Abstract
Zinc is a vital micronutrient to all organisms and a potential toxicant to aquatic animals. It is therefore of importance to understand the mechanism of zinc regulation. In the present study, we molecularly cloned and functionally characterized a zinc transporter of the SLC39A family [commonly referred to as the ZIP (Zrt- and Irt-related protein) family] from the gill of zebrafish (Danio rerio) (DrZIP1). DrZIP1 protein was found to localize at the plasma membrane and to function as a zinc uptake transporter when being expressed in either chinook salmon (Oncorhynchus tshawytscha) embryonic 214 cells or Xenopus laevis oocytes. In comparison with pufferfish transporter proteins (FrZIP2 and FrECaC) that are known to facilitate cellular zinc uptake, DrZIP1 appears to have high affinity to bind and transport zinc, suggesting that it may be a high-affinity zinc uptake transporter (Km<0.5 μM) in fish. Orthologues of DrZIP1 were also identified in both freshwater and seawater pufferfish (Tetraodon nigroviridis and Takifugu rubripes), indicating that these proteins may be functionally conserved among different fish species. DrZIP1 mRNA is expressed in all the tissues examined in the present study and thus DrZIP1 may be a constitutive zinc uptake transporter in many cell types of zebrafish.
Keywords: chinook salmon (Oncorhynchus tshawytscha) embryonic 214 cells (CHSE-214 cells), DrZIP1, molecular cloning, pufferfish (Tetraodon nigroviridis and Takifugu rubripes), transport, zebrafish (Danio rerio)
Abbreviations: Caco-2 cells, human Caucasian colon adenocarcinoma cells; CHSE-214 cells, chinook salmon (Oncorhynchus tshawytscha) embryonic 214 cells; ECaC, epithelial calcium channel; EGFP, enhanced green fluorescent protein; ZIP, Zrt- and Irt-related protein; LZT, LIV-1 subfamily of ZIP zinc transporters; MDCK cells, Madin–Darby canine kidney cells; MT, metallothionein; ORF, open reading frame; PKC, protein kinase C; RACE, rapid amplification of cDNA ends; RT–PCR, reverse transcriptase–PCR; TM domain, transmembrane domain
INTRODUCTION
Many macromolecules in cells require zinc as a vital structural cofactor for their functions in processes such as growth control, apoptosis, development and differentiation [1,2]. Due to its functional importance, even moderate zinc deficiency can cause problems including anaemia, loss of appetite, immune system defects, developmental problems and teratogenesis [3]. Conversely, excessive zinc accumulation can be toxic and has been linked to neurodegeneration [4,5]. Furthermore, zinc may be an environmental threat to aquatic organisms and is one among the elements with the highest frequency of water quality criteria violation in Europe and U.S.A. Thus zinc balance has to be precisely controlled to ensure that a proper amount of zinc is kept for the normal physiological functions in organisms.
Zinc uptake across the apical membrane of absorptive epithelial cells is an important step to regulate zinc homoeostasis in vertebrates [6,7]. In fish, there are two routes of metal acquisition: from the diet through the intestine and from water through the gill. Teleost fish gill is an ion transporting epithelium that has long been used as a model to study ionoregulation [8]. Zinc uptake through the fish gill is considered to take place primarily in discrete ‘mitochondria-rich’ ionocytes, called chloride cells [6,9], and is a carrier-mediated transcellular process that follows Michaelis–Menten kinetics [6,10]. Kinetic studies suggest that there may be two different pathways for cellular zinc uptake in fish gill [10–12]. One of them is thought to be a lanthanum-sensitive Ca2+-channel, partly on the basis of the observation that zinc and calcium compete for their entry to gill cells [10–12]. Similarly, competitive uptake of zinc and calcium was also described in the brush border membrane of rat and piglet intestines [13–15]. Recently, we molecularly cloned the ECaC (epithelial calcium channel) genes from pufferfish (Takifugu rubripes) and zebrafish (Danio rerio) gills [16,17]. These fish ECaC genes seem to be mainly expressed in the gill and function to mediate the uptake of both calcium and zinc when expressed in both MDCK cells (Madin–Darby canine kidney cells) and Xenopus laevis oocytes [16,17]. Thus fish ECaC seems to be the candidate protein mediating the competitive pathway of zinc and calcium uptake in the gill. The facts that branchial zinc uptake cannot be completely inhibited by increased waterborne calcium and is less efficiently blocked by lanthanum compared with calcium [6,10], suggest the existence of another unknown zinc uptake pathway in fish gill. This assumption is further supported by the observation that there seems to be no obvious zinc uptake mediated by ECaC in fish intestine [18], as only a very low level of ECaC mRNA could be detected in pufferfish intestine and almost negligible ECaC mRNA was present in zebrafish and rainbow-trout (Oncorhynchus mykiss, formerly Salmo gairdneri) intestines [16,17]. As fish intestine appears to act as the bulk pathway for zinc uptake [19], there must be some unknown specific zinc uptake pathway mediating the intestinal zinc acquisition in fish. However, the molecular nature of the unknown zinc uptake pathways in fish gill and intestine remains unknown.
