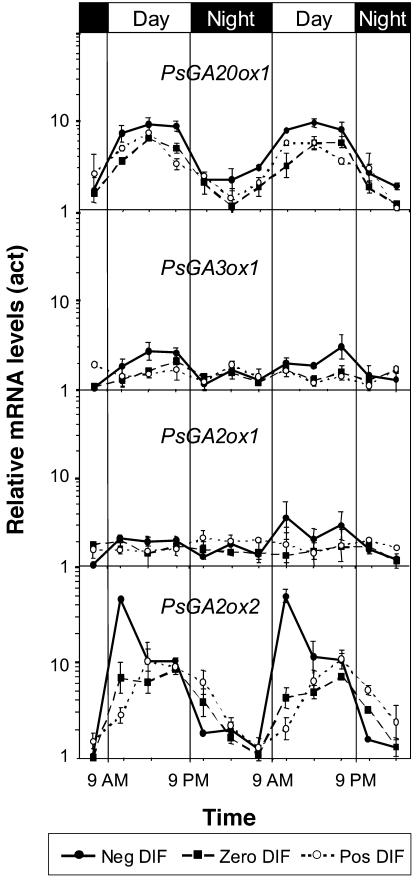
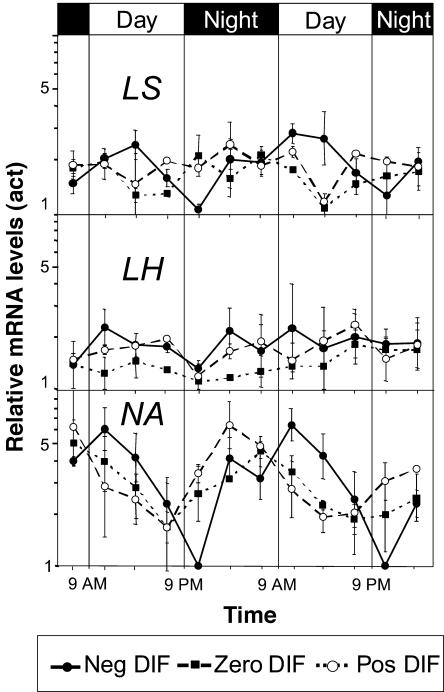
Figure 4.
Effects of different DT/NT combinations on expression of late-stage GA metabolism genes in pea. Diurnal variation of PsGA20ox1, PsGA3ox1, PsGA2ox1, and PsGA2ox2 mRNA levels after 12 d of negative DIF, zero DIF, and positive DIF treatment. The mRNA levels are normalized to mRNA of actin. Results are average of two independent experiments ±se. In each independent experiment, the sample at each time point consisted of 18 plants. Note that the scale is logarithmic.
Figure 5.
Effects of different DT/NT combinations on expression of GA biosynthesis genes in pea. Diurnal variation of LS, LH, and NA mRNA levels after 12 d of negative DIF, zero DIF, and positive DIF treatment. The mRNA levels are normalized to mRNA of actin. Results are average of two independent experiments ±se. In each independent experiment, the sample at each time point consisted of 18 plants. Note that the scale is logarithmic and different from Figure 4.
The major effect of the DIF treatments was to regulate steady-state expression of the GA deactivation gene PsGA2ox2 (Fig. 4). This fit with earlier evidence that GA deactivation is important for thermoperiodic regulation of stem elongation (Grindal et al., 1998a). Under zero DIF (17°C/17°C), there was a clear diurnal rhythm in PsGA2ox2 steady-state transcription. A 7- to 8-fold increase in the steady-state transcript level of PsGA2ox2 from the night period to the day period was observed under zero DIF. However, under positive DIF (21°C/13°C), there was a delayed up-regulation of PsGA2ox2 mRNA levels compared to zero DIF in the morning, as measured at 11 am (Fig. 4). At the same time point under negative DIF, PsGA2ox2 steady-state expression was up-regulated and was on average 8 and 19 times higher than under zero and positive DIF, respectively. However, this marked difference was much reduced at 3 pm, and, by the end of the light period at 7 pm, there was virtually no difference in steady-state expression between the DIF treatments. Two hours into the dark period, when the temperature had increased to 21°C with negative DIF treatment, the mRNA level dropped to levels below those observed under zero and positive DIF treatments. Thus, the mRNA level of PsGA2ox2 under negative DIF increased when the temperature dropped and was reduced when the temperature increased, as compared to zero DIF. However, the most dramatic effect on PsGA2ox2 mRNA levels was caused by a simultaneous temperature drop and onset of light.
The steady-state expression of PsGA2ox1 under zero DIF was quite stable and showed no sign of any diurnal rhythm (Fig. 4). However, steady-state expression appeared to be slightly higher during the light period and lower during the dark period under negative DIF. The opposite pattern was observed under positive DIF. The amplitude of these measurements was not more than 2-fold.
The steady-state expression of the GA 20-oxidase gene, PsGA20ox1, showed a diurnal rhythm correlating with the light cycle. A 5- to 6-fold higher mRNA level during the day than during the night was observed (Fig. 4). The DIF treatments did not affect the phase or amplitude of the rhythm, but at negative DIF there was a slight increase in steady-state expression as compared to zero and positive DIF in the dark period as well as in the light period. It was also clear that PsGA20ox1 steady-state expression correlated with the rhythm of PsGA2ox2 steady-state expression.
