Abstract
Establishing a reliable genotyping protocol is a critical matter in postsequence genetics. In this article, we describe a highly sensitive electrochemical detection of complementary DNAs (up to 43-mer) based on hole transport with molecular-scale, “wire-like” DNA probes. The presence of a single-base mismatch in the DNA duplexes caused a dramatic decrease in the electrochemical response. We applied this method to detect all of the possible transition and transversion SNPs and achieved “on–off”-type discrimination of fully complementary DNAs from their SNPs. Furthermore, naturally occurring polymorphisms, “hot spots” from the p53 gene, could clearly be distinguished from wild type by using our methodology.
Keywords: electrochemistry, hole transport, ferrocene, gold electrode
SNPs of genes characterize personality of individuals, the medicinal importance of which includes risks of various diseases and differences in drug response (pharmacogenomics) (1, 2). Consequently, various enzymeless, chip-based assay systems, so-called DNA-chips, have been proposed for large-scale analysis of SNPs (3). The hybridization events on the chips can be read out from the inherent changes, such as optical, micro-gravimetric, and electrochemical responses. Among them, electrochemical detection promises to be far superior to existing sensory formats with respect to sensitivity, rapidity, and availability of low-cost portable devices.
DNA is electrochemically silent unless highly negative and positive potentials are applied, at which time, solvent decomposition occurs and interferes with the signals from DNA. Thus, electrochemically active DNA probes have been developed (4–6), including solution-borne ferrocene-modified DNAs (7), redox-active intercalators (8), surface-bound molecular beacon-like DNAs (9), etc. (10–14). In these electrochemical assays, the through-space distance between the electrodes and the redox labels determines the electron-transfer efficiency. Therefore, discrimination of fully matched DNAs from their SNPs by using these probes depends directly or indirectly on a difference in pairing energies between a probe sequence and a fully matched sequence vs. a mismatched one, as in many other assays used in DNA chips. These differences, however, sometimes can be very small and can largely vary based on sequence context, necessitating complex and sophisticated experimental manipulations to discriminate the signals resulting from matched and mismatched complements (15).
Barton and coworkers (16) and Okamoto et al. (17) have reported a conceptually different approach for electrochemical detection of SNPs. They exploited a method that relied on charge transport through the π -stack of duplexes (18–22) on gold electrodes by combining redox-active intercalators with exogenous electrocatalytic species. Generally, single-base mismatches may not cause substantial changes in duplex stability and structure but may have a large influence on the electronic coupling nature within the base pair stacks. In this assay, the current flow from the intercalator to the electrode through the π -stack is diminished even to single-base mismatches. Nevertheless, the existing methodologies are not quite satisfactory, particularly for identifying SNPs of each individual patient. The ultimate assay protocol for this purpose must have high sensitivity and keen discernment for probes and should be a reagentless method with unmodified, native target DNAs.
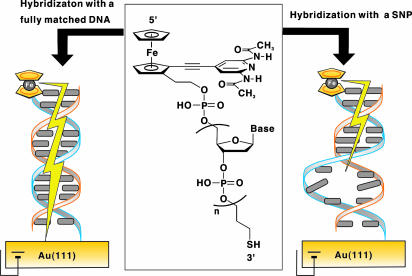
Our strategy is to use a completely π -conjugated ferrocene-modified nucleoside analogue, the structure of which is inspired from our previous studies on molecular recognition for ferrocene-modified synthetic receptors (23, 24). We thought that if this nucleoside analogue were connected at the 5′ end of a single-stranded oligonucleotide, the modified DNA would behave as a molecular-scale, “wire-like” electrochemical probe (25). When the hybridized duplex with a complementary DNA is attached onto a gold electrode at the 3′ end of the probe DNA by means of gold-thiol chemistry, “π -way” will be fully opened from the ferrocene tag of the nucleoside residue to the gold electrodes by means of the conjugated acetylene–synthetic base–stacked base pair bridges. If a mismatched base pair presents in the duplex, hole transport from the electrodes will be insulated at that position like a blockade of the π -way (Fig. 1). This straightforward methodology will meet the criteria in future chip-based genetic analysis for identifying SNPs of individuals.
Fig. 1.
A concept of our research. (Center) Chemical structure of HS-DNA-Fc. In the case of HS-DNA-Fc2, an ethane linker (-CH2CH2-) is substituted for the acetylene linker (-C≡C-). (Left and Right) Illustration for electrochemical discrimination of SNPs with a ferrocene-modified, wire-like DNA probe.
