Abstract
Transcriptional and posttranscriptional regulation are well-established mechanisms for circadian gene expression. Among the latter, differential messenger RNA (mRNA) stability has been hypothesized to control gene expression in response to the clock. However, direct proof that the rate of mRNA turnover can be regulated by the clock is lacking. Previous microarray expression data for unstable mRNAs in Arabidopsis (Arabidopsis thaliana) revealed that mRNA instability is associated with a group of genes controlled by the circadian clock. Here, we show that CCR-LIKE (CCL) and SENESCENCE ASSOCIATED GENE 1 transcripts are differentially regulated at the level of mRNA stability at different times of day. In addition, the changes in CCL mRNA stability continue under free-running conditions, indicating that it is controlled by the Arabidopsis circadian clock. Furthermore, we show that these mRNAs are targets of the mRNA degradation pathway mediated by the downstream (DST) instability determinant. Disruption of the DST-mediated decay pathway in the dst1 mutant leads to aberrant circadian mRNA oscillations that correlate with alterations of the half-life of CCL mRNA relative to parental plants in the morning and afternoon. That this is due to an effect on the circadian control is evidenced by mRNA decay experiments carried out in continuous light. Finally, we show that the defects exhibited by dst mutants are reflected by an impact on circadian regulation at the whole plant level. Together, these results demonstrate that regulation of mRNA stability is important for clock-controlled expression of specific genes in Arabidopsis. Moreover, these data uncover a connection between circadian rhythms and a sequence-specific mRNA decay pathway.
Plants, like many other organisms, have internal clocks that command biological rhythms with a period close to 24 h. These rhythms provide selective advantages because they allow anticipation of the daily changes in environmental conditions (Ouyang et al., 1998; Green et al., 2002). Examples of processes that can exhibit circadian rhythms in plants are leaf movement, hypocotyl elongation, stomatal opening, and floral induction (for review, see McClung, 2001). At the molecular level, DNA microarray experiments have shown that 2% to 6% of Arabidopsis (Arabidopsis thaliana) mRNAs can oscillate (Harmer et al., 2000; Schaffer et al., 2001). For Arabidopsis and other circadian clocks studied to date, the core circadian oscillator is comprised of transcriptional feedback loops. The underlying master oscillator in Arabidopsis is believed to include the LATE ELONGATED HYPOCOTYL (LHY; Schaffer et al., 1998), the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1; Wang and Tobin, 1998), and the TIMING OF CAB EXPRESSION 1 (TOC1; Millar et al., 1995; Strayer et al., 2000) genes. Similar to what is observed in other systems (Glossop et al., 1999), the clock components form a regulatory loop in which TOC1 positively regulates LHY and CCA1 gene expression, and LHY and CCA1 proteins in turn repress TOC1 gene expression (Alabadí et al., 2001). This loop then outputs cyclic positive or negative expression commands to the downstream targets.
It is clear that transcriptional control plays an important role in the circadian expression of LHY, CCA1, TOC1, and other clock-controlled genes (CCGs; for review, see Harmer et al., 2001). For example, recently, a novel sequence element, termed the evening element, was identified in the promoter region of 31 Arabidopsis circadian genes, 30 of which peaked at the end of the subjective day (Harmer et al., 2000). This element was shown to be important for cycling of the COLD CIRCADIAN RHYTHM AND RNA BINDING 2 (CCR2) and TOC1 promoter activities and has been proposed as the binding site for LHY and CCA1 transcription factors (Alabadí et al., 2001). In addition to the body of evidence for circadian control of transcription, results in various systems suggest that posttranscriptional regulatory mechanisms at the RNA level are also required for clock function (Edery, 1999; Harmer et al., 2001).
Perhaps the best-characterized example corresponds to the period (per) gene, one of the components of the central oscillator in Drosophila melanogaster (Panda et al., 2002), where comparison of per transcription rates and mRNA levels implicated a temporal regulation of mRNA half-life (So and Rosbash, 1997). Regulatory elements located in both promoter and transcribed regions of the per gene are necessary to replicate wild-type cycling (So and Rosbash, 1997; Stanewsky et al., 1997). The functional significance of this posttranscriptional level of regulation has been well documented (Stanewsky, 2002). A per-transgene completely devoid of its promoter sequences but including parts of the first intron is capable of restoring rhythmic behavior in per01 mutant flies solely through posttranscriptional cycling of its mRNA (Frisch et al., 1994; So and Rosbash, 1997). Moreover, disruption of the 3′ untranslated region of the per gene affects circadian behavioral rhythms in Drosophila (Chen et al., 1998). Hence, posttranscriptional mechanisms are not only important for normal circadian gene expression but also for proper function of per. Posttranscriptional regulation of per is thought to be involved in the adaptation of Drosophila to cold as well. An alternatively spliced form of the per transcript is generated at lower temperature, causing an advance in the phases of both the mRNA and protein cycles (Majercak et al., 1999). Another example is the Crg-1 gene, for which the lack of cycling transcriptional activity in run-on experiments suggests that posttranscriptional mechanisms could contribute to the circadian expression of the gene (So and Rosbash, 1997). This indicates that posttranscriptional control might be of significance for the expression of additional CCGs in Drosophila.
Posttranscriptional mechanisms affecting mRNA levels have been invoked to explain circadian oscillation of plant genes as well. Transcript stability has been hypothesized to be partially responsible for the oscillations of the CHLOROPHYLL A/B BINDING 1 mRNA in Arabidopsis (Millar and Kay, 1991). Increased transcript stability has also been implicated in the accumulation of high steady-state levels of CATALASE 3 mRNA in continuous dark (Zhong et al., 1997). Furthermore, based on nuclear run-on assays, the cycling of NITRATE REDUCTASE 2 (NIA2) is thought to occur through posttranscriptional regulation (Pilgrim et al., 1993). In rice (Oryza sativa), the circadian regulation of CATALASE A expression has been postulated to be at the level of pre-mRNA stability (Iwamoto et al., 2000). Finally, two clock-regulated RNA-binding proteins have been reported in Arabidopsis, at least one of which has been proposed to be a slave oscillator downstream of the central clock (Staiger, 2001).
Although no examples of changes in mRNA stability regulated by the circadian clock have been reported, recent evidence suggests a more prominent role of control of mRNA stability in CCG expression in Arabidopsis. Microarray analysis has shown that a subset of unstable transcripts in Arabidopsis is controlled by the circadian clock (Gutierrez et al., 2002). Additional microarray studies led to the finding that an unexpectedly high percentage of transcripts that were changed in abundance in a mutant deficient in downstream element (DST)-mediated decay, dst1, were circadian regulated, indicating that the biological significance of the DST-mediated mRNA decay pathway may be associated with the circadian clock (Pérez-Amador et al., 2001). Together, these data suggest that rapid mRNA turnover, perhaps at specific times of the day, might be important for the specific circadian oscillation of CCGs in Arabidopsis. To explore the relationship between mRNA stability and circadian gene expression, we analyzed the stability of selected CCGs that have unstable transcripts in Arabidopsis plants at different times during the day. We present direct evidence that circadian regulation of mRNA stability occurs in Arabidopsis and is altered in the dst1 mutant. Furthermore, our data indicate that the control of mRNA stability has a significant effect on circadian processes at the whole plant level.