The PsGA3ox1 gene product regulates the conversion of GA20 to GA1 in shoot tissue. The steady-state expression of this gene was 2- to 2.5-fold higher during the light period than during the dark period under negative DIF (Fig. 4). Steady-state expression of PsGA3ox1 in plants grown at zero and positive DIF were relatively constant, with no indications of any diurnal rhythm.
The LS gene product regulates the conversion of geranyl geranyl diphosphate to ent-copalyl diphosphate in proplastids. The LS mRNA levels were quite stable and not much affected by the DIF treatments (Fig. 5). However, there appears to be a weak rhythm in steady-state expression under negative DIF, with the highest level in the beginning/midday and the lowest level in the beginning of the night. The amplitude of this rhythm was 2-fold. However, based on average diurnal steady-state expression, there was no difference in mRNA levels between the DIF treatments. The LH gene encodes a multifunctional enzyme that converts ent-kaurene to ent-kaurenol and then ent-kaurenal to ent-kaurenoic acid. mRNA levels of LH were quite stable, with no tendencies of any rhythm or difference in steady-state expression (Fig. 5). The NA gene encodes another multifunctional enzyme that converts ent-kaurenoic acid to GA12 aldehyde in three consecutive oxidation steps (Fig. 1). NA steady-state expression showed a diurnal rhythm. Under zero and positive DIF, steady-state expression was highest in the middle or at the end of the night. During the day, steady-state expression gradually declined, and the lowest level was reached by the end of the light period. The amplitude of this rhythm was a 3-fold change in relative steady-state expression. However, at negative DIF, both phase and amplitude of the rhythm in steady-state expression of NA was changed (Fig. 5). The highest mRNA level was measured at the beginning of the day and the lowest value was measured at the beginning of the night. The phase shift in steady-state expression under negative DIF made the mRNA levels higher than zero and positive DIF during the daytime and lower during the nighttime. However, the average relative expression level was not significantly different between the DIF treatments.
Based on average critical threshold (ΔCt) values, which give a rough estimate of relative mRNA levels (ΔCt is Ct of gene of interest minus Ct of endogenous reference gene), we concluded that there was more mRNA of LH and PsGA20ox1 than of NA, PsGA3ox1, and LS. The two GA 2-oxidase genes had the lowest mRNA levels of the genes investigated in apical stem tissue (data not shown). Comparing steady-state expression of the GA 2-oxidase genes, the mRNA levels of PsGA2ox2 were consistently lower than PsGA2ox1 during the night, but higher during the day (data not shown).
Thermoperiodic Regulation of GA Levels
We also investigated whether the temperature-mediated changes in mRNA levels of genes encoding enzymes involved in late-stage GA metabolism were accompanied by corresponding changes in GA levels. Analyses of GA53, GA44, and GA19 under the different DIF treatments indicate a diurnal rhythm in the levels of these metabolites, with the highest levels in the beginning of the light period and the lowest levels in the middle of the dark period (Fig. 6). The different DIF treatments did not appear to affect the levels of these metabolites nor their rhythmic behavior. Apparently, there is a correlation between both PsGA20ox1 and NA steady-state expression (Figs. 4 and 5) and the level of GA53, GA44, and GA19 (Fig. 6).
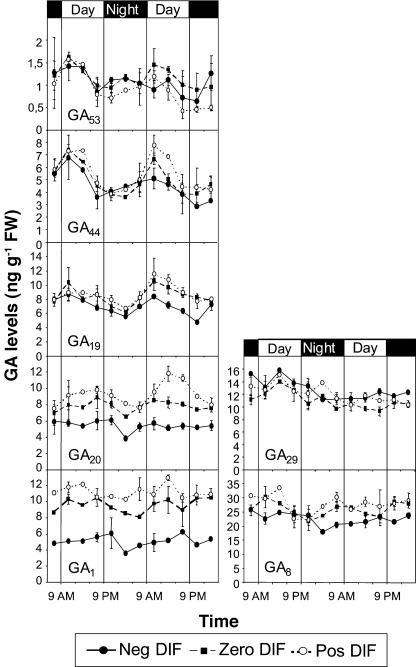
Figure 6.
Effects of different DT/NT combinations on GA levels. Comparisons of diurnal variation of GA levels in pea after 12 d of negative, zero, and positive DIF treatments. Results are average of two independent experiments ±se, except from the third and sixth measurement in each series, where n = 1. In each independent experiment, the sample at each time point consisted of 18 plants.
The level of GA20 was affected by the different temperature treatments (Fig. 6). A negative DIF (13°C/21°C) treatment resulted in a 40% reduction in the levels of GA20 as compared to the positive DIF treatment. This reduction correlated with the higher levels of PsGA3ox1, PsGA2ox1, and PsGA2ox2 mRNA during the light period under a negative DIF temperature regime. Compared to a positive DIF treatment, a zero DIF (17°C/17°C) treatment resulted in an average of 80% of the levels of GA20. The diurnal rhythm observed for GA44 and GA19 was not observed for GA20 under negative DIF, but was observed in the positive DIF treatment with the highest values in the middle of the light period (average of 10.5 ng g−1 fresh weight) and lowest values in the middle of the dark period (average of 8 ng g−1 fresh weight). GA1 levels under negative DIF were on average only 40% to 45% of that under the positive DIF temperature regime, and GA1 under a constant temperature was almost 90% of the level found under positive DIF (Fig. 6). There were no indications of a diurnal rhythm in GA1 levels, and therefore no correlation between stem elongation rhythms and GA1. GA29 was not affected by the DIF treatments, but GA8 was slightly reduced under negative DIF as compared to zero and positive DIF. However, the ratio of GA8/GA1 and GA29/GA20 was significantly increased under a negative DIF treatment as compared to zero and positive DIF (Table I).