Materials and Methods
Materials. The corresponding phosphoramidite of the nucleoside analogue for solid-phase DNA synthesis was easily prepared from known (pS)-2-iodoferrocenecarboxaldehyde 25 (26), and the procedures are shown in Scheme 1 and Supporting Text, which are published as supporting information on the PNAS web site. Other materials were all commercially available.
Synthesis of Wire-Like DNA Probes. A single-stranded 15-mer DNA possessing a C3-alkylthiol linker (3′ end was masked as -phosphate-C3-SS-C3-O-succinyl-lcaa-CPG) was coupled with the corresponding phosphoramidite at the 5′ end on solid-phase DNA synthesis and released from the CPG support. Nihon Gene Research Laboratories (Sendai, Japan) performed these operations. The resulting 5′-modified alkyldisulfide-linked DNA (3′end was -phosphate-C3-SS-C3-OH) was converted into the free thiol probe with DTT and purified by reverse-phase HPLC as follows. A Milli-Q (100 μl) solution of the protected probe (200 nmol) and DTT (1.54 mg) was stirred at 25°C for 6 h with a thermomixer (24 × g). The reaction mixture was prepurified with a short octadecylsilyl column (Sep-Pack Cartridges, Waters) for exclusion of DTT. The deprotected probe was further purified by HPLC using a Cosmosil 5C18-AR-II column (4.6 × 150 mm, Nacalai Tesque, Kyoto) with an eluent of 0.05 M ammonium formate and 10–13% acetonitrile linear gradient (0–40 min) at a flow rate of 1.0 ml/min. The probe DNAs were characterized by MALDI-TOF MS (Fig. 4, which is published as supporting information on the PNAS web site).
Preparation of Duplex-Modified Gold Electrodes. Commercially available gold (111) surfaces (≥80%) prepared by vapor deposition of gold onto polyethylene terephthalate were used (Tanaka Kikinzoku, Tokyo). The gold electrode (the deposition area of 1 mm2) was cleaned as follows. Sonication was performed for 5 min in Milli-Q water with an ultrasonic generator, and electrochemical etching was performed by cycling the electrode potential between –0.1 and +1.5 V vs. Ag/AgCl in 0.05 M H2SO4 at a scan rate of 0.1 V/s 10 times. A mixture of 0.5 μl of the probe DNA solution (200 μM in 1 M NaClO4) and 0.5 μl of a target DNA solution (220 μM in 1 M NaClO4) was heated to 65°C, followed by slow cooling to 25°C over 90 min. All of the hybridized solution was placed on the gold electrode and kept in a closed container under high humidity for 90 min at room temperature. The modified electrode was then rinsed with 1 M NaClO4 several times.
Electrochemical Measurements. Cyclic voltammetry and square wave voltammetry (SWV) were carried out on the electrodes by using a Bioanalytical Systems (BASi) (West Lafayette, IN) model 620A electrochemical analyzer. Used was a normal three-electrode configuration consisting of the gold working electrode, a saturated Ag/AgCl reference electrode (BASi), and a platinum wire auxiliary electrode. The working compartment of the electrochemical cell was separated from the reference compartment by a glass frit. All measurements were performed at 25.0 ± 0.1°C in 1 M NaClO4 that had been thoroughly degassed with N2. SWV was conducted by scanning from 0 to +0.6 V with an amplitude of 25 mV and a step potential of 4 mV at 8 Hz.