RESULTS
Stability of CCR-LIKE and SENESCENCE ASSOCIATED GENE 1 mRNAs Changes during the Day
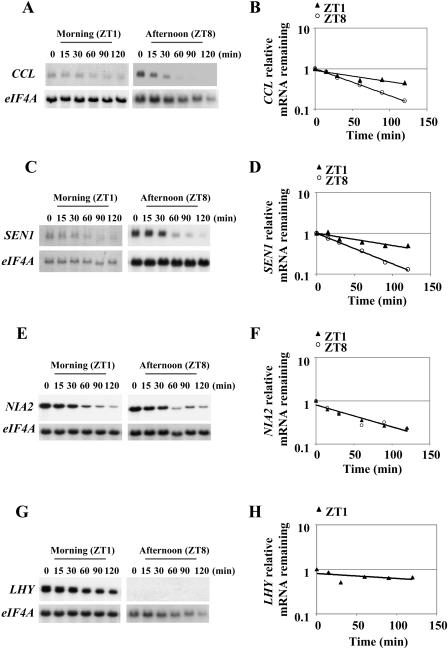
Microarray experiments showed that a group of Arabidopsis genes that encode highly unstable mRNAs is also regulated by the circadian clock (Gutierrez et al., 2002). To determine whether regulation of mRNA stability contributes to the oscillatory expression of these genes, we measured mRNA decay rates at two times during the day for the clock-controlled CCR-LIKE (CCL) gene (At3g26740; Harmer et al., 2000; Schaffer et al., 2001), the SENESCENCE ASSOCIATED GENE 1 (SEN1; At4g35770), the NIA2 gene (At1g37130; Pilgrim et al., 1993), and the light-regulated homeodomain transcription factor ATHB6 (At2g22430). The well-characterized clock gene LHY was used to control for timing of the experiments. Two-week-old plants grown in 16-h-light/8-h-darkness cycles (16/8 LD) were used for time-course experiments following transcriptional inhibition 1 h after dawn (zeitgeber time 1 [ZT1]) as detailed in “Materials and Methods.” Another set of time-course experiments was performed 8 h after dawn (ZT8). As shown in Figure 1, A and B, CCL mRNA stability changed during the day. The transcript for this gene was significantly more stable in the morning, ZT1, as compared to the afternoon (ZT8; P-value < 0.0001). Similarly, SEN1 mRNA turnover was differentially regulated during the day as well (Fig. 1, C and D), with the transcript being more stable in the morning as compared to the afternoon (P-value = 0.0053). In contrast, no evidence was found that the turnover rates of the cycling NIA2 mRNA changes during the day (Fig. 1, E and F; P-value = 0.79). Similarly, no evidence was found that the mRNA stability of the diurnally regulated ATHB6 transcript changes during the day (P-value = 0.81; data not shown). The mRNA for these genes decayed at similar rates when measured in the morning or in the afternoon under the conditions tested. LHY mRNA has a circadian expression pattern with a peak soon after dawn and with very low levels throughout most of the day (Schaffer et al., 1998). As expected, LHY was easily detectable in the morning and was near background levels in the afternoon (Fig. 1, G and H). LHY mRNA was relatively stable in the morning (t1/2 > 130 min; Fig. 1G), but low levels precluded accurate determination of its half-life in the afternoon experiments. These data indicate that the half-life changes observed are not the result of differences in the global cellular mRNA turnover rates in the morning and afternoon. Furthermore, they indicate that regulation at the level of mRNA stability is not a general property but rather specific to some CCGs in Arabidopsis.
Figure 1.
CCL and SEN1 but not NIA2 mRNA stability is regulated during the day. Representative northern-blot analysis of half-life experiments carried out in the morning (1 h after dawn; ZT1) and in the afternoon (8 h after dawn; ZT8) for CCL (A), SEN1 (C), NIA2 (E), and LHY (G) mRNAs. Samples consisted of 10 μg of total RNA isolated from the indicated time points. The signal for the stable eIF4A transcript was used as a reference for equal loading. Quantitation of the decrease in abundance for CCL (B), SEN1 (D), NIA2 (F), and LHY (H) mRNAs (ZT1 only). Average half-life and se values were calculated for CCL (ZT1 = 175 ± 65 min; ZT8 = 35 ± 5 min), SEN1 (ZT1 = 99 ± 13 min; ZT8 = 37 ± 3 min), and NIA2 (ZT1 = 49 ± 5 min; ZT8 = 43 ± 8 min) from at least two independent half-life experiments. The graph for the quantitation of half-life corresponds to the RNA gel blot shown.
CCL mRNA Stability Changes Are Dictated by the Circadian Clock
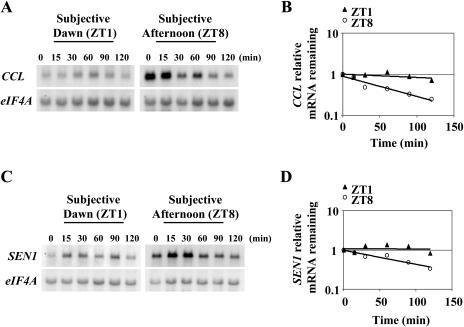
Two possible mechanisms could explain the changes in mRNA stability observed during the day for CCL and SEN1 genes. Signaling pathways activated by the changes in light patterns during the normal day/night cycle could be responsible. Alternatively, the circadian clock could promote the change. To discriminate between these two possibilities, mRNA half-lives were determined under free-running conditions, i.e. in the absence of light/dark changes. Arabidopsis plants were grown for 12 d in 16/8 LD cycles. On the morning of the 12th day, they were transferred to continuous light. Half-lives were then determined 1 (ZT1) and 8 (ZT8) h after the subjective dawn of the 14th day. As shown in Figure 2A, regulation of CCL mRNA stability occurs under continuous light conditions in the same manner as seen previously in the day/night cycles (Fig. 1B). The transcript was significantly more stable in the subjective morning (ZT1) as compared to the subjective afternoon (ZT8; P-value = 0.0004), indicating that the change in mRNA turnover is controlled by the Arabidopsis circadian clock. These data suggest that posttranscriptional control of mRNA stability is important for the circadian expression of CCL gene. A change in the stability of SEN1 mRNA in the circadian experiments was also observed in each time course as exemplified in Figure 2B but was not as great as for CCL and cannot be argued for on a statistical basis (P-value = 0.28). However, it is possible, and perhaps likely, that circadian regulation of SEN1 mRNA stability also occurs in vivo given that we consistently observed a decrease in mRNA half-life in the subjective afternoon. In any event, our results with CCL clearly show that regulation of mRNA stability by the circadian clock occurs in Arabidopsis.
Figure 2.