Table I.
The ratio between average endogenous levels of GA29 to GA20 and of GA8 to GA1 in pea cv Torsdag grown under a 12-h photoperiod and the following DT/NT combinations: negative DIF (21°C/13°C), zero DIF (17°C/17°C), and positive DIF (13°C/21°C)
| DT/NT | GA29/GA20 | GA8/GA1 |
|---|---|---|
| 21°C/13°C | 1.47 | 2.42 |
| 17°C/17°C | 1.56 | 2.59 |
| 13°C/21°C | 2.51 | 4.31 |
The sln Mutant Has a Clear DIF Response
Based on gene expression analysis, we hypothesized that increased deactivation of GA1 under negative DIF did not involve PsGA2ox1, as steady-state expression of this gene was quite stable. To test this hypothesis and investigate further the relative roles of PsGA2ox2 and PsGA2ox1 in thermoperiodic responses of stem elongation, we studied the effects of positive and negative DIF temperature regimes on the slender sln pea mutant, which has a nonfunctional PsGA2ox1 gene product (Lester et al., 1999; Martin et al., 1999). After 12 d of growth under positive and negative DIF, we observed a clear effect of DIF treatments on stem elongation in the sln mutant (Fig. 7). Stem elongation under negative DIF treatment was reduced by 27% as compared to positive DIF treatment. This result is comparable to the effect of negative and positive DIF treatment on the wild type, where stem elongation in plants grown under negative DIF was 30% lower than plants grown under positive DIF (Figs. 2 and 7). This result further supports the conclusion that PsGA2ox2 is important in mediating thermoperiodic responses in pea as well as that any significant contribution of PsGA2ox1 in mediating these responses on stem elongation can be excluded.
Figure 7.
Effects of different DT/NT combinations on stem elongation of the sln mutant. Seedlings were grown for 6 d at constant temperature of 17°C prior to the start of DIF treatments. Negative DIF, 13°C/21°C; zero DIF, 17°C/17°C; and positive DIF, 21°C/13°C. Results are average ±sd of 12 plants.
DISCUSSION
Based on gene expression analyses and quantitative GA analyses, this study shows that a GA deactivation gene, PsGA2ox2, is involved in thermoperiodic regulation of stem elongation in pea. Furthermore, gene expression data and growth responses of the sln mutant reveal important differences regarding both regulation and relative importance of the two GA deactivation genes (PsGA2ox1 and PsGA2ox2) in their contribution in mediating thermoperiodic regulation of stem elongation.
The reduced levels of GA1 in plants grown under negative DIF (Fig. 6) seems to be linked to the temperature drop that occurs in the morning because, as a response to the temperature drop, steady-state expression of PsGA2ox2 was up-regulated (Fig. 4). When the temperature dropped in the beginning of the dark period (positive DIF), there was no response on PsGA2ox2 steady-state expression as compared to zero DIF, and the mRNA levels decreased in parallel throughout the night. This suggests that a temperature drop in the transition between light/dark is qualitatively different from a temperature drop in the transition between dark/light. The opposite situation, a rise in temperature from 13°C to 21°C in the morning (positive DIF) or evening (negative DIF) resulted in reduced steady-state expression of PsGA2ox2 compared to zero DIF, indicating that a change to a favorable growth temperature (e.g. 21°C in our experiment) could lead to an increase in the amount of active GA1 through reduced GA1 deactivation. Thus, there is a good correlation between steady-state expression of PsGA2ox2 and stem elongation rate (Figs. 3 and 4).
The diurnal rhythm of PsGA2ox2 expression indicates that most of the GA1 inactivation of which this gene is responsible occurs in the light period. At constant temperature, the expression pattern of PsGA2ox2 followed the rhythm of PsGA20ox1 and the light/dark alternation (Fig. 4). It could be that the expression rhythm of PsGA2ox2 observed in our experiment at zero DIF is a consequence of a feed-forward regulation caused by an increased flux of 20C-GAs in the light period. A feed-forward mechanism at the transcriptional level has previously been reported in pea (Thomas et al., 1999; Elliott et al., 2001). However, if the variations in PsGA2ox2 mRNA levels are the results of a feed-forward regulation, temperature most certainly modifies this feed-forward regulation, indicating that changes in ambient growth temperature (at least in the light period) is important in controlling GA levels by control of deactivation of GA1. In contrast, the other GA-deactivation gene investigated, PsGA2ox1, showed no sign of being feed-forward regulated or much regulated by temperature (Fig. 4).
The response to a negative DIF treatment of the sln mutant, which has a nonfunctional PsGA2ox1 gene product, was comparable to the wild type (Figs. 2 and 7). In contrast, the la crys mutant (Potts et al., 1985), which is assumed to be a loss-of-function mutant and appears to be GA-saturated, shows no or a very poor DIF response on stem elongation (J.A. Stavang, R.I. Pettersen, J.E. Olsen, and R. Moe, unpublished data; Grindal et al., 1998a). This demonstrates that thermoperiodic regulation of stem elongation is dependent on a functional GA metabolism pathway, but that PsGA2ox1 is not important in mediating thermoperiodic responses on stem elongation.