Results and Discussion
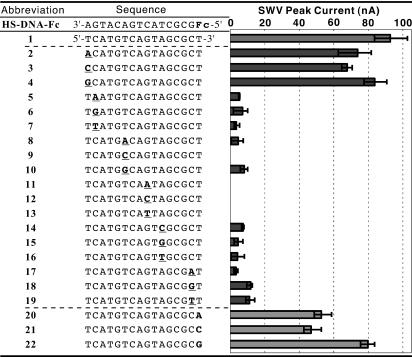
The sequence of a 16-mer DNA probe (3′-HS(CH2)3-AGT ACA GTC ATC GCG Fc-5′) is selected on the basis of the report of Barton and colleagues (16). In the sequence, the residue of the ferrocene-modified nucleoside analogue is abbreviated as Fc (a ferrocene unit with an acetylene linker), and the probe is referred to as HS-DNA-Fc. Table 1 shows the sequences of a fully matched complement 1 and its SNPs 2–19 examined in this study. To discuss unambiguously the ability of the probe for hole transport, the duplexes must definitely be formed not only from fully matched complements but also from single-base mismatched ones. In advance, a thiol-free version of HS-DNA-Fc, DNA-Fc (3′-AGT ACA GTC ATC GCG Fc-5′) was synthesized and examined for the duplex formation in 1 M NaClO4. The melting temperature values of 63°C and 50°C were determined for the duplexes from the fully matched complement 1 and the complement 12, respectively (Fig. 5, which is published as supporting information on the PNAS web site). The duplex from 12 is anticipated to have the lowest melting temperature value in all of the duplexes examined because of the mismatched position at the middle of the sequence and the mismatched pattern of C–C (15). The duplex formation was also corroborated on the basis of CD in 1 M NaClO4 at 25°C. A double-stranded DNA is polymorphic in secondary structure that readily changes with solvent conditions, and the technique of CD is exquisitely sensitive to the secondary structure. The CD spectrum in the wavelength region of ≈200–300 nm was monitored to determine the secondary structure for duplexes. Because Cotton effects in this region are attributed to the sum of many π–π * transitions of four chromophoric bases in DNA, the sequences of the duplexes must be similar when making comparisons. Thus, we used duplexes formed by combination between DNA-Fc and 1 or 12 and found that both of the duplexes did reveal almost the same CD spectra characteristic of a typical double-stranded B-form DNA (27) (Fig. 6, which is published as supporting information on the PNAS web site).
Table 1. The sequences of HS-DNA-Fc and complementary strands 1–22 and a summary of SWV peak current for the duplexes from HS-DNA-Fc with strands 1–22.
All complementary strands are given in the 5′ to 3′ orientation. Underlined residues are the mismatched bases. The error bar was determined from three measurements.
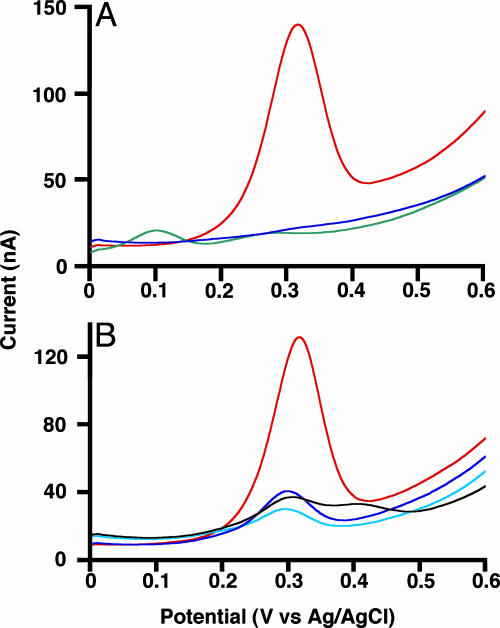
Gold electrodes modified with duplexes were interrogated by the SWV method scanning from 0 to +0.6 V vs. Ag/AgCl in 1 M NaClO4. No SWV peak appeared in the cases for bare gold electrodes and gold electrodes modified with a ferrocene-free duplex in this potential range (data not shown). When applying the duplex from HS-DNA-Fc and the fully matched complement 1, we observed a strong anodic peak at +0.31 V vs. Ag/AgCl caused by the oxidation of the ferrocene moiety (Fig. 2A). This potential falls within the potential range for redox of various ferrocene tags so far reported (4). In cyclic voltammetry with the duplex-modified electrode, the peak currents are directly proportional to scan rates, implying electrochemical reactions of surface-bound redox species (18) (Fig. 7, which is published as supporting information on the PNAS web site). Therefore, one might imagine that the redox conversion of the ferrocene tag, even being spatially far apart from the electrode, indeed occurs through a newly developed π -way in the duplex.
Fig. 2.