CCL and SEN1 mRNA stability is regulated by the circadian clock. Half-life analysis was carried out as mentioned in “Materials and Methods” in the subjective morning, 1 h after subjective dawn (ZT1), and in the subjective afternoon, 8 h after subjective dawn (ZT8). Representative northern-blot analysis of half-life experiments for CCL (A) and SEN1 (C). Representative quantitation of the decrease in mRNA abundance for CCL (B) and SEN1 (D) mRNAs. Average half-life and se values were calculated for CCL (ZT1 = 410 ± 150 min; ZT8 = 76 ± 18 min) and SEN1 (ZT1 = 380 ± 110 min; ZT8 = 95 ± 12 min) from at least two independent half-life experiments. The graph for the quantitation of half-life corresponds to the RNA gel blot shown.
Decay of CCL and SEN1 mRNAs Is Altered in the dst1 Mutant at Different Times of Day
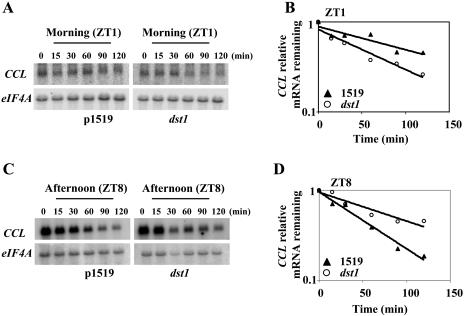
Recently it was shown that CCL and SEN1 mRNA levels are altered in dst1 (Pérez-Amador et al., 2001), an Arabidopsis mutant that exhibits defective mRNA degradation mediated by the DST instability sequence (Johnson et al., 2000). Because SEN1 and CCL contain DST elements in their 3′ untranslated regions (UTRs), it seemed likely that they might be degraded by the DST-mediated decay pathway. To examine this, mRNA turnover rates were measured in the dst1 mutant and 1519 parental lines (analogous to wild type; Johnson et al., 2000) as described previously. As shown in Figure 3A, northern-blot analysis of time-course experiments following transcriptional inhibition indicated that CCL mRNA decayed faster in the mutant as compared to 1519 parental plants when measured 1 h after dawn (P-value = 0.0003). Similarly, the SEN1 transcript was also degraded more rapidly in the mutant as compared to the parental plants (P-value <0.0019; Supplemental Fig. 1A). These data are consistent with the microarray and northern-blot experiments of Pérez-Amador et al. (2001), which showed that both transcripts were diminished in dst1 in samples harvested in the morning.
Figure 3.
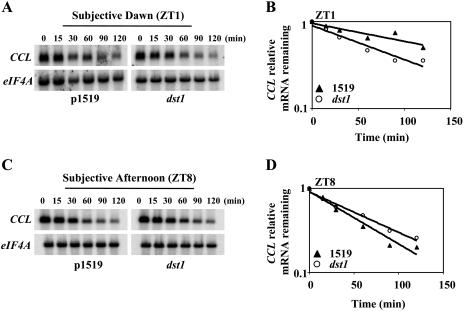
Regulation of CCL mRNA stability is altered in the dst1 mutant during the day. Half-life analysis was carried out as mentioned in “Materials and Methods.” Representative northern-blot analysis of half-life experiments for CCL in the morning (A) and in the afternoon (C). Representative quantitation of the decrease in mRNA abundance for CCL in the morning (B) and in the afternoon (D), respectively. Average half-life and se values were calculated at ZT1 and ZT8 for parental 1519 and dst1 mutant plants, respectively, from at least two independent half-life experiments (B) 106 ± 11 min and 62 ± 5 min (B); 48 ± 0 min and 86 ± 9 min (D). The graph for the quantitation of half-life corresponds to the RNA gel blot shown.
Interestingly, when mRNA decay rates for CCL were measured in the afternoon (ZT8), the effects of the mutant were reversed (Fig. 3B) compared to the morning (Fig. 3A). Figure 3B shows that the CCL mRNA was more stabilized in the afternoon in the mutant relative to the parental plants (P-value < 0.0001). A similar effect was observed for SEN1 mRNA decay kinetics with the transcript being more stable in the afternoon relative to the parental plants (P-value = 0.0061) in contrast to the morning (Supplemental Fig. 1B). Statistical analysis, as indicated above, showed that the observed differences in half-lives seen for CCL and SEN1 mRNAs in the mutant and parental plants are significant. These results suggest that normal DST1 function is required for the proper timing of degradation of CCL and SEN1 transcripts under diurnal conditions.
Disruption of the DST-Mediated mRNA Decay Pathway Leads to Altered CCL Diurnal Oscillation
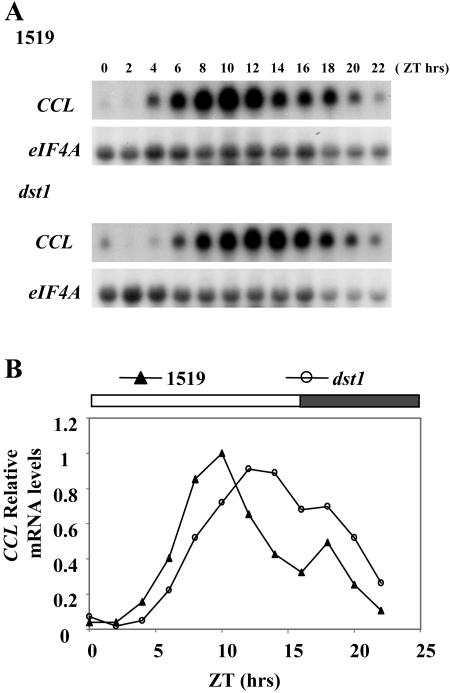
It has been theoretically calculated that a cycling mRNA with a longer half-life will show a lagging phase when compared with an mRNA with a shorter half-life, assuming that the transcription rates are similar (Wuarin et al., 1992). To test whether the dst1 mutant also alters diurnal oscillation of the CCL transcript, mRNA levels were examined throughout the day in the dst1 mutant and 1519 parental lines (analogous to wild type; Johnson et al., 2000). Two-week-old Arabidopsis plants grown on 16/8 LD cycles were harvested every 2 h after dawn. Total RNA was isolated, and mRNA levels were examined by northern blotting. As shown in Figure 4, CCL peaked late during the day in the parental 1519 line. In contrast, in the dst1 mutant, CCL mRNA started to accumulate later and peaked at least 2 h later than in the 1519 line (Fig. 4). This lagging nature of the expression peak in the mutant could be explained by the increased stability of the CCL transcript in the mutant relative to parental plants in the afternoon (Fig. 3B) leading to the slower decline in mRNA levels at later time points (Fig. 4). The impact of the dst1 mutation on SEN1 mRNA oscillation was less dramatic, but a small reproducible change in the diurnal pattern of expression was observed (data not shown). These data indicate that the dst1 mutation disrupts the normal oscillation of CCL and SEN1 mRNAs.
Figure 4.