The diurnal rhythm of PsGA20ox1 expression in stem tissue (Fig. 4) indicates that most of the production of C20-GAs in this tissue in pea probably is coupled to the light period. In Sorghum bicolor, phytochrome B seems to control the daily regulation of GA20 biosynthesis (Foster and Morgan, 1995). Also, in potato, phytochrome B is involved in the regulation of transcript levels of one GA20-oxidase gene (Jackson et al., 2000). A rhythm of PsGA20ox1 steady-state expression has also been reported in pea leaves with higher steady-state expression in light than in dark (Garcia-Martinez and Gil, 2002). However, the rhythm disappeared when the pea plants, after an 8-h dark period, were kept in continuous light, as the high level of PsGA20ox1 expression was maintained. This indicated that the observed rhythm in PsGA20ox1 steady-state expression was not the result of an endogenous circadian rhythm. Thus, evidence accumulated so far suggests that light perception is important for the regulation of the expression of this gene. It is therefore likely that the expression rhythm we report here in green apical stem tissue is established by light and dark alternation. It should be mentioned, however, that in the process of de-etiolation in pea, PsGA20ox1 is apparently not involved in any rapid light regulation (Ait-Ali et al., 1999; Reid et al., 2002).
Compared to the situation with PsGA2ox2, steady-state expression of PsGA3ox1 was only slightly (2- to 3-fold) up-regulated under negative DIF (Fig. 4). However, GA1 levels in pea plants grown under negative DIF were only 45% of those grown under positive DIF (Fig. 6). It might be that the low levels of GA1 under negative DIF lead to a feedback up-regulation of both PsGA20ox1 and PsGA3ox1 expression. However, due to the strong up-regulation of PsGA2ox2 expression (and maybe genes upstream in the GA metabolic pathway that are not yet characterized in pea), the total effect on GA1 levels is a 55% reduction.
Neither LS nor LH are subjected to feedback regulation by the activity of the GA response pathway (Hedden and Phillips, 2000). Our study shows that neither gene is substantially regulated by daily temperature alternations (Fig. 5). However, the rhythm and amplitude of NA steady-state expression was affected by DIF treatments, showing that daily temperature alternations affect synchronization of this rhythm (Fig. 5). The gene product of NA, ent-kaurene acid oxidase, is localized in the endoplasmic reticulum membrane (for review, see Olszewski et al., 2002). The biological significance of the rhythm observed and the effect of DIF on NA steady-state expression are at this point unclear.
GA44 and GA19 are under the control of the PsGA20ox1 gene product (Garcia-Martinez et al., 1997), and, similar to the PsGA20ox1 steady-state expression, the levels of GA44 and GA19 were not affected by the DIF treatments. The level of these metabolites, as well as GA53, shows trends of a rhythm with the highest levels in the beginning of the light period and the lowest levels in the dark period in all DIF treatments, thus correlating with both NA and PsGA20ox1 steady-state expression rhythms. Rhythms of GA metabolites have previously been reported in S. bicolor and spinach (Talon et al., 1991; Foster and Morgan, 1995). The level of GA53 was not affected by the DIF treatments and this is in contrast to the findings of Grindal et al. (1998a), who found reduced levels of GA53 at negative DIF. In the experiment of Grindal et al. (1998a), the plants were harvested a week later and this might suggest that if plants are subjected to negative DIF for a longer time period, production of GAs are also affected. However, since our experiment differs from theirs with respect to light intensity and quality, daylength, average temperature, and diurnal temperature variations, comparison may not be straightforward.
GA20 was reduced by 40% at negative DIF as compared to positive DIF. Different steady-state expression of PsGA3ox1, PsGA2ox1, and PsGA2ox2 at negative and zero DIF, as compared to positive DIF, might cause the observed reduction of GA20 in these treatments as well as the disappearance of the rhythm under negative DIF.
There were no or only weak indications of any diurnal rhythm in GA1 levels. Since there is no indication of any rhythm in GA1 levels despite the large variation in stem elongation between dark and light periods, stem elongation rate in a short-term perspective is obviously much more dependent on ambient growth temperature than on GA1 levels (Figs. 3 and 6). In a study of wheat leaves grown at different temperatures, Tonkinson et al. (1997) suggested that GA functions as a stimulus for continued cell extension by preventing cell maturation in the extension zone. Thus, higher GA levels in plants will increase the size of the extension zone and thus the ability of a plant to elongate. Furthermore, they proposed that low temperatures increase the sensitivity threshold for GA action and reduce maximum stem elongation within the extension zone. This model could explain why wheat grown at 10°C, having the same level of active GA as wheat grown at 20°C, was much shorter. To explain the growth patterns observed in our experiment, the sensitivity threshold for GA action must also be elevated by light, as stem elongation rate was reduced 20% to 30% by light under zero DIF (Fig. 3). Under the positive DIF treatment, it was apparent that, by increasing the temperature, the inhibition of stem elongation by light was counteracted. Lowering the temperature in the light period as under negative DIF, however, further inhibited stem elongation. Thus, even though GA1 levels do not change diurnally, it appears that sensitivity does, and it appears that sensitivity to GA and thus stem elongation is dependent on both light and temperature. Further support of this view is the fact that the la crys mutant, which is a saturated GA response mutant, shows similar diurnal stem elongation rate rhythms as wild type (although the stem elongation rate is generally higher; J.A. Stavang, R.I. Pettersen, J.E. Olsen, and R. Moe, unpublished data), and the demonstration that light reduces tissue responsiveness to GAs (Weller et al., 1994; Lopezjuez et al., 1995; Reed et al., 1996; O'Neill et al., 2000).