Electrochemical SNPs detection. Uncorrected SWV profiles at the gold working electrodes modified with HS-DNA-Fc·1 (fully matched duplex, red line), HS-DNA-Fc·12 (mismatched duplex, blue line), and HS-DNA-Fc2·1 (fully matched duplex, green line) (A) and the duplexes from HS-DNA2-Fc with fully matched DNA (red line), 248A SNP (light blue line), 249 SNP (black line), and 248T SNP (blue line) (B). Sequences are 5′-TGA ACC GGA GGC CCA T-3′ for fully matched DNA, 5′-TGA ACC AGA GGC CCA T-3′ for 248A SNP, 5′-TGA ACC GGA GTC CCA T-3′ for 249 SNP, and 5′-TGA ACT GGA GGC CCA T-3′ for 248T SNP (residues set in italics are the mismatched bases).
In this hole transport, with our wire-like DNA probes, the electron of the ferrocene moiety is expected to flow to the top of the base pair in duplexes through a conjugated π -orbital of the acetylene linker (reverse of hole transport). Other possible pathways contain a tunneling mechanism through space and/or through a σ-bonded linker. A DNA probe HS-DNA-Fc2 possessing the same sequence for HS-DNA-Fc was designed to shed light on this aspect. In the Fc2 residue of the probe, the ferrocene tag is connected to the synthetic base by means of an ethane linker of σ-bond without π -conjugation (Fig. 1 Center). The duplex from HS-DNA-Fc2 and the fully matched complement 1 was subjected to SWV analysis in the same manner as described for HS-DNA-Fc. For the duplex from HS-DNA-Fc2, we observed only a weak anodic peak at +0.10 V vs. Ag/AgCl (Fig. 2 A). Because this potential is appropriate for a dialkylferrocene skeleton (4), the faint peak results from the oxidation for the ferrocene tag of the Fc2 residue. The insulation of the anodic peak current must be ascribed to the lack of π -conjugation between the ferrocene tag and the base pairs in the duplex. This finding demonstrates the role of the acetylene linker that behaves as a “small wire” connecting electrically the ferrocene and the stacked base pairs.
Various SNPs 2–19 mismatched at a different position with a different base were examined to explore the specificity of the HS-DNA-Fc probe (Fig. 2 A and Table 1). Each mismatched base is placed on most positions in the sequence, covering all of the possible mismatched patterns (A–A, A–G, A–C, T–T, T–G, T–C, G–G, and C–C). Table 1 summarizes the SWV analyses for the duplexes from HS-DNA-Fc. The duplexes from 5–19 showed only small or no electrochemical responses. Indeed, the prominent discernment of HS-DNA-Fc is demonstrated by this nearly perfect discrimination of all of the types of the mismatched duplexes. The only exceptional cases were for the complements 2–4 possessing a mismatched base at their 5′ ends. The mismatched duplexes from the 16-mer 2–4, however, can also be regarded as fully matched duplexes of 15-mer because of their mismatched position. Furthermore, the mismatched base pair is located at the closest position from the electrode surface. Thus, we can assume that in the cases of 2–4, the pseudo fully matched duplexes of 15-mer are connected to the electrode by means of a somewhat longer (with one extra nucleotide unit) thiol linker. This interpretation may explain the slightly small oxidation currents for these duplexes compared with the “true” value for the duplex from 1 (28).
At the present stage, it is difficult to clearly envisage the molecular basis for the dramatic decrease of the hole transport efficiency in these mismatched duplexes. Giese and Wessely (29) demonstrated that a hole transport in duplexes is interrupted by the presence of mismatched G bases because of a loss of the cation charge induced by a proton transfer from the developed, nonpaired G cation radical to the surrounding water. In our duplexes from 8, 9, 11, 12, 13, 14, and 16, however, mismatched base pairs without a G base also revealed the strong interruption for the hole transport. Thus, although the mechanism suggested by Giese and Wessely will play a partial role in some of the present cases, another possibility must be considered. This situation still remains obscure.
The synthetic base in the Fc residue was originally designed to bind a thymine base but is expected to allow at least partial hydrogen bonds with any base. Complements 20–22 have a different base at each 3′ end, in which the base should bind to the synthetic base of the Fc residue in HS-DNA-Fc. All of the duplexes from the complements 20–22 displayed substantially strong electrochemical responses. Given the circumstance, the selectivity is an obstacle rather than a necessity. The tolerance of the synthetic base for binding to any base is an additional advantage for our detection method.