In recent years, the molecular characterization of zinc uptake transporters from evolutionarily diverse organisms (yeast, plants and mammals) has been achieved [20] and these proteins are from the large ZIP (Zrt- and Irt-related protein) family of metal transporters [21] that has been recently assigned the HUGO name, solute carrier family 39, with the approved symbol SLC39A [22]. The SLC39A family contains four separate subfamilies of metal transporter proteins [23]. The subfamily I is mainly composed of fungal and plant sequences, whereas the subfamily II consists of mammalian, nematode and insect genes [23]. The subfamily III (gufA subfamily) is a group of prokaryote and eukaryote proteins related to the gufA gene of Myxococcus xanthus, whose function is unknown. The fourth subfamily, LZT (LIV-1 subfamily of ZIP zinc transporters), consists of proteins related to the oestrogen-regulated gene, LIV-1, that facilitates cellular zinc uptake in glandular tissues [24]. LIV-1 also exists in the zebrafish along with another LZT member KE4 [23], suggesting that just like their mammalian counterparts, they may be involved in zinc homoeostasis in zebrafish as well. Interestingly, the zebrafish LIV-1 orthologue, LZT-Dr3 (SLC39A6), was recently shown to control the epithelia–mesenchymal transition in the gastrula organizer [25], underscoring the importance of characterizing the function of SLC39A proteins in zebrafish. Although members in the other ZIP subfamilies have not been identified from fish so far, it is probable that these ZIP homologues also exist in teleosts and participate in cellular zinc uptake. In the present study, to investigate the molecular mechanisms of zinc uptake in fish, we molecularly cloned and functionally characterized a ZIP homologue (DrZIP1) from zebrafish gill and this ZIP protein functioned as a high-affinity zinc uptake transporter when expressed in the heterologous expression systems, X. laevis oocytes and CHSE-214 cells [chinook salmon (Oncorhynchus tshawytscha) embryonic 214 cells].
EXPERIMENTAL
Cloning of the ZIP homologues from zebrafish (DrZIP1) and pufferfish (FrZIP1)
Total RNA was isolated from zebrafish gill with TRI Reagent (Sigma–Aldrich, Poole, Dorset, U.K.) and total RNA of pufferfish gill was obtained from HGMP-RC (Human Genome Mapping Project Resource Centre) of the MRC (Medical Research Council, Cambridge, U.K.). Reverse transcription was performed with powerscript reverse transcriptase (ClonTech Laboratories, Basingstoke, U.K.) using the primer ADP1 (Table 1).
Table 1. Primers used in RT–PCR.
| Primer name | Nucleotide sequence (5′→3′) |
|---|---|
| DRF | TGAGCTGTTTCGCAGGAG |
| DRR | CCYTCRAANACNGAGTG |
| DR5GSP | CGCAAGCCATGATAAACTCTG |
| DR5NGSP | AAGTGGGAAGCCATCATCAAG |
| DR3GSP | CTCAGTCAATGATCTGGAGGG |
| DR3NGSP | GGAGTGGACATCACGTTCATGT |
| DRSQF | GGGGTAGTGGAGTGTCAC |
| DRSQR | CTACAGATACTTGATGTCCTGGTC |
| DRORFF | CGCCGAATTCGTCACGAATGGATTACCTGCTTC |
| DRORFR | ATATGGATCCTCAATGGTGATGGTGATGAGGCCCAGAAAACACAGAGC |
| FRF | ATGGAATATCTGCTCCAGG |
| FRR | TCAGCCCAGAAACGACAG |
| FR5GSP | AGCCATGATAAACTCTGGAAG |
| FR5NGSP | GACGCCTCCAGCGAAACAGC |
| FR3GSP | ATGGAATATCTGCTCCAGG |
| FR3NGSP | CAGCTGTTTCGCTGGAGGC |
| UAP1 | TGATCTAGAATTCGCGAAGC |
| UAP2 | AAGCAGTGGTATCAACGCAGAGT |
| ADP1 | CTGATCTAGAATTCGCGAAGC(T)17V |
| DRRTF | GTCACGATGGATTACCTGCTTC |
| DRRTR | GTTCAGCCCAGAAAACACAGAG |
Partial cDNA of DrZIP1 was PCR-amplified from zebrafish gill with primers DRF and DRR (Table 1), and the unknown 5′- and 3′-ends of DrZIP1 cDNA were subsequently amplified by RACE–PCR (where RACE stands for rapid amplification of cDNA ends). For 5′-RACE–PCR, the SMART oligonucleotide II (5′-AAGCAGTGGTATCAACGCAGAGTACGCGGG-3′; ClonTech Laboratories) was used in reverse transcription to produce the first-strand cDNA for 5′-RACE–PCR, and semi-nested 5′-RACE–PCR reactions were performed with gene-specific primers (DR5GSP and DR5NGSP; Table 1) and universal primer 2 (UAP2; Table 1). Semi-nested 3′-RACE–PCR was performed with gene-specific primers (DR3GSP and DR3NGSP; Table 1) and universal primer 1 (UAP1; Table 1). The full DrZIP1 ORF (open reading frame) was then amplified by PCR with primers, DRSQF and DRSQR (Table 1) and a Platinum™ Pfx Taq polymerase (Invitrogen), and its sequence was verified through DNA sequencing on a capillary DNA sequencer (CEQ2000; Beckman, Coulter, High Wycombe, Bucks., U.K.).