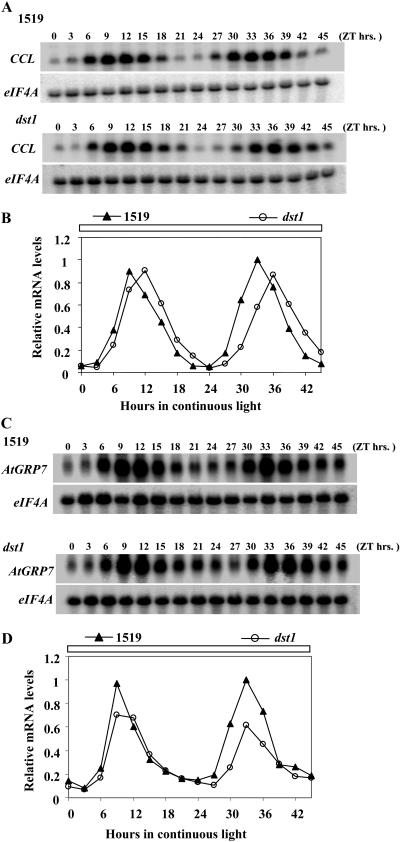
Diurnal oscillation of CCL mRNA is altered in the dst1 mutant. A, Representative northern-blot analysis of time courses performed throughout an entire day for CCL mRNA in dst1 mutant and parental 1519 plants. Samples consisted of 10 μg of total RNA isolated from the indicated times of the day after dawn (ZT = 0). The signal for eIF4A was used as a reference for equal loading. B, Quantitation of mRNA levels for CCL mRNA. All values are representative of at least two independent experiments and are made relative to the highest mRNA accumulation in either of the two genetic backgrounds.
dst1 Affects Circadian Control of mRNA Stability
We hypothesized that the stabilization of CCL mRNA caused by dst1 in the afternoon under diurnal conditions would also occur in the subjective afternoon under free-running conditions. To test this hypothesis, mRNA half-lives were measured in Arabidopsis seedlings that were transferred to continuous light for 2 d. Transcription was inhibited 1 and 8 h after the subjective morning, and mRNA decay rates were monitored thereafter. The impact of dst1 was recapitulated for CCL in continuous light with the transcript being more unstable in dst1 in the subjective morning (Fig. 5A; P-value < 0.0001) and more stable in dst1 in the subjective afternoon (Fig. 5B; P-value = 0.0171) relative to the parental plants. Also, relative to itself, the dst1 mutant does not show a dramatic difference in mRNA decay rates measured in the subjective morning versus the subjective afternoon in contrast to the parental plants. Even though some dampening in mRNA half-lives was seen under circadian conditions relative to the diurnal conditions, the differences in mRNA decay rates between the mutant and parental plants were statistically significant. These results suggest that normal DST1 function is required for normal circadian control of CCL mRNA stability.
Figure 5.
Circadian regulation of CCL mRNA stability is altered in the dst1 mutant. Half-life analysis was carried out as mentioned in “Materials and Methods” in the subjective morning, 1 h after subjective dawn (ZT1), and in the subjective afternoon, 8 h after subjective dawn (ZT8). Representative northern-blot analysis of half-life experiments for CCL in the subjective morning (A) and in the subjective afternoon (C). Representative mRNA decay curves for CCL in the subjective morning (B) and in the subjective afternoon (D), respectively, are shown. Average half-life and se values were calculated at ZT1 and ZT8 for parental 1519 and dst1 mutant plants, respectively, from at least two independent half-life experiments were 129 ± 15 min and 70 ± 7 min (B); 51 ± 3 min and 64 ± 2 min (D). The graph for the quantitation of half-life corresponds to the RNA gel blot shown.
Circadian Oscillation of CCL mRNA Is Altered in the dst1 Mutant
To evaluate the impact of the dst1 mutation on the circadian oscillation of CCL, mRNA levels were examined under free-running conditions. Arabidopsis seedlings were grown for 12 d in 16/8 LD cycles and on the morning of the 12th day transferred to continuous light. Seedling tissue for mRNA isolation was harvested every 3 h starting on the morning of the 12th day (ZT0) up to the 14th day (ZT45). As shown in Figure 6, A and B, CCL mRNA peaked approximately 3 h later in the dst1 mutant than in the parental plants. This effect of the mRNA peak lagging in dst1 was apparent on both days in constant light. The oscillation pattern under free-running conditions was comparable to that seen previously under diurnal conditions (Fig. 4, A and B), so circadian control appears to be the primary component. In order to gain insight into the effect of dst1 on general CCG expression, circadian oscillation of GLYCINE RICH PROTEIN 7 (AtGRP7)/CCR2, which functions downstream of the master clock (Staiger, 2001), was tested. The oscillation phase of AtGRP7 mRNA was unchanged in the dst1 mutant compared to the parental plants (Fig. 6, C and D). Taken together, this indicates that the circadian oscillation of a subset of CCGs is dependent on DST1 function.
Figure 6.
Circadian oscillation of CCL, but not AtGRP7, mRNA is altered in the dst1 mutant. A and C, Representative northern-blot analysis of time courses performed throughout 2 d in continuous light for CCL and AtGRP7 mRNA, respectively, in dst1 mutant and parental 1519 plants. RNA samples were loaded as in Figure 4 from the indicated times of the day after subjective dawn (ZT = 0). B and D, Quantitation of mRNA levels for CCL and AtGRP7, respectively. All values are representative of at least two independent experiments and are made relative to the highest mRNA accumulation in either of the two genetic backgrounds.
Impact at the Whole Plant Level: A Classical Circadian Response Is Altered in dst Mutants
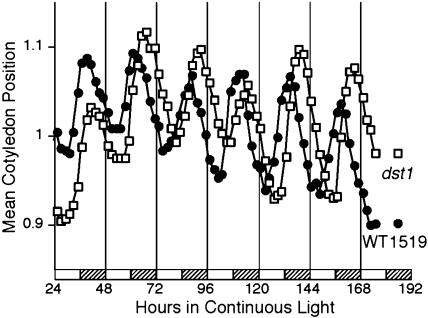
To address whether the link between the DST-mediated decay pathway and the circadian clock impacted Arabidopsis at the whole plant level, cotyledon movement, a classical circadian phenotype, was monitored in the dst mutants. The oscillation in the position of cotyledons can be monitored by video imaging, and this technique has been used to study circadian clock mutants (Millar et al., 1995). Seedlings were grown for 5 d in 12 h light/12 h dark, transferred to 24-well plates, and released into continuous light. Cotyledon movement was then recorded for 7 d. The results correlate very well with the observation that CCGs in dst1 peak slightly later (i.e. phase lagging) at the RNA level (Fig. 6A). Indeed cotyledon movement in the dst1 mutant lags behind that of parental plants (Fig. 7). Interestingly, cotyledon movement in another independently isolated dst mutant (dst2; Johnson et al., 2000) was phase leading as compared to parental plants (Supplemental Fig. 3). dst2 also is known to have an opposite effect on the accumulation of CCL and SEN1 mRNAs as compared to dst1 (Pérez-Amador et al., 2001). In contrast to the reduction of SEN1 and CCL transcripts in dst1, dst2 exhibits higher levels of both transcripts compared to parental plants (Pérez-Amador et al., 2001). Thus, we found a clear parallel between the molecular phenotypes of these mutants and the phenotypes observed at the whole organism level.