GA29 was not significantly affected by the temperature treatments (Fig. 6). The level of GA8 was slightly reduced under the negative DIF temperature regime. This implies that the ratios between endogenous levels of GA29 to GA20 and of GA8 to GA1 are increased under negative DIF (Table I). However, the expression of PsGA2ox1 was not affected to any extent (Fig. 4), and since the level of GA20 was reduced while GA29 was unaffected under negative DIF, the higher GA29/GA20 relationship is probably not caused by increased 2-oxidation of GA20. The increased GA29 to GA20 ratio is therefore most likely a consequence of reduced GA20 levels only.
The GA8 level under negative DIF was slightly reduced as compared to zero and positive DIF despite the higher levels of PsGA2ox2 steady-state expression (Figs. 4 and 6). However, the ratio of GA8 to GA1 was significantly higher under negative DIF than at zero and positive DIF (Table I). Thus, our results support the findings of Grindal et al. (1998b), who showed that in pea plants dwarfed with paclobutrazol, the response to GA1 was more strongly reduced by negative DIF compared to positive DIF than the response to GA3. GA3 is protected from enzymatic deactivation due to an extra double bond and it differs structurally from GA1. Grindal et al. (1998b) therefore concluded that the reduced response to GA1 compared to GA3 under negative DIF was caused by a higher rate of 2-oxidation of GA1 into GA8 under negative DIF than positive DIF. Recent studies of the process of de-etiolation in pea have shown that GA8 levels increase upon irradiation with white light as a consequence of increased inactivation activity (Gil and Garcia-Martinez, 2000; Reid et al., 2002). This did not occur in our study, suggesting that a step in the biosynthesis of GA1 is down-regulated. However, none of the five GA biosynthesis genes investigated in this study seems to be involved in such down-regulation, at least at the level of transcription, and further studies are needed to elucidate the complete role of thermoperiodic regulation of GA metabolism.
In this article, we have shown that the GA deactivation gene PsGA2ox2 is involved in mediating thermoperiodic stem elongation by regulating GA1 levels in pea. In contrast, the other GA deactivation gene characterized in pea, PsGA2ox1, does not seem to contribute in mediating thermoperiodic stem elongation. Furthermore, we have shown that NA, PsGA20ox1, and PsGA2ox2 all are expressed in diurnal rhythms. However, while PsGA20ox1 steady-state expression was only slightly affected by the DIF treatments, DIF treatments affected both amplitude and phase of the rhythmic steady-state expression of NA and PsGA2ox2. Still, when comparing average diurnal steady-state expression of the GA metabolism genes investigated as affected by DIF treatments, major changes were observed in PsGA2ox2 steady-state expression only.
MATERIALS AND METHODS
Plant Materials and Experimental Conditions
Three seeds per pot of Pisum sativum L. wild-type line 107 (cv Torsdag) or the sln mutant were sown in fertilized peat (Floralux; Nittedal Torvindustrier, Norway) and grown under controlled environmental conditions (Conviron growth chambers; Controlled Environments, Winnipeg, Manitoba, Canada). The humidity was adjusted to give 0.47 ± 0.03 kPa water vapor deficit. The daily light period was 12 h with a photon flux density of 170 ± 10 μmol m−2 s−1 at 400 to 700 nm (F96T12/CW/1500 fluorescent tubes; General Electric, Fairfield, CT), enriched with light from incandescent lamps (Osram, Munich). The red/far-red ratio was 1.7 ± 0.1. The seedlings were watered daily with a complete nutrient solution of EC = 1.5 mS cm−1. The temperature was kept at 17°C until the hypocotyls had straightened (6 d), then the plants were transferred to three different combinations of DT and NT, all at a daily average temperature of 17°C in separate growth chambers. The effect of DT/NT of 13°C/21°C (negative DIF) was compared to 17°C/17°C (zero DIF) and 21°C/13°C (positive DIF). The DIF treatments started on day 6, when the light was turned on. In each DIF treatment, the height of 18 plants in six pots randomly placed in each growth chamber was measured daily.
After 12 d of DIF treatment, the uppermost 5 to 6 cm of the stem that included the apex was harvested. All leaves were removed, except the smallest ones surrounding the apex. The harvested stem tissue was under active growth and GA1 levels should determine the capacity of these internodes to elongate. In total, 18 randomly chosen seedlings from each chamber were harvested into liquid nitrogen every 4 h during a 48-h period. Upon analyses of GAs and transcripts of the GA biosynthetic pathway, each sample, containing material from 18 plants, was homogenized in liquid nitrogen and kept at −80°C until use in the analyses. In total, 12 samples were harvested during a 48-h period in each DIF treatment, giving a total of 36 samples. The experiment was repeated once.
Stem Elongation Rate Recordings
For fine-scale recording of stem elongation in each of the two replicate experiments, three plants from each temperature regime were transferred 2 d before the start of the harvest period to separate transducer cabinets with temperature and light conditions as described above. In these chambers, the stem elongation rate was continuously measured every 10 s for 2 d according to Torre and Moe (1998) by an angular displacement transducer, series 604 (Trans-Tec, Ellington, CT) connected to a data logger, type CR10-AM416 (Campbell Scientific, Shepshed, Loughborough, UK). The water vapor deficit could not be precisely controlled in these chambers and relative humidity varied from 45% to 65%. These plants were not harvested for further analyses.