MRC HGMP Fugu genomic database [26] was mined with the predicted DrZIP1 protein sequence. Two genomic scaffolds, S000956 and M000624, were identified as they contain the same segments with high homology to DrZIP1 protein sequence. Partial FrZIP1 cDNA containing the predicted ORF was then amplified from pufferfish gill cDNA with primers, FRF and FRR (Table 1), and the 3′- and 5′-ends of FrZIP1 cDNA were amplified by RACE–PCR as described by Dolphin et al. [27]. The 5′-end of FrZIP1 cDNA was amplified by polyadenylic acid tailing and subsequent 5′-RACE–PCR with gene-specific primers (FR5GSP and FR5NGSP; Table 1) and universal primers (ADP1 and UAP2; Table 1). 3′-RACE–PCR was performed to amplify the 3′-end of FrZIP1 cDNA with gene-specific primers (FR3GSP and FR3NGSP; Table 1) and UAP1 (Table 1). The full-length FrZIP1ORF was amplified with Platinum™ Pfx Taq polymerase (Invitrogen) and both of its strands were thoroughly sequenced on a capillary DNA sequencer (CEQ2000; Beckman) to verify its sequence information.
The nucleotide sequences of DrZIP1 and FrZIP1 cDNA have been submitted to GenBank® database (NCBI) with accession numbers AY314992 and AY529485 respectively.
Computational analysis
The alignment of the ZIP proteins and the resulting prediction of phylogenetic relationship were generated with the CLUSTAL W method [28]. Hydrophobicity of DrZIP1 and FrZIP1 was analysed as described by Moller et al. [29]. Potential phosphorylation sites in DrZIP1 and FrZIP1 proteins were predicted with PhosphoBase v2.0 [30].
Expression constructs
For functional expression of DrZIP1, the full-length DrZIP1 ORF was subcloned into the EcoRI and SmaI sites of mammalian expression plasmid vectors, pIRES2EGFP (ClonTech) and pCI-neo (Promega, Chilworth, Southampton, U.K.), to generate pIRES2EGFP-DrZIP1 and pCI-neo-DrZIP1 respectively. The full ORF of DrZIP1 was epitope-tagged by PCR at its C-terminus with a His6 epitope and primers DRORFF and DRORFR (Table 1). The tagged DrZIP1 ORF was then cloned into the EcoRI and BamHI sites of pIRES2EGFP to generate pIRES2EGFP-DrZIP1(H)6. Overlapping PCR was performed to tag the DrZIP1 ORF at its C-terminus with the EGFP (enhanced green fluorescent protein) gene. The produced hybrid gene, DrZIP1EGFP, was then inserted into the EcoRI and NotI sites of the mammalian expression plasmid vector, pCI-neo (Promega), to generate pCI-neo-DrZIP1EGFP. All the inserted genes in the plasmid constructs above were thoroughly sequenced to verify the reading directions and sequence information.
Cell culture
CHSE-214 cells (obtained from A.T.C.C., Manassas, VA, U.S.A.), MDCK and Caco-2 cells (human Caucasian colon adenocarcinoma cells; a gift from Dr A. T. Mckie, King's College, London), were grown in minimum essential medium supplemented with 10% (v/v) fetal bovine serum (Sigma–Aldrich), 1% non-essential amino acids and 2% (v/v) penicillin/streptomycin (Gibco BRL, Paisley, Renfrewshire, Scotland, U.K.). MDCK and Caco-2 cells were cultured in six-well plates (Corning, Corning, NY, U.S.A.) at 37 °C and 5% CO2, whereas CHSE-214 cells were grown at 20 °C and 5% CO2.