Figure 7.
The dst1 mutant displays a lagging circadian phase in cotyledon movement. Seedlings were grown for 5 d in 12 h light/12 h dark, transferred to 24-well plates, and released into continuous light. Cotyledon movement was recorded for 7 d. Each trace is the average of 5 to 14 individual seedlings. The experiment was repeated three times with very similar results. Hatched bars indicate subjective night.
DISCUSSION
Although it has become clear that posttranscriptional mechanisms contribute significantly to CCG expression, prior to this work, the evidence for regulation at the level of mRNA stability by the clock was indirect. In this study, we identified two genes that have different mRNA decay rates at different times of day. For one of these, the CCL gene, this difference was maintained robustly in continuous light, demonstrating that the circadian clock controls the stability of the mRNA. The CCL gene was also interesting because the transcript contains DST-like elements and is diminished and less stable in the sequence-specific mRNA decay mutant dst1. These are the expected characteristics of a transcript that is a direct target of the DST-mediated mRNA decay pathway. More importantly, we observed that the diurnal and circadian control of CCL mRNA stability was altered in dst1 in a manner consistent with the phase lagging nature of the overall circadian oscillation of the transcript. Not only is the CCL RNA phase lagging in the mutant, but a classical circadian response, that of cotyledon movement, is phase lagging as well. These results indicate that DST-mediated mRNA decay can have a significant impact on circadian control.
Potential roles of mRNA stability in circadian control of gene expression had been hypothesized previously in other systems, such as for control of the Drosophila per gene highlighted earlier. Recently, entrainment of the circadian clock in Neurospora crassa was shown to be controlled at least partly by an RNA that is antisense to the transcript encoded by the clock gene frequency (frq; Kramer et al., 2003). Levels of this antisense RNA oscillate in antiphase to that of frq mRNA in the dark and are inducible by light. Mutants with altered regulation of the expression of the antisense RNA exhibit clock phenotypes. Although the exact mechanism of action of this antisense transcript remains to be elucidated, it is plausible that it binds to sense frq RNA and triggers rapid frq mRNA decay via an RNA silencing pathway (Tijsterman et al., 2002). In another recent example, a Xenopus deadenylase, nocturnin, was shown to be expressed in a rhythmic manner (Baggs and Green, 2003). A nocturnin homolog is also present in Arabidopsis (Dupressoir et al., 2001). Since nocturnin is expressed in a circadian fashion, it is possible that this enzyme is responsible for the deadenylation of clock-controlled mRNAs. A potential target is the vasopressin transcript in mammals, where two species of mRNAs with differences in their poly(A) tail length are present at different times of day (Robinson et al., 1988). The implications of these studies, together with our work on the circadian control of CCL mRNA stability, argue that mRNA decay mechanisms likely influence clock responses in a variety of organisms.
An unexpected observation was the differences in mRNA half-lives measured under diurnal versus circadian conditions. For example, in Figure 1, the half-life analysis was carried out in the morning (1 h after dawn; ZT1), while in Figure 2, mRNA half-lives were determined at the same relative time of day but under free-running conditions, i.e. in the absence of light/dark changes. CCL and SEN1 mRNAs were more stable in continuous light as compared to the equivalent times of the day under regular 16/8 LD cycles. It is possible that this moderate stabilization is caused by the circadian clock promoting mRNA degradation, and the clock may be dampened in continuous light. Alternatively, continuous light or the absence of a dark period might in some way slow down the function of the mRNA decay machinery involved in CCL and SEN1 transcript degradation.
SEN1 shares many of the characteristics of CCL, such as circadian regulation (Supplemental Figs. 4 and 5), the presence of DST-like elements, and a transcript that is more stable in the morning than the afternoon in wild-type plants. In dst1, SEN1 mRNA, similar to CCL, was less stable than the parental plants in the morning and more stable than the parental plants in the afternoon. Although we also observed this pattern under circadian conditions (Supplemental Fig. 2), the magnitude of the effect was modest in the subjective morning and not observed in the subjective afternoon. Nevertheless, the trends were similar to that observed for CCL in the subjective morning (Fig. 5A; Supplemental Fig. 2A), and the results with both genes indicate that a functional DST-mediated decay pathway is required for proper circadian and/or diurnal regulation of these DST-containing transcripts.
CCL and SEN1 mRNAs belong to a unique category of probable targets of the DST-mediated decay pathway. In RNA gel blots of samples harvested in the morning, both transcripts were decreased in abundance in the dst1 mutant but increased in the dst2 mutant (Pérez-Amador et al., 2001). The instability of both CCL and SEN1 mRNAs in the morning in dst1 correlates with the decreased abundance. In the case of CCL, the stabilization of the mRNA in the mutant compared to the parental plants in the afternoon could explain why the RNA oscillation cycle is phase lagging in dst1. Indeed, this phase lagging characteristic of dst1 is also evident during oscillations of leaf movement. Strikingly, an opposite effect was seen with dst2 at the whole plant level, i.e. it was phase leading in the cotyledon movement studies (Supplemental Fig. 3). This is consistent with the opposite effect of dst2 relative to dst1 for CCL and SEN1 mRNA abundance that we previously observed (Pérez-Amador et al., 2001).
Intriguingly, not all probable targets of the DST-mediated decay pathway seem to be regulated in a diurnal fashion. Stability measurements for RAP2.4 mRNA, first identified in the dst1 microarray experiments (Pérez-Amador et al., 2001), indicated that although the half-life of the mRNA is consistently altered in the dst1 mutant, this trend is insensitive to the time of day (data not shown). This data also suggests that the dst1 mRNA decay phenotype is not a consequence of altered sensitivity of the mutant to cordycepin. The abundance of the eIF4A transcript, used as a non-DST-containing control, is also equivalent in the mutant and parental plants. In addition, other transcripts, encoding a subunit of RNA polymerase II (AtRPB15.9) and aconitase, which lack DST elements, are equally abundant in the mutant and parental plants (Johnson et al., 2000). The reason for this selectivity is not clear at this point. No obvious bias in the 3′UTR sequences could be detected. In addition, two copies of the consensus DST sequence element failed to confer diurnal oscillation to a reporter mRNA (data not shown). However, previous studies have suggested different modes of recognition of the DST sequence in different cell types (Sullivan and Green, 1996; Feldbrugge et al., 2002). There is also precedent for differences in DST element function depending upon the sequence context in which it is present (Newman et al., 1993). Therefore, it is possible that a specific arrangement of subdomains and/or additional sequence requirements is important for the regulation of mRNA stability by the DST-mediated decay pathway.