Analyses of Transcripts of GA Biosynthesis Genes
In total, 72 samples from the two independent time course experiments were analyzed. mRNA was extracted from 150 to 200 mg of homogenized tissue per sample using Dynal beads (Dynal Beads kit 610.12; Dynal Biotech, Oslo). Any DNA was removed with DNA-free (Ambion, Austin, TX). Concentration and integrity of the mRNA were analyzed with an Agilent 2100 bioanalyzer (Agilent Technologies, Palo Alto, CA). Ribosomal RNA contamination was subtracted before a total of 300 ng mRNA from each sample was reverse transcribed using TaqMan reverse transcription reagents (PE-Applied Biosystems, Foster City, CA).
Primers and gene-specific probes (TAMRA probes; Applied Biosystems) were designed using Primer Express 1.5 software (Applied Biosystems). Primers and probes are listed in Table II. Transcript levels were analyzed using a real-time PCR machine (ABI Prism 7700 sequence detection system; Applied Biosystems). All chemicals used in the PCR reactions followed the recommendations as specified in “The PCR Master Mix Protocol” (part no. 4304449 Rev. C; Applied Biosystems). However, instead of using a 50-μL reaction volume in each tube, we used a 25-μL reaction volume. The following primer concentrations were used: actin, 150 nm forward and reverse; α-tubulin, 150 nm forward and reverse; LS, LH, NA, PsGA3ox1, PsGA2ox1, and PsGA2ox2, 900 nm forward and 900 nm reverse; PsGA20ox1, 900 forward and 300 reverse.
Table II.
The genes, their GenBank accession numbers, and the primers and the probes (TAMRA; Applied Biosystems) used for the real-time RT-PCR analysis of mRNA levels from genes of the late step of the GA biosynthesis in pea cv Torsdag
F, Forward primer sequence; R, reverse primer sequence; P, probe sequence.
| Gene (GenBank Association) | Primers and Probe Sequences |
|---|---|
| LS (AY245442) | F: TTATTTGAACATATTTGGGTGGTTGA |
| R: CAATCTTTGATCTCATGTCGAAAAA | |
| P: CGTCTCGAACGCCTTGGAATATCTCGA | |
| LH (U63652) | F: TGGATAGCAACTTGTGGGAAAA |
| R: CCGCTTGGGCATATTTCTCAT | |
| P: CCAGACCAGTGGATCCCAGAGAGATTTCTT | |
| NA (AF537321) | F: CTTAATCATGGAGTTAGAGCTATGCAA |
| R: TTCCTAGCCTTGAGCGCTTTA | |
| P: TCAATGTTCCTGGATTTGCATACT | |
| PsGA20ox1 (U70471) | F: CATTCCATTAGGCCAAATTTCAAT |
| R: TGCCCTATGTAAACAACTCTTGTATCTC | |
| P: CAATATTGGTGACACCTTCATGGCTCTTTCA | |
| PsGA3ox1 (AF001219) | F: CATTCCATTAGGCCAAATTTCAAT |
| R: ATGTTCCTGCTAACTTTTTCATGGT | |
| P: TCATCATATTGCACGACAATATCACAGAATCTGG | |
| PsGA2ox1 (AF056935) | F: CATAGCTCCTTCTTTATCAATGTT |
| R: TGCCATTTGCCAAAACTCTATGT | |
| P: ACTTTTGAACCTCCCATTAGTCATAACCTGAAGA | |
| PsGA2ox2 (AF100954) | F: GGTTGATAAGCCCGTTATCGAA |
| R: GGCCCATGTAAAGGGCCTATAT | |
| P: TGGTGACGGCCCATAGCCCATG | |
| PsACTIN (U81049) | F: ACACTGTCCCTATCTACGAGGGTTA |
| R: CGCGACCAGCCAGATCA | |
| P: CCCTTCCACATGCCATCCTTCGTC | |
| Psα-TUBULIN (U12589) | F: TGAGGGAGTGCATTTCGATTC |
| R: AGCTCCCAGCAGGCGTTT | |
| P: CATCGGTCAAGCCGGTATCCAGGTC |
Relative mRNA levels were determined using separate tubes and the comparative Ct method for LS, LH, NA, PsGA20ox1, PsGA3ox1, and PsGA2ox1 and the relative standard curve method for PsGA2ox2 according to the User Bulletin 2 (ABI PRISM sequence detection system; PE-Applied Biosystems). The PCR amplification of PsGA2ox2 cDNA was slightly less effective than the other genes (on average, it took 3.7 cycles to increase the amplicon 10-fold, as compared to 3.3–3.4 cycles for the other genes). To avoid overestimation of the amplitudes in mRNA levels within and between treatments, the relative standard curve method was used in estimating mRNA levels of PsGA2ox2. Actin, as well as α-tubulin, was tested as endogenous reference genes since the expression of both genes has been shown to be relatively stable under different environmental conditions, such as varying temperatures (Chu et al., 1993). The relative values of steady-state gene expression in all figures were normalized to actin and the lowest mRNA level for each gene (which is set to 1).