SDS-urea/PAGE and Western blotting
pIRES2EGFP-DrZIP1(H)6 and pIRES2EGFP were transiently transfected into CHSE-214, MDCK and Caco-2 cells with Lipofectamine™ reagent (Invitrogen) respectively. After 48 h, these transfected cells were lysed in lysis buffer (10 mM Tris/HCl, pH 7.5, 150 mM NaCl, 2% (w/v) CHAPS, 20 μg·ml−1 leupeptin, 10 μg·ml−1 pepstatin, 10 μg·ml−1 aprotinin, 2 mM PMSF and 0.5 mM 4-[2-aminoethyl[1-benzenesulphonyl]-fluoride]) and subsequently sonicated. Urea was then added to the cell lysate to a final concentration of 8 M.
Protein samples (20 μg total proteins/sample) were resolved by SDS-Urea/PAGE (8 M urea added to SDS/PAGE gel). The proteins in the gel were then blotted on to nitrocellulose membrane (Schleicher and Schuell, Elquevilly, France) and His6-tagged DrZIP1 proteins were immunodetected by mouse anti-His antibody solution (1/2000 dilution) and secondary rabbit-anti-mouse IgG/horseradish peroxidase-conjugate (1/8000 dilution). ECL® Western blotting system (Amersham Biosciences, Little Chalfont, Bucks., U.K.) was used for the luminol-based detection of horseradish peroxidase-conjugated anti-mouse antibodies on the membrane.
Subcellular localization of DrZIP1 proteins
Recombinant expression plasmid, pCI-neo-DrZIP1EGFP, was transfected into CHSE-214 cells with Lipofectamine™ reagent (Invitrogen). Sub-localization of the expressed DrZIP1EGFP fusion proteins in transfected CHSE-214 cells was examined with an inverted phase-contrast fluorescence microscope (Nikon Eclipse 200, Nikon, Tokyo, Japan) 72 h after the start of transfection.
Monitoring of intracellular weakly bound Zn2+ level and luciferase assay
The zinc reporter plasmid pMT-793 [Rainbow trout MT-A promoter (793 bp) with six metal-responsive elements driving luciferase gene; kindly provided by Dr P. E. Olsson] was co-transfected together with expression plasmid constructs with Lipofectamine™ reagent (Invitrogen). After 72 h, these transfected cells were treated with 30 μM ZnSO4 in minimum essential medium (Sigma–Aldrich) for 24 h. Then, the cells were lysed with report lysis buffer (Promega) and the supernatant of the cell lysate was collected for luciferase assay. Luciferase activities were assayed with luciferase assay reagent (Promega) in an LKB 1251 Luminometer. Protein concentration was quantified with Bradford reagent (Sigma–Aldrich) to normalize the luciferase activities.
Stable transfection and 65Zn2+ accumulation assay in CHSE-214 cells
CHSE-214 cells were transfected with expression plasmids, pIRES2EGFP-DrZIP1 and pIRES2EGFP with Lipofectamine™ reagent (Invitrogen) and clonally derived stable transfectant CHSE-214 cell lines were generated by the combined selection of G418 and green fluorescence.
The cells with 80% confluence in six-well plates were washed with prewarmed uptake buffer (150 mM KCl, 100 mM glucose and 10 mM Hepes, pH 7.0) [31] three times and then preincubated in prewarmed uptake buffer for 10 min at 20 °C before assays. For time-dependent 65Zn2+ accumulation, CHSE-214 cells were incubated in 1 ml of prewarmed uptake buffer (150 mM KCl, 100 mM glucose and 10 mM Hepes, pH 7.0), containing 0.5 μM ZnCl2 and 0.033 μCi of 65Zn2+ at 20 °C for 1, 5, 10, 20, 40, 60 and 90 min respectively. Concentration-dependent 65Zn2+ accumulation was measured by incubating CHSE-214 cells in 1 ml of prewarmed uptake buffer containing ZnCl2 at concentrations of 0.033, 3, 10 or 20 μM plus 65Zn2+ tracer at 20 °C for 40 min. Assays were stopped by removing the uptake buffer and washing the cells three times in ice-cold washing buffer (150 mM KCl, 100 mM glucose, 10 mM Hepes and 5 mM EDTA, pH 7.0). The cells in each well were then lysed in 1 ml of 1 M NaOH. After a thorough mixing, 50 μl of cell lysate from each well was diluted 50 times for protein quantification with Bradford reagents and 0.9 ml was taken for radioactivity counting in an LKB1282 CompuGamma counter.