The circadian clock can persist in the absence of environmental cues but can be reset by them. This allows rhythmic activities to occur at particular times during the day, thus endowing plants with enhanced fitness and adaptive ability (Green et al., 2002; Michael and McClung, 2003). Even though many plant mRNAs oscillate, not all proteins encoded by these mRNAs demonstrate circadian rhythmicity. For example, in Arabidopsis, phytochrome B transcription is rhythmic, but the bulk of phytochrome B protein does not oscillate (Bognar et al., 1999). As a result, a molecular defect at the RNA level does not always have an obvious impact at the whole plant level. Remarkably, in the dst1 mutant, a molecular change at the level of mRNA stability is reflected by a correlative influence on cotyledon movement and thus is clearly relevant to the physiology of the plant, although the precise mechanism is not yet known. CCL and/or SEN1 may be involved in the cotyledon response, but it is equally possible that other targets of clock-gated mRNA destabilization contribute to cotyledon movement. More detailed studies are needed to understand the role of CCL and SEN1, if any, in affecting cotyledon movement.
It is evident that cis-acting elements as well as trans-acting factors are involved in the posttranscriptional regulation of clock controlled mRNAs. Both CCL and SEN1 contain DST-like elements in their 3′UTR, which are distinct from instability determinants such as AU-rich elements that act in plants and other eukaryotes (Newman et al., 1993; Ohme-Takagi et al., 1993; Chen and Shyu, 1995). Furthermore, our results indicate that these mRNAs are likely to be primary targets of the dst1 mutation. The exact nature of the DST1 mutation is not known as yet, but it is possible that it corresponds to a sequence-specific RNA-binding protein. Clock-controlled RNA-binding proteins have been identified in the algae Gonyaulax polyedra (Morse et al., 1989) and Chlamydomonas reinhardtii (Mittag et al., 1994) that bind to the 3′UTRs of several mRNAs that contain a UG-repeat region (Waltenberger et al., 2001). The circadian clock also controls the binding activity of these proteins, which are hypothesized to function as translational suppressors (Mittag, 2003). Another example is the putative RNA-binding LARK protein of Drosophila that oscillates in abundance and regulates adult eclosion (McNeil et al., 1998). Precedence for clock-controlled RNA-binding proteins in Arabidopsis has also been documented. AtGRP7 mRNA as well as the protein undergoes circadian oscillations with slight delay of the protein peak relative to the RNA peak (Heintzen et al., 1997). Overexpression studies have shown that the transcript and protein are linked in a negative autoregulatory circuit (Staiger, 2001). This negative autoregulation was shown to be mediated through the binding of the protein to its own pre-mRNA, resulting in the formation of an alternatively spliced transcript with a premature stop codon that is rapidly degraded (Staiger et al., 2003).
Additionally, CCGs such as AtGRP7 show normal oscillation patterns in dst1 plants (Fig. 6, C and D), suggesting that the mutant plants do not exhibit a global effect on the clock at the molecular level. This hypothesis is further supported by the fact that DST1 maps to an interval on chromosome 1 that does not contain any genes known to be involved in clock function. Our data predict that DST1 probably functions downstream of the master clock and affects a subset of CCGs at the level of mRNA stability. Therefore, the cloning of the DST1 gene should identify a new component that will help elucidate the precise relationship between the DST-mediated decay machinery and the circadian clock.
The connection between a sequence-specific mRNA decay pathway and circadian rhythms described here provides a fine-tuning mechanism to achieve the precise oscillatory patterns of expression controlled by the clock. Our results also strengthen the idea that DST sequences are more functionally versatile than previously anticipated. At this time, it is difficult to predict how many CCGs are impacted by the DST-mediated mRNA decay pathway. A recent study by Michael and McClung (2003) proposes that the number of CCGs estimated by DNA microarrays could be an underestimate, and using enhancer trapping, the authors showed that 36% of the Arabidopsis genome is under transcriptional control by the circadian clock. Even though insertional mutagenesis has different sensitivity and constraints as opposed to DNA microarrays, this result suggests that additional clock genes might be regulated at the level of stability by the DST-mediated mRNA decay pathway. New technologies that allow global turnover rates at different times of the day to be measured in plants (Gutierrez et al., 2002) and other systems (Bernstein et al., 2002; Wang et al., 2002) should help to identify genes with similar regulatory strategies and thus serve as a powerful tool for unraveling circadian clock mechanisms at the posttranscriptional level.
MATERIALS AND METHODS
Arabidopsis Strains and Growth Conditions
All Arabidopsis (Arabidopsis thaliana) plants described are from the accession Columbia. dst1 mutant (from the second backcross to the parental line) and 1519 parental seeds were plated on agar plates containing 1× Murashige and Skoog salts, 1× Gamborg's vitamins, 1% Suc, and 50 μg/mL kanamycin and grown for 2 weeks in an incubator set at 16 h light (60–70 μmol/s/m2)/8 h dark and 21°C. For the circadian (free-running) experiments, Arabidopsis seedlings were grown for 12 d in 16 h light/8 h dark and on the morning of the 12th day were transferred to continuous light (40–50 μmol/s/m2) for 48 h. Lighting was provided by fluorescent light bulbs.
Half-Life Measurements, RNA Preparation, and Analysis
Half-lives were determined as described by Seeley et al. (1992) with the following modifications. Two-week-old Arabidopsis seedlings were transferred to a flask with incubation buffer. After a 30-min incubation, 3′-deoxyadenosine (cordycepin) was added to a final concentration of 0.6 mm (time 0). Tissue samples were harvested at regular intervals thereafter and frozen in liquid nitrogen. Total RNA was isolated using standard techniques. RNA was analyzed by electrophoresis on 2% formaldehyde/1.2% agarose gels and blotted onto nylon membrane (Nytran Plus; Schleicher & Schuell, Keene, NH). DNA probes were labeled with [α-32P]dCTP by the random primer method (Feinberg and Vogelstein, 1983) and purified from unincorporated nucleotides using probe purification columns (NucTrap; Stratagene, La Jolla, CA). The RNA blots were hybridized as described previously (Taylor and Green, 1991), using the indicated 32P-labeled probes. For a loading control, RNA blots were hybridized with a 32P-labeled cDNA probe for the Arabidopsis translation initiation factor eIF4A (Taylor et al., 1993). The same probe and hybridization conditions were used for the RNA gel blots showing the morning and afternoon data.
SEN1 is a single gene in the genome of Arabidopsis (Oh et al., 1996). CCL protein has no similarity to other protein sequences in the Arabidopsis genome as determined by BLASTCLUST. In addition, BLASTN analysis (nucleotide versus nucleotide comparison) using SEN1 and CCL transcribed sequences as query revealed no significant similarity to other transcribed sequences in the Arabidopsis genome. Hence, northern-blot probes used in this study were made by PCR using EST clones as template, 111D3T7 and 170N19T7 for SEN1 and 245H16T7 for CCL, and SP6 and T7 vector primers.