Analyses of GAs
In the quantitative analyses of GAs, we used plant material from which mRNA extractions were also performed. In total, 72 samples from two independent experiments were analyzed. The samples were extracted at 4°C in 75 mL of cold methanol containing 0.02% (w/v) disodium diethyl-dithiocarbamate as an antioxidant. [17, 17-2H]GA44, [17, 17-2H]GA53, [17, 17-2H]GA19, [17, 17-2H]GA20, [17, 17-2H]GA29, [17, 17-2H]GA1, [17, 17-2H]GA8 (L.N. Mander, Australian National University, Canberra, Australia), and [13C6]-indole-3-acetic acid (Cambridge Isotope Laboratories, Woburn, MA) were used as internal standards, and the ratio of internal standards to endogenous GA was kept near 1:1. Purification of samples and gas chromatography-mass spectrometry-selected ion monitoring analysis were performed according to Olsen et al. (1994, 1995) and Olsen and Junttila (2002). This included partition against ethyl acetate, use of QAE-Sephadex A25 (Pharmacia, Uppsala) anion-exchange columns combined with 0.5 g Sep-pak Vac C18 cartridges (Varian, Harbor City, CA), methylation and purification on 0.1-g bond elute aminopropyl cartridges (Varian), followed by reverse-phase HPLC. Combined HPLC fractions were trimethyl silylated and subjected to gas chromatography-selected ion monitoring analysis.
Acknowledgments
We thank Marit Siira for technical assistance, Professor John Ross for providing us with seeds from the sln mutant, and Dr. Peter Hedden for useful comments on the manuscript.
This work was supported by the Norwegian Research Council (grant no. 140322/110).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.063149.
References
- Ait-Ali T, Frances S, Weller JL, Reid JB, Kendrick RE, Kamiya Y (1999) Regulation of gibberellin 20-oxidase and gibberellin 3β-hydroxylase transcript accumulation during de-etiolation of pea seedlings. Plant Physiol 121: 783–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ait-Ali T, Swain SM, Reid JB, Sun TP, Kamiya Y (1997) The LS locus of pea encodes the gibberellin biosynthesis enzyme ent-kaurene synthase A. Plant J 11: 443–454 [DOI] [PubMed] [Google Scholar]
- Chu B, Snustad DP, Carter JV (1993) Alteration of β-tubulin gene expression during low-temperature exposure in leaves of Arabidopsis thaliana. Plant Physiol 103: 371–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson SE, Elliott RC, Helliwell CA, Poole AT, Reid JB (2003) The pea gene NA encodes ent-kaurenoic acid oxidase. Plant Physiol 131: 335–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson SE, Smith JJ, Helliwell CA, Poole AT, Reid JB (2004) The pea gene LH encodes ent-kaurene oxidase. Plant Physiol 134: 1123–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott RC, Ross JJ, Smith JL, Lester DR, Reid JB (2001) Feed-forward regulation of gibberellin deactivation in pea. J Plant Growth Regul 20: 87–94 [Google Scholar]
- Erwin JE, Heins RD (1995) Thermomorphogenic responses in stem and leaf development. Hortscience 30: 940–949 [Google Scholar]
- Erwin JE, Heins RD, Karlsson MG (1989) Thermomorphogenesis in Lilium longiflorum. Am J Bot 76: 47–52 [Google Scholar]
- Foster KR, Morgan PW (1995) Genetic regulation of development in Sorghum bicolor. IX. The ma3R allele disrupts diurnal control of gibberellin biosynthesis. Plant Physiol 108: 337–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Martinez JL, Gil J (2002) Light regulation of gibberellin biosynthesis and mode of action. J Plant Growth Regul 20: 354–368 [DOI] [PubMed] [Google Scholar]
- Garcia-Martinez JL, Lopez-Diaz I, Sanchez-Beltran MJ, Phillips AL, Ward DA, Gaskin P, Hedden P (1997) Isolation and transcript analysis of gibberellin 20-oxidase genes in pea and bean in relation to fruit development. Plant Mol Biol 33: 1073–1084 [DOI] [PubMed] [Google Scholar]
- Gil J, Garcia-Martinez JL (2000) Light regulation of gibberellin A1 content and expression of genes coding for GA 20-oxidase and GA 3β-hydroxylase in etiolated pea seedlings. Physiol Plant 108: 223–229 [Google Scholar]
- Grindal G, Ernstsen A, Reid JB, Junttila O, Lindgard B, Moe R (1998. a) Endogenous gibberellin A(1) levels control thermoperiodic stem elongation in Pisum sativum? Physiol Plant 102: 523–531 [Google Scholar]
- Grindal G, Junttila O, Reid JB, Moe R (1998. b) The response to gibberellin in Pisum sativum grown under alternating day and night temperature. J Plant Growth Regul 17: 161–167 [Google Scholar]
- Hedden P, Phillips AL (2000) Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci 5: 523–530 [DOI] [PubMed] [Google Scholar]
- Ihlebekk H, Eilertsen S, Junttila O, Grindal G, Moe R (1995) Control of plant height in Campanula by temperature alternations, involvement of GAs. Acta Hortic 394: 347–355 [Google Scholar]
- Ingram TJ, Reid JB, Murfet IC, Gaskin P, Willis CL, Macmillan J (1984) Internode length in Pisum—the Le gene controls 3β-hydroxylation of gibberellin A20 to gibberellin A1. Planta 160: 455–463 [DOI] [PubMed] [Google Scholar]
- Jackson SD, James PE, Carrera E, Prat S, Thomas B (2000) Regulation of transcript levels of a potato gibberellin 20-oxidase gene by light and phytochrome B. Plant Physiol 124: 423–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen E, Eilertsen S, Ernsten A, Juntilla O, Moe R (1996) Thermoperiodic control of stem elongation and endogenous gibberellins in Campanula isophylla. J Plant Growth Regul 5: 167–171 [Google Scholar]