DrZIP1 expression and 65Zn2+ flux in X. laevis oocytes
The full-length DrZIP1 ORF was excised from pIRES2EGFP-DrZIP1 and inserted into pPSPT-18 plasmid vector (Roche Biochemicals, Welwyn Garden City, U.K.) to generate pSPT-18-DrZIP1, in which transcription of DrZIP1 is under the control of the T7 promoter. The capped DrZIP1 mRNA was synthesized by in vitro transcription using mMACHINE™ T7 Ultra kit® (Ambion, Austin, TX, U.S.A.).
X. laevis oocytes at developmental stage IV were isolated and the capped DrZIP1 mRNA (∼30 ng/oocyte) were injected into them. Some oocytes were injected with nuclease-free water as control. After 72 h, 65Zn2+ influx was assayed by incubating 12 injected oocytes of each group in 500 μl of influx buffer (73 mM NaCl, 1 mM KCl, 1 mM MgCl2, 10 mM NaHCO3 and 15 mM Hepes, pH 7.0) containing 1.26 μM 65Zn2+ (6 μCi·ml−1) at 18 °C for 2 h. The flux of oocytes were washed thoroughly six times in ice-cold stop buffer (influx buffer supplemented with 100 μM ZnCl2) and dispersed individually into each 0.5 ml tube. The radioactivity of each oocyte was counted in an LKB1282 CompuGamma counter.
The statistical significance of differences between groups was determined by Student's t test (SPSS software, SPSS, San Rafael, CA, U.S.A.) and groups were considered significantly different at P<0.05.
RT–PCR (reverse transcriptase–PCR)
Total RNA was isolated from gill, brain, intestine, kidney, eye, ovary, skin, fin, heart and muscle of ten zebrafish individuals. RT–PCR reactions were performed to amplify DrZIP1 fragments by using DrZIP1-specific primers, DRF and DRR (Table 1). The cycle conditions were: an initial cycle of 2 min at 94 °C, 30 cycles of 0.5 min at 94 °C, 0.5 min at 60 °C and 1 min at 72 °C, followed by a final extension cycle of 2 min at 72 °C. House-keeping gene, zebrafish β-actin (GenBank® accession no. AF057040), was analysed by RT–PCR (25 cycles) as a control.
Total RNA of pufferfish gill, gut, kidney and ovary was obtained from HGMP-RC of the MRC. The level of FrZIP1 mRNA was analysed by RT–PCR using FrZIP1-specific primers, FRF and FRR (Table 1). Cycle parameters: an initial cycle of 2 min at 94 °C, 30 cycles of 94 °C for 0.5 min, 55 °C for 0.5 min and 72 °C for 2 min, followed by a single cycle of 72 °C for 2 min. As a control, a 0.5 kb fragment of β-actin (Ensembl ID: SINFRUG00000081338) was amplified by RT–PCR (25 cycles) using primers which are specific to sequences in exons three and five respectively of pufferfish β-actin.
The expression levels of DrZIP1 and FrZIP1 mRNA were normalized to β-actin mRNA by using SigmaGel software (Jandel Scientific, Madera, CA, U.S.A.).
RESULTS
Structural characterization of DrZIP1
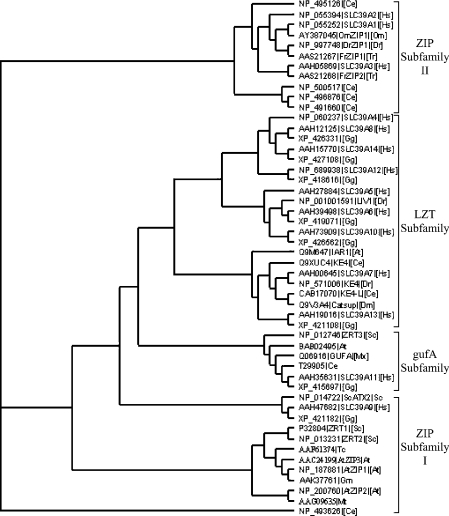
A zebrafish cDNA, which is predicted to encode a 302 amino acid protein of 33 kDa with obvious homology to mammalian ZIP transporters (Figure 1), was molecularly cloned by the combination of database mining and degenerate PCR, and we termed it DrZIP1. The predicted DrZIP1 protein shows significant identity with members of the ZIP family, for example, approx. 38% with the human ZIP1 and ZIP2 transporters (SLC39A1 and SLC39A2) (Figure 1). Furthermore, when plotted in a phylogenetic tree of the SLC39A family, DrZIP1 is found to be more similar to human SLC39A1, SLC39A2 and SLC39A3 proteins compared with members of other ZIP clades, including zebrafish LIV-1 and KE4 in the LZT subfamily and fungal and plant ZIPs in the ZIP subfamily I. DrZIP1 forms a cluster together with human SLC39A1-3, rainbow trout ZIP1 (OmZIP1) and pufferfish ZIP1 (FrZIP1) and ZIP2 (FrZIP2) proteins in ZIP subfamily II (Figure 2).