We used the Statistical Analysis System (http://www.sas.com) package to perform analysis of variance under a repeated measures design for all half-life experiments. To determine the effect of time of day in the half-lives of the genes analyzed, we employed the PROC MIXED procedure. PROC MIXED fits a variety of mixed linear models and produces appropriate statistics to enable one to make statistical inferences about the data. The model we used for the analysis allowed evaluation of the effect of time, time of day, and genotype on the response observed (mRNA levels) in each experiment. The procedures used to calculate the effect of the variables analyzed take into account all time-course data points simultaneously; hence, the presence of two measures for some data points does not interfere with proper statistical analysis. For further details, please see Sahai and Ageel (2000). The standard error is reported in Figures 1, 2, 3, and 5. Standard error calculation takes into account the number of replicates available. This value is reported as a reference for the reader, but it was not used to determine statistical significance.
Statistical comparison of the mRNA half-lives measured at the different times of the day was performed using a repeated measures model with the Statistical Analysis System software.
Cotyledon Movement Assay
Assessment of rhythmicity in cotyledon movement was carried out as described (Millar et al., 1995; Salomé et al., 2002). Seeds were surface-sterilized by the vapor-phase method (Clough and Bent, 1998) and then plated on Murashige and Skoog salts + 2% Suc. For light entrainment, seedlings were grown under white light (70–80 μmol/s/m2) for 5 d in a 12-h-light/12-h-dark photoperiod. For temperature entrainment, seedlings were grown under white light (30–40 μmol/s/m2) for 7 d in a 12 h 22°C/12 h 12°C temperature regime. On the 5th or 7th day, seedlings were transferred to 24-well cloning plates (Greiner Labortechnik, Frickenhausen, Germany), and the plates were released into continuous white light (30–40 μmol/s/m2) and constant temperature of 22°C. Leaf movement was recorded every 20 min over 7 d by Panasonic CCTV cameras, model WV-BP120 (Matsushita Communications Industrial, Laguna, Philippines). Post-run analysis was performed using the Kujata software program (Millar et al., 1995), and traces were analyzed by fast Fourier transform-nonlinear least squares (Plautz et al., 1997).
Supplementary Material
Acknowledgments
We would like to thank Lan Xue (Statistical Consulting Service, Department of Statistics, Michigan State University) for help with the statistical analysis of mRNA half-lives.
This work was funded by grants from the U.S. Department of Agriculture (2000–1419 and 2002–1272) and the Department of Energy (FG002–91ER20021) to P.J.G. and the National Science Foundation (MCB–0091008) to C.R.M.
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.060368.
References
- Alabadí D, Oyama T, Yanovsky MJ, Harmon FG, Más P, Kay SA (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 [DOI] [PubMed] [Google Scholar]
- Baggs JE, Green CB (2003) Nocturnin, a deadenylase in Xenopus laevis retina: a mechanism for posttranscriptional control of circadian-related mRNA. Curr Biol 13: 189–198 [DOI] [PubMed] [Google Scholar]
- Bernstein JA, Khodursky AB, Lin PH, Lin-Chao S, Cohen SN (2002) Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc Natl Acad Sci USA 99: 9697–9702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bognar LK, Hall A, Adam E, Thain SC, Nagy F, Millar AJ (1999) The circadian clock controls the expression pattern of the circadian input photoreceptor, phytochrome B. Proc Natl Acad Sci USA 96: 14652–14657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Shyu AB (1995) AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci 20: 465–470 [DOI] [PubMed] [Google Scholar]
- Chen Y, Hunter-Ensor M, Schotland P, Sehgal A (1998) Alterations of per RNA in noncoding regions affect periodicity of circadian behavioral rhythms. J Biol Rhythms 13: 364–379 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dupressoir A, Morel AP, Barbot W, Loireau MP, Corbo L, Heidmann T (2001) Identification of four families of yCCR4- and Mg2+-dependent endonuclease-related proteins in higher eukaryotes, and characterization of orthologs of yCCR4 with a conserved leucine-rich repeat essential for hCAF1/hPOP2 binding. BMC Genomics 2: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edery I (1999) Role of posttranscriptional regulation in circadian clocks: lessons from Drosophila. Chronobiol Int 16: 377–414 [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B (1983) A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem 132: 6–13 [DOI] [PubMed] [Google Scholar]
- Feldbrugge M, Arizti P, Sullivan ML, Zamore PD, Belasco JG, Green PJ (2002) Comparative analysis of the plant mRNA-destabilizing element, DST, in mammalian and tobacco cells. Plant Mol Biol 49: 215–223 [DOI] [PubMed] [Google Scholar]
- Frisch B, Hardin PE, Hamblen-Coyle MJ, Rosbash M, Hall JC (1994) A promoterless period gene mediates behavioral rhythmicity and cyclical per expression in a restricted subset of the Drosophila nervous system. Neuron 12: 555–570 [DOI] [PubMed] [Google Scholar]
- Glossop NR, Lyons LC, Hardin PE (1999) Interlocked feedback loops within the Drosophila circadian oscillator. Science 286: 766–768 [DOI] [PubMed] [Google Scholar]
- Green RM, Tingay S, Wang ZY, Tobin EM (2002) Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiol 129: 576–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez RA, Ewing RM, Cherry JM, Green PJ (2002) Identification of unstable transcripts in Arabidopsis by cDNA microarray analysis: Rapid decay is associated with a group of touch- and specific clock-controlled genes. Proc Natl Acad Sci USA 99: 11513–11518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290: 2110–2113 [DOI] [PubMed] [Google Scholar]
- Harmer SL, Panda S, Kay SA (2001) Molecular bases of circadian rhythms. Annu Rev Cell Dev Biol 17: 215–253 [DOI] [PubMed] [Google Scholar]
- Heintzen C, Nater M, Apel K, Staiger D (1997) AtGRP7, a nuclear RNA-binding protein as a component of a circadian-regulated negative feedback loop in Arabidopsis thaliana. Proc Natl Acad Sci USA 94: 8515–8520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto M, Higo H, Higo K (2000) Differential diurnal expression of rice catalase genes: the 5′-flanking region of CatA is not sufficient for circadian control. Plant Sci 151: 39–46 [Google Scholar]
- Johnson MA, Pérez-Amador MA, Lidder P, Green PJ (2000) Mutants of Arabidopsis defective in a sequence-specific mRNA degradation pathway. Proc Natl Acad Sci USA 97: 13991–13996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer C, Loros JJ, Dunlap JC, Crosthwaite SK (2003) Role for antisense RNA in regulating circadian clock function in Neurospora crassa. Nature 421: 948–952 [DOI] [PubMed] [Google Scholar]
- Majercak J, Sidote D, Hardin PE, Edery I (1999) How a circadian clock adapts to seasonal decreases in temperature and day length. Neuron 24: 219–230 [DOI] [PubMed] [Google Scholar]