The discrimination between fully matched and single-base mismatched complements with our wire-like DNA probes will be independent of DNA sequence context and composition. To illustrate this aspect, we chose naturally occurring polymorphisms, “hot spots” from the p53 gene, as a target sequence (30, 31). In the p53 tumor suppressor gene, serious tumor-inducing mutations have predominantly been discovered in regions that encode the DNA-binding domain (in codons 100–293). Among the mutations, representative two-transition SNPs (T from C and A from G in codon 248) and a one-transversion SNP (T from G in codon 249) were chosen. For this purpose, a wire-like DNA probe with a completely different sequence was synthesized, and the 16-mer probe of 3′-HS(CH2)3-ACT TGG CCT CCG GGT Fc-5′ is referred to as HS-DNA2-Fc. The detection protocol is identical with that described for the HS-DNA-Fc probe. As shown in Fig. 2B, the three SNPs were clearly differentiated from a fully base-paired wild type by using the probe that we synthesized. This experiment proves that the wire-like DNA probes can detect mutations within natural sequences of duplexes and that the method should be applied to other genes of clinical importance.
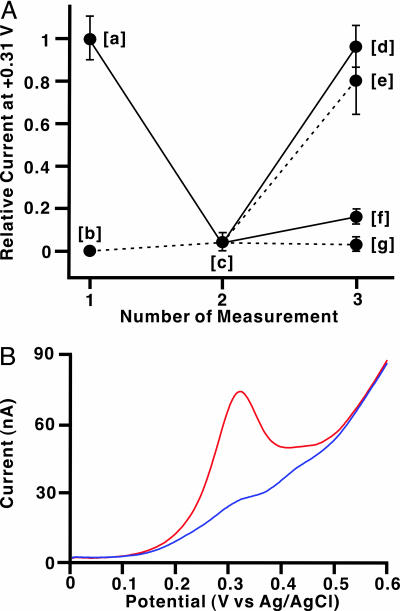
Our DNA probe can be regarded as a “signal-on” sensor in that hybridization produces an electrochemical response. Signal-on sensors are preferred to “signal-off” sensors (9) because in the latter, false-positive responses might be induced by unpredictable accidents (e.g., the degradation of a fully complementary DNA by sample contaminants). Another important point for considering device application is that discrimination of SNPs should also be performed with sequences hybridized reversibly on the electrode surface (in situ detection; Fig. 3A). The well responded electrode modified with HS-DNA-Fc and 1 was thoroughly rinsed with Milli-Q water to remove 1. The resulting electrode showed no SWV signal, resembling the electrode modified with a single-base mismatched duplex. It is worth noting that the electrode modified with only the probe reveals the “off” state. Thus, the “on” state of the electrode is achieved only in the presence of a fully matched complement, not in the presence of a single-base mismatched complement, nor in the absence of complements. Rehybridization with the fully matched 1 on the electrode restored the SWV signal to nearly the same level of the previous on state. In contrast, hybridization with the single-base mismatched 12 produced only a small signal under the same conditions. This exchange operation also succeeded starting from the single-base mismatched duplex of 12 and can be cycled repeatedly.
Fig. 3.
In situ detection on the electrode and electrochemical SNPs detection for longer target. (A) Relative current (I/I0) plots at 0.31 V from SWV measurements for in situ detection (I0 is the average current of HS-DNA-Fc·1 at 0.31 V). The first SWV measurements were carried out as described in Materials and Methods (point [a] for the target 1 and point [b] for the target 12). After rinsing three times with Milli-Q water (1 ml) at room temperature, the electrodes were subjected to the second SWV analyses under the same conditions (point [c]). Then, the electrodes were soaked in a fresh 50-μl target solution (200 μM 1 or 12), stirred at 25°C for 90 min with a thermomixer (24 × g), and rinsed with 1 M NaClO4 (300 μl). The third measurements of the electrodes were carried out in 1 M NaClO4 (points [d] and [e] for 1 and points [f] and [g] for 12). (B) SNP discrimination for long target strands (43-mer). Uncorrected SWV profiles at the gold working electrodes modified with HS-DNA-Fc·23 (fully matched duplex, red line) and HS-DNA-Fc·24 (mismatched duplex, blue line).