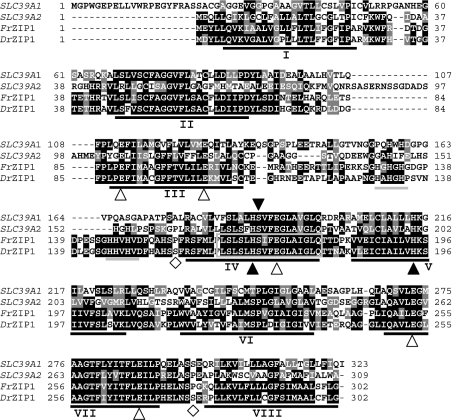
Figure 1. Structural relationship of DrZIP1 and FrZIP1 with human SLC39A1 and SLC39A2.
The predicted DrZIP1 and FrZIP1 proteins were aligned with human SLC39A1 and SLC39A2. The eight predicted TM domains are underlined and numbered I–VIII. Shaded residues represent positions of identity or similarity among the sequences as compared with DrZIP1 and FrZIP1. Two potential histidine-rich metal binding motifs between TM III and TM IV are underlined in grey. The signature sequences are highlighted in grey. ▲, conserved histidine and serine residues; △, glutamate residues conserved between DrZIP1, FrZIP1, SLC39A1 and SLC39A2; ◇, potential phosphorylation sites of PKC.
Figure 2. Phylogenetic relationship of DrZIP1 and FrZIP1 with some representative members of the SLC39 family.
Four subfamilies of the SLC39 family are named by Taylor and Nicholson [23]. DrZIP1 and FrZIP1 are members of the ZIP subfamily II. At, Arabidopsis thaliana (thale cress); Ce, the nematode worm Caenorhabditis elegans; Dm, Drosophila melanogaster (fruitfly); Dr, Danio rerio (zebrafish); Gg, Gallus gallus (chicken); Gm, Glycine max (soybean); Hs, Homo sapiens (human); Mt, Medicago truncatula (barrel medic); Mx, the Gram-negative bacterium M. xanthus; Om, Oncorhynchus mykiss (rainbow trout); Sc, S. cerevisiae (baker's yeast); Tc, Thlaspi caerulescens (stone cress); and Tr, Ta. rubripes (seawater pufferfish).
Like the functionally characterized ZIP proteins (Zrt1, Zrt2, human SLC39A1 and SLC39A2, etc.), DrZIP1 protein is predicted to have eight TM (transmembrane) domains and both the N- and C-terminus located on the extracellular surface of the cell (Figure 1). A number of other structural features in DrZIP1 are conserved in other functional ZIPs. Two potential metal binding motifs containing three histidine residues (…HA/GHGH… and …HHVHV…) are found in the histidine-rich variable region between TM III and IV (Figure 1). These motifs are conserved in many of the characterized ZIP members including human SLC39A1 and SLC39A2 (Figure 1) and are proposed to play important roles in metal binding. Just like the functionally characterized ZIP proteins, TM IV and V of DrZIP1 protein are amphipathic and each possesses a central, fully conserved histidine residue on its hydrophilic side (Figure 1). These histidine residues are considered to be providing an intramembranous zinc-binding site and to be of functional importance [32]. TM IV of DrZIP1 proteins contain a serine residue that is conserved in most of the characterized ZIP family members (Figure 1). The conserved histidine and serine residues are predicted to lie on the polar face of amphipathic α-helices with a possible role in metal ion transport. Additionally, in the DrZIP1 protein, there are correspondingly two aspartate residues in TM II, two glutamate residues in TM III and VII, and one aspartate residue in TM IV and V respectively (Figure 1). The aspartate residues in TM III, IV and VII are also conserved in human SLC39A1 and SLC39A2 proteins (Figure 1). Aspartate and glutamate residues are moderately strong biological ligands for zinc ions [33]. The DrZIPl protein has very short, cytoplasmic N- and C-termini, which also feature the functionally characterized ZIP proteins (Figure 1). Importantly, a typical signature sequence of the SLC39A family is identified in the TM IV of DrZIP1 (LSLSLHSVFEGLAIG) protein (Figure 1). All these structural key features together with the overall amino acid sequence suggest that DrZIP1 belongs to the SLC39 family and may function as a zinc uptake transporter just like the functionally characterized ZIP proteins in yeast, plant and mammals.
As shown in Figure 1, DrZIP1 protein has two potential PKC (protein kinase C) phosphorylation sites (residues 153–159 between TM III and IV and 275–281 between TM II and VIII), suggesting the possible existence of post-translational regulation on DrZIP1 functions. However, there is no predictable N-glycosylation site in DrZIP1 protein (Figure 1).