- McClung CR (2001) Circadian rhythms in plants. Annu Rev Plant Physiol Plant Mol Biol 52: 139–162 [DOI] [PubMed] [Google Scholar]
- McNeil GP, Zhang X, Genova G, Jackson FR (1998) A molecular rhythm mediating circadian clock output in Drosophila. Neuron 20: 297–303 [DOI] [PubMed] [Google Scholar]
- Michael TP, McClung CR (2003) Enhancer trapping reveals widespread circadian clock transcriptional control in Arabidopsis. Plant Physiol 132: 629–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AJ, Carré IA, Strayer CA, Chua NH, Kay SA (1995) Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science 267: 1161–1163 [DOI] [PubMed] [Google Scholar]
- Millar AJ, Kay SA (1991) Circadian control of cab gene transcription and mRNA accumulation in Arabidopsis. Plant Cell 3: 541–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittag M (2003) The function of circadian RNA-binding proteins and their cis-acting elements in microalgae. Chronobiol Int 20: 529–541 [DOI] [PubMed] [Google Scholar]
- Mittag M, Lee DH, Hastings JW (1994) Circadian expression of the luciferin-binding protein correlates with the binding of a protein to the 3′ untranslated region of its mRNA. Proc Natl Acad Sci USA 91: 5257–5261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse D, Pappenheimer AM Jr, Hastings JW (1989) Role of a luciferin-binding protein in the circadian bioluminescent reaction of Gonyaulax polyedra. J Biol Chem 264: 11822–11826 [PubMed] [Google Scholar]
- Newman TC, Ohme-Takagi M, Taylor CB, Green PJ (1993) DST sequences, highly conserved among plant SAUR genes, target reporter transcripts for rapid decay in tobacco. Plant Cell 5: 701–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SA, Lee SY, Chung IK, Lee CH, Nam HG (1996) A senescence-associated gene of Arabidopsis thaliana is distinctively regulated during natural and artificially induced leaf senescence. Plant Mol Biol 30: 739–754 [DOI] [PubMed] [Google Scholar]
- Ohme-Takagi M, Taylor CB, Newman TC, Green PJ (1993) The effect of sequences with high AU content on mRNA stability in tobacco. Proc Natl Acad Sci USA 90: 11811–11815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Y, Andersson CR, Kondo T, Golden SS, Johnson CH (1998) Resonating circadian clocks enhance fitness in cyanobacteria. Proc Natl Acad Sci USA 95: 8660–8664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S, Hogenesch JB, Kay SA (2002) Circadian rhythms from flies to human. Nature 417: 329–335 [DOI] [PubMed] [Google Scholar]
- Pérez-Amador MA, Lidder P, Johnson MA, Landgraf J, Wisman E, Green PJ (2001) New molecular phenotypes in the dst mutants of Arabidopsis revealed by DNA microarray analysis. Plant Cell 13: 2703–2717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilgrim ML, Caspar T, Quail PH, McClung CR (1993) Circadian and light-regulated expression of nitrate reductase in Arabidopsis. Plant Mol Biol 23: 349–364 [DOI] [PubMed] [Google Scholar]
- Plautz JD, Straume M, Stanewsky R, Jamison CF, Brandes C, Dowse HB, Hall JC, Kay SA (1997) Quantitative analysis of Drosophila period gene transcription in living animals. J Biol Rhythms 12: 204–217 [DOI] [PubMed] [Google Scholar]
- Robinson BG, Frim DM, Schwartz WJ, Majzoub JA (1988) Vasopressin mRNA in the suprachiasmatic nuclei: daily regulation of polyadenylate tail length. Science 241: 342–344 [DOI] [PubMed] [Google Scholar]
- Sahai H, Ageel MI (2000) The Analysis of Variance: Fixed, Random and Mixed Models. Birkhauser, Boston
- Salomé PA, Michael TP, Kearns EV, Fett-Neto AG, Sharrock RA, McClung CR (2002) The out of phase 1 mutant defines a role for PHYB in circadian phase control in Arabidopsis. Plant Physiol 129: 1674–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer R, Landgraf J, Accerbi M, Simon V, Larson M, Wisman E (2001) Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis. Plant Cell 13: 113–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carre IA, Coupland G (1998) The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93: 1219–1229 [DOI] [PubMed] [Google Scholar]
- Seeley KA, Byrne DH, Colbert JT (1992) Red light-independent instability of oat phytochrome mRNA in vivo. Plant Cell 4: 29–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- So WV, Rosbash M (1997) Post-transcriptional regulation contributes to Drosophila clock gene mRNA cycling. EMBO J 16: 7146–7155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger D (2001) RNA-binding proteins and circadian rhythms in Arabidopsis thaliana. Philos Trans R Soc Lond B Biol Sci 356: 1755–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger D, Zecca L, Wieczorek Kirk DA, Apel K, Eckstein L (2003) The circadian clock regulated RNA-binding protein AtGRP7 autoregulates its expression by influencing alternative splicing of its own pre-mRNA. Plant J 33: 361–371 [DOI] [PubMed] [Google Scholar]
- Stanewsky R (2002) Clock mechanisms in Drosophila. Cell Tissue Res 309: 11–26 [DOI] [PubMed] [Google Scholar]
- Stanewsky R, Jamison CF, Plautz JD, Kay SA, Hall JC (1997) Multiple circadian-regulated elements contribute to cycling period gene expression in Drosophila. EMBO J 16: 5006–5018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Más P, Panda S, Kreps JA, Kay SA (2000) Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289: 768–771 [DOI] [PubMed] [Google Scholar]
- Sullivan ML, Green PJ (1996) Mutational analysis of the DST element in tobacco cells and transgenic plants: identification of residues critical for mRNA instability. RNA 2: 308–315 [PMC free article] [PubMed] [Google Scholar]
- Taylor CB, Bariola PA, delCardayre SB, Raines RT, Green PJ (1993) RNS2: a senescence-associated RNase of Arabidopsis that diverged from the S-RNases before speciation. Proc Natl Acad Sci USA 90: 5118–5122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CB, Green PJ (1991) Genes with homology to fungal and S-gene RNases are expressed in Arabidopsis thaliana. Plant Physiol 96: 980–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijsterman M, Ketting RF, Plasterk RH (2002) The genetics of RNA silencing. Annu Rev Genet 36: 489–519 [DOI] [PubMed] [Google Scholar]
- Waltenberger H, Schneid C, Grosch JO, Bareiss A, Mittag M (2001) Identification of target mRNAs for the clock-controlled RNA-binding protein Chlamy 1 from Chlamydomonas reinhardtii. Mol Genet Genomics 265: 180–188 [DOI] [PubMed] [Google Scholar]
- Wang Y, Liu CL, Storey JD, Tibshirani RJ, Herschlag D, Brown PO (2002) Precision and functional specificity in mRNA decay. Proc Natl Acad Sci USA 99: 5860–5865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Tobin EM (1998) Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93: 1207–1217 [DOI] [PubMed] [Google Scholar]
- Wuarin J, Falvey E, Lavery D, Talbot D, Schmidt E, Ossipow V, Fonjallaz P, Schibler U (1992) The role of the transcriptional activator protein DBP in circadian liver gene expression. J Cell Sci Suppl 16: 123–127 [DOI] [PubMed] [Google Scholar]
- Zhong HH, Resnick AS, Straume M, McClung CR (1997) Effects of synergistic signaling by phytochrome A and cryptochrome1 on circadian clock-regulated catalase expression. Plant Cell 9: 947–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.