In all of the experiments described above, the length of the target DNAs was made to be compatible with that of the probe DNAs to exactly assess the performance of the probes. However, a further criterion for realizing clinical applications is that any length of DNAs should be subjected to this assay protocol. In advance, gold surfaces modified by the duplexes were analyzed by cyclic voltammetry (CV) on the basis of diffusion-controlled electrochemical reactions of Fe(CN)64– (see Supporting Text and Fig. 8, which are published as supporting information on the PNAS web site). The data for the CV revealed that the electrode surface should be covered partially but not entirely with the duplex. In other words, some space presents between the duplexes in contrast to the surface structure reported by Barton and coworkers (32). Therefore, even when the wire-like DNA probe is hybridized with any region of longer target DNAs, the resulting duplexes possessing nonhybridized extra residues will bind onto the electrode surface without interference from each other. This speculation encouraged us to apply the probes to detect 43-mer DNA 23 (5′-CTG CAT GGG CGG CAT CAT GTC AGT AGC GCT CCT CAC CAT CAT C-3′), in which the targeted 16-mer exists at the middle of the sequence (residues set in italics). The gold electrode modified with the duplex from HS-DNA-Fc and 23 did show a similar SWV peak to that from the 16-mer target 1 (Fig. 3B). In contrast, no peak appeared in the case of the SNP of 43-mer DNA 24 (5′-CTG CAT GGG CGG CAT CAT GTC ACT AGC GCT CCT CAC CAT CAT C-3′). The discrimination of SNPs was easily achieved not only with sequences hybridized reversibly on the electrode but also with longer ones. These successes demonstrate that our wire-like DNA probes can provide a practical method of genetic screening, being compatible with a chip-based assay.
In our experiments, no optimization has been carried out for sensitivity of the DNA probes. However, by coupling the hole transport to electrocatalytic cycle and/or by using gold surface on ultramicroelectrodes, the sensitivity may increase to a sufficient level that allows direct pathogen detection. Furthermore, from the viewpoint of the manipulation protocol, the reagentless detection of unmodified target DNAs is suitable for applying our method to chip-based genetic analysis.
Supplementary Material
Acknowledgments
We thank Dr. Shigeori Takenaka (Kyushu Institute of Technology, Fukuoka, Japan), Ms. Shinobu Sato (Kyushu University, Fukuoka), and Mr. Hiroshi Okamura (Toyo Kohan, Tokyo) for technical assistance and useful suggestions. This work was supported by a grant from Japan Science and Technology Agency.
Author contributions: M.I. designed research; M.I., R.I., M.T., T.T., and J.C. performed research; M.I., R.I., M.T., and T.T. contributed new reagents/analytic tools; M.I., R.I., M.T., and J.C. analyzed data; and M.I. and J.C. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Fc, ferrocene unit with acetylene linker; Fc2, ferrocene unit with ethane linker; SWV, square wave voltammetry.
References
- 1.Brookes, A. J. (1999) Gene 234, 177–186. [DOI] [PubMed] [Google Scholar]
- 2.McCarthy, J. J. & Hilfiker, R. (2000) Nat. Biotechnol. 18, 505–508. [DOI] [PubMed] [Google Scholar]
- 3.Wang, J. (2000) Nucleic Acids Res. 28, 3011–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Staveren, D. R. & Metzler-Nolte, N. (2004) Chem. Rev. 104, 5931–5985. [DOI] [PubMed] [Google Scholar]
- 5.Drummond, T. G., Hill, M. G. & Barton, J. K. (2003) Nat. Biotechnol. 21, 1192–1199. [DOI] [PubMed] [Google Scholar]