On the basis of mining of pufferfish genomic database (http://fugu.hgmp.mrc.ac.uk), genes orthologous to DrZIP1 can also be identified in seawater and freshwater pufferfish, Takifugu rubripes (Contig FS: S000956) and Tetraodon nigroviridis (FS_CONTIG_7691_2). Thus ZIP1 seems to be present in both freshwater and seawater fish. The molecularly cloned full-length FrZIP1 cDNA contains an ORF of 909 bp encoding a protein of 302 amino acids with a predicted mass of 33.4 kDa (Figure 1). It is therefore not surprising that the predicted FrZIP1 protein shares approx. 73% identity with DrZIP1 (Figure 1).
Protein expression of DrZIP1 and its subcellular localization
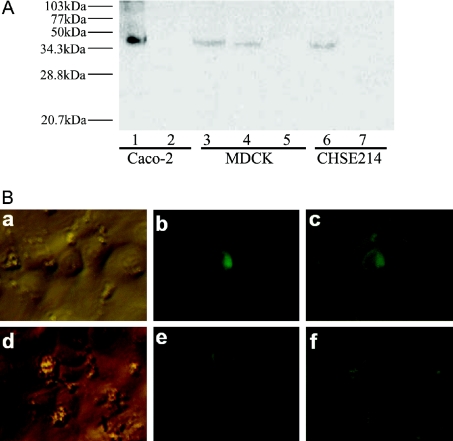
To examine if DrZIP1 protein can be expressed in cultured cells, we tagged the DrZIP1 ORF with a His6 epitope at its C-terminus to generate DrZIP1(H)6, which was subsequently inserted into pIRES2EGFP to create pIRES2EGFP-DrZIP1(H)6 for expression from the cytomegalovirus promoter. CHSE-214, MDCK and Caco-2 cells were transfected with pIRES2EGFP-DrZIP1(H)6, and DrZIP1(H)6 protein was subsequently examined by SDS-Urea/PAGE and Western blotting. As shown in Figure 3(A), a single protein band was detected in lanes containing proteins from pIRES2EGFP-DrZIP1(H)6-transfected Caco-2, MDCK and CHSE-214 cells, but there is no such band in other lanes containing proteins from vector-only transfected Caco-2, MDCK and CHSE-214 cells. When compared with protein markers, the molecular mass of the detected DrZIP1(H)6 protein is approx. 40 kDa, which is a little bigger than the 33.6 kDa of the predicted DrZIP1 protein (Figure 3A).
Figure 3. Expression of DrZIP1 protein in cultured cells and its subcellular localization in CHSE-214 cells.
(A) Detection of DrZIP1(H)6 protein expressed in Caco-2, MDCK and CHSE-214 cells by SDS-Urea/PAGE and Western blotting. Lanes 1, 3 and 6: DrZIP1(H)6-expressed cells; lane 4: stable MDCK-DrZIP1(H)6 cells; and lanes 2, 5 and 7: negative control cells transfected with pIRES2EGFP. (B) Subcellular localization of DrZIP1 proteins expressed in CHSE-214 cells. The plasmid construct, pCI-neo-DrZIP1EGFP, was transfected into CHSE-214 cells, and the subcellular localization of the expressed DrZIP1EGFP protein in CHSE-214 cells was examined with an inverted fluorescent microscope. a and d, bright light; b and e, blue light; c and f, overlapping of pictures from both bright and blue light.
The immunodetection of DrZIP1(H)6 protein suggests that DrZIP1(H)6 can be successfully expressed as a full-size protein in Caco-2, MDCK and CHSE-214 cells. As the only difference between expression constructs pIRES2EGFP-DrZIP1(H)6 and pIRES2EGFP-DrZIP1 is the His6 epitope, which was tagged at the C-terminus of DrZIP1 ORF, it can be supposed that a full-length DrZIP1 protein can also be produced in these cells.
As a hypothesized zinc uptake transporter, DrZIP1 proteins were predicted to function mainly at the plasma membrane. To test this hypothesis, we tagged the DrZIP1 ORF at the C-terminus with an EGFP gene to produce hybrid gene, DrZIP1EGFP, which was subsequently cloned into pCI-neo to generate recombinant expression plasmid pCI-neo-DrZIP1EGFP. After 3 days from the transfection with pCI-neo-DrZIP1EGFP, a bright rim of green fluorescence was observed at the periphery of CHSE-214 cells that were expressing DrZIP1EGFP, whereas the other cells without DrZIP1EGFP expression showed no fluorescence (Figure 3B). This result suggested plasma membrane localization of the EGFP-tagged DrZIP1 proteins in CHSE-214 cells. As EGFP itself is a soluble protein [34] and DrZIP1 is predicted to be a membrane protein on the basis of computational analysis, the sorting of DrZIP1EGFP proteins in cultured cells may mainly depend on the structural properties of DrZIP1. Thus the subcellular localization of DrZIP1EGFP at plasma membrane of CHSE-214 cells probably represents the real location of DrZIP1 proteins expressed in CHSE-214 cells. In the cell shown in Figure 3(B), panels b and c, there appears to be a concentrated spot of potentially intracellular fluorescence in addition to the pericellular rim. It is not known whether or not this perhaps represents a pool of intracellular DrZIP1.