- Lester DR, Ross JJ, Davies PJ, Reid JB (1997) Mendel's stem length gene (Le) encodes a gibberellin 3β-hydroxylase. Plant Cell 9: 1435–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester DR, Ross JJ, Smith JJ, Elliott RC, Reid JB (1999) Gibberellin 2-oxidation and the SLN gene of Pisum sativum. Plant J 19: 65–73 [DOI] [PubMed] [Google Scholar]
- Lopezjuez E, Kobayashi M, Sakurai A, Kamiya Y, Kendrick RE (1995) Phytochrome, gibberellins, and hypocotyl growth. A study using the cucumber (Cucumis sativus L.) long hypocotyl mutant. Plant Physiol 107: 131–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DN, Proebsting WM, Hedden P (1997) Mendel's dwarfing gene: cDNAs from the Le alleles and function of the expressed proteins. Proc Natl Acad Sci USA 94: 8907–8911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DN, Proebsting WM, Hedden P (1999) The SLENDER gene of pea encodes a gibberellin 2-oxidase. Plant Physiol 121: 775–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moe R (1990) Effect of day and night temperature alternations and of plant-growth regulators on stem elongation and flowering of the long-day plant Campanula isophylla Moretti. Sci Hortic 43: 291–305 [Google Scholar]
- Moe R, Heins RD (2000) Thermo- and photomorphogenesis in plants. In E Stroemme, ed, Advances in Floriculture Research, Report no. 6/2000. Agricultural University of Norway, Aas, Norway, pp 52–64
- Myster J, Moe R (1995) Effect of diurnal temperature alternations on plant morphology in some greenhouse crops—a mini review. Sci Hortic 62: 205–215 [Google Scholar]
- O'Neill DP, Ross JJ, Reid JB (2000) Changes in gibberellin A1 levels and response during de-etiolation of pea seedlings. Plant Physiol 124: 805–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JE, Jensen E, Junttila O, Moritz T (1995) Photoperiodic control of endogenous gibberellins in seedlings of Salix pentandra. Physiol Plant 93: 639–644 [Google Scholar]
- Olsen JE, Junttila O (2002) Far red end-of-day treatment restores wild type-like plant length in hybrid aspen overexpressing phytochrome A. Physiol Plant 115: 448–457 [DOI] [PubMed] [Google Scholar]
- Olsen JE, Moritz T, Jensen E, Junttila O (1994) Comparison of endogenous gibberellins in roots and shoots of elongating Salix pentandra sedlings. Physiol Plant 90: 378–381 [Google Scholar]
- Olszewski N, Sun TP, Gubler F (2002) Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell 14: S61–S80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts WC, Reid JB, Murfet IC (1985) Internode length in Pisum. Gibberellins and the slender phenotype. Physiol Plant 63: 357–364 [Google Scholar]
- Reed JW, Foster KR, Morgan PW, Chory J (1996) Phytochrome B affects responsiveness to gibberellins in Arabidopsis. Plant Physiol 112: 337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid JB, Botwright NA, Smith JJ, O'Neill DP, Kerckhoffs LHJ (2002) Control of gibberellin levels and gene expression during de-etiolation in pea. Plant Physiol 128: 734–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid JB, Ross JJ, Swain SM (1992) Internode length in Pisum—a new, slender mutant with elevated levels of C19 gibberellins. Planta 188: 462–467 [DOI] [PubMed] [Google Scholar]
- Ross JJ, Davidson SE, Wolbang CM, Bayly-Stark E, Smith JJ, Reid JB (2003) Developmental regulation of the gibberellin pathway in pea shoots. Funct Plant Biol 30: 83–89 [DOI] [PubMed] [Google Scholar]
- Ross JJ, Reid JB, Gaskin P, Macmillan J (1989) Internode length in Pisum—estimation of GA1 levels in genotypes Le, le and led. Physiol Plant 76: 173–176 [Google Scholar]
- Talon M, Zeevaart JAD, Gage DA (1991) Identification of gibberellins in spinach and effects of light and darkness on their levels. Plant Physiol 97: 1521–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangerås H (1979) Modifying effects of ancymidol and gibberellins on temperature induced elongation in Fuchsia x hybrida. Acta Hortic 91: 411–417 [Google Scholar]
- Thomas SG, Phillips AL, Hedden P (1999) Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc Natl Acad Sci USA 96: 4698–4703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkinson CL, Lyndon RF, Arnold GM, Lenton JR (1997) The effects of temperature and the Rht3 dwarfing gene on growth, cell extension, and gibberellin content and responsiveness in the wheat leaf. J Exp Bot 48: 963–970 [Google Scholar]
- Torre S, Moe R (1998) Temperature, DIF and photoperiod effects on the rhythm and rate of stem elongation in Campanula isophylla Moretti. Sci Hortic 72: 123–133 [Google Scholar]
- Weller JL, Ross JJ, Reid JB (1994) Gibberellins and phytochrome regulation of stem elongation in pea. Planta 192: 489–496 [Google Scholar]
- Went FW (1944) Plant growth under controlled conditions. II. Thermoperiodicity in growth and fruiting of tomato. Am J Bot 31: 135–150 [Google Scholar]
- Zieslin N, Tsujita MJ (1988) Regulation of stem elongation of lilies by temperature and the effect of gibberellin. Sci Hortic 37: 165–169 [Google Scholar]