- 6.Gooding, J. J. (2002) Electroanalysis 14, 1149–1156. [Google Scholar]
- 7.Nakayama, M., Ihara, T., Nakano, K. & Maeda, M. (2002) Talanta 56, 857–866. [DOI] [PubMed] [Google Scholar]
- 8.Yamashita, K., Takagi, M., Kondo, H. & Takenaka, S. (2002) Anal. Biochem. 306, 188–196. [DOI] [PubMed] [Google Scholar]
- 9.Fan, C., Plaxco, K. W. & Heeger, A. J. (2003) Proc. Natl. Acad. Sci. USA 100, 9134–9137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu, C. J., Wan, Y., Yowanto, H., Li, J., Tao, C., James, M. D., Tan, C. L., Blackburn, G. F. & Meade, T. J. (2001) J. Am. Chem. Soc. 123, 11155–11161. [DOI] [PubMed] [Google Scholar]
- 11.Umek, R. M., Lin, S. W., Vielmetter, J., Terbrueggen, R. H., Irvine, B., Yu, C. J., Kayyem, J. F., Yowanto, H., Blackburn, G. F., Farkas, D. H., et al. (2001) J. Mol. Diagn. 3, 74–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim, K., Yang, H., Park, S. H., Lee, D., Kim, S., Lim, Y. T. & Kim, Y. T. (2004) Chem. Commun., 1466–1467. [DOI] [PubMed]
- 13.Kerman, K., Saito, M., Morita, Y., Takamura, Y., Ozsoz, M. & Tamiya, E. (2004) Anal. Chem. 76, 1877–1884. [DOI] [PubMed] [Google Scholar]
- 14.Liu, G., Lee, T. M. H. & Wang, J. (2005) J. Am. Chem. Soc. 127, 38–39. [DOI] [PubMed] [Google Scholar]
- 15.Peyret, N., Seneviratne, P. A., Allawi, H. T. & SantaLucia, J., Jr. (1999) Biochemistry 38, 3468–3477. [DOI] [PubMed] [Google Scholar]
- 16.Boon, E. M., Ceres, D. M., Drummond, T. G., Hill, M. G. & Barton, J. K. (2000) Nat. Biotechnol. 18, 1096–1100. [DOI] [PubMed] [Google Scholar]
- 17.Okamoto, A., Kamei, T., Tanaka, K. & Saito, I. (2004) J. Am. Chem. Soc. 126, 14732–14733. [DOI] [PubMed] [Google Scholar]
- 18.Long, Y. T., Li, C. Z., Sutherland, T. C., Chahma, M., Lee, J. S. & Kraatz, H. B. (2003) J. Am. Chem. Soc. 125, 8724–8725. [DOI] [PubMed] [Google Scholar]
- 19.Giese, B. (2002) Curr. Opin. Chem. Biol. 6, 612–618. [DOI] [PubMed] [Google Scholar]
- 20.Lewis, F. D., Letsinger, R. L. & Wasielewski, M. R. (2001) Acc. Chem. Res. 34, 159–170. [DOI] [PubMed] [Google Scholar]
- 21.Schuster, G. B. (2000) Acc. Chem. Res. 33, 253–260. [DOI] [PubMed] [Google Scholar]
- 22.Delaney, S. & Barton, J. K. (2003) J. Org. Chem. 68, 6475–6483. [DOI] [PubMed] [Google Scholar]
- 23.Inouye, M. & Takase, M. (2001) Angew. Chem. Int. Ed. 40, 1746–1748. [PubMed] [Google Scholar]
- 24.Takase, M. & Inouye, M. (2001) Chem. Commun., 2432–2433. [DOI] [PubMed]
- 25.Creager, S., Yu, C. J., Bamdad, C., O'Connor, S., MacLean, T., Lam, E., Chong, Y., Olsen, G. T., Luo, J., Gozin, M., et al. (1999) J. Am. Chem. Soc. 121, 1059–1064. [Google Scholar]
- 26.Riant, O., Samuel, O., Flessner, T., Taudien, S. & Kagan, H. B. (1997) J. Org. Chem. 62, 6733–6745. [Google Scholar]
- 27.Johnson, W. C. (2000) in Circular Dichroism: Principles and Application, eds. Berova, N., Nakanishi, K. & Woody, R. W. (Wiley–VCH, New York), 2nd Ed., pp. 703–718.
- 28.Sumaoka, J., Pan, F., Nonaka, A., Takeuchi, O., Shigekawa, H. & Komiyama, M. (2004) Chem. Lett. 33, 700–701. [Google Scholar]
- 29.Giese, B. & Wessely, S. (2001) Chem. Commun. 2108–2109. [DOI] [PubMed]
- 30.Kirby, G. M., Batist, G., Fotouhi-Ardakani, N., Nakazawa, H., Yamasaki, H., Kew, M., Cameron, R. G. & Alaoui-Jamali, M. A. (1996) Int. J. Cancer 68, 21–25. [DOI] [PubMed] [Google Scholar]
- 31.Ryberg, D., Kure, E., Lystad, S., Skaug, V., Stangeland, L., Mercy, I., Børresen, A. L. & Haugen, A. (1994) Cancer Res. 54, 1551–1555. [PubMed] [Google Scholar]
- 32.Kelly, S. O., Barton, J. K., Jakson, N. M. & Hill, M. G. (1997) Bioconjugate Chem. 8, 31–37. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.