DrZIP1 as a zinc-binding protein of high affinity
The hypothesized role of DrZIP1 as a zinc uptake transporter suggests that DrZIP1 protein can bind to Zn2+ and enable it to cross the plasma membrane into cells, and therefore increase the cellular zinc accumulation. To examine the assumed zinc-binding and transporting properties of DrZIP1, we chose a zinc reporter system to detect the effects of DrZIP1 expression on intracellular weakly bound Zn2+ concentration, and this system includes a zinc-sensitive reporter plasmid, pMT793, and CHSE-214 cells. pMT793 contains a luciferase reporter gene driven by a rainbow trout MT (metallothionein)-A promoter (793 bp) [35]. The MT-A promoter contains six metal-responsive elements as transcription enhancers that are very sensitive to intracellular weakly bound Zn2+ and can be greatly activated by elevated intracellular weakly bound Zn2+ through MTF-1 (metal-responsive transcription factor-1) [35,36]. Thus the fluctuation of intracellular weakly bound Zn2+ can be represented as the change of luciferase activities in the pMT793-transfected cells. CHSE-214 cells are chosen due to their deficiency of functional expression of zinc-binding proteins and MT, and consequently high sensitivity to zinc [37,38].
To express DrZIP1 in CHSE-214 cells, the full-length ORF of DrZIP1 was inserted into the mammalian expression vector, pIRES2EGFP, to generate pIRES2EGFP-DrZIP1. As a control, pufferfish ECaC (FrECaC) and ZIP2 (FrZIP2) were also cloned into pIRES2EGFP to generate pIRES2EGFP-FrZIP2 and pIRES2EGFP-FrECaC respectively. Both FrECaC and FrZIP2 have been shown to stimulate zinc uptake when individually expressed in MDCK cells.
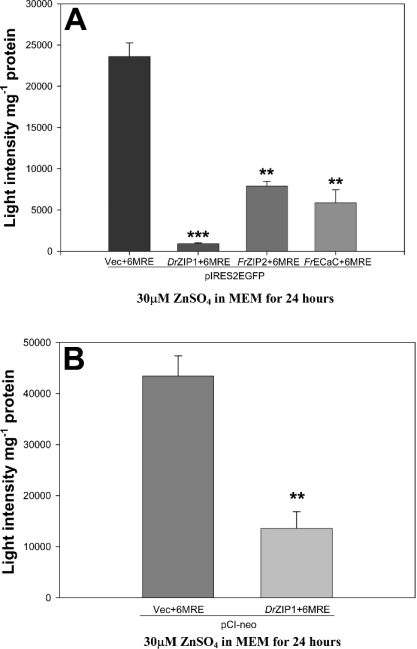
pIRES2EGFP-DrZIP1, pIRES2EGFP-FrZIP2, pIRES2EGFP-FrECaC or pIRES2EGFP was co-transfected with pMT793 into CHSE-214 cells respectively. After 72 h, the transfected cells were treated with 30 μM ZnSO4 for 24 h and luciferase activity was subsequently assayed to examine the effects of DrZIP1, FrZIP2 and FrECaC expression on the intracellular weakly bound Zn2+ concentration of CHSE-214 cells. As shown in Figure 4(A), luciferase activities were 28 times lower in pIRES2EGFP-DrZIP1-transfected CHSE-214 cells compared with pIRES2EGFP-transfected CHSE-214 cells, suggesting that the expression of DrZIP1 largely decreased the weakly bound Zn2+ concentration in CHSE-214 cells. Just like DrZIP1, expression of FrECaC and FrZIP2 also exhibited similar effects on the weakly bound Zn2+ level when expressed in CHSE-214 cells by lowering luciferase activities approx. four and three times in comparison with pIRES2EGFP-transfected CHSE-214 cells (Figure 4A). Comparatively, DrZIP1 showed approx. eight and six times higher ability to decrease the luciferase activity compared with FrZIP2 and FrECaC in CHSE-214 cells respectively (Figure 4A). To exclude the possibility of the pIRES2EGFP effects on the luciferase activity in CHSE-214 cells, we used another mammalian expression plasmid vector, pCI-neo, to replace pIRES2EGFP for the overexpression of DrZIP1 from cytomegalovirus promoter. Concordantly, expression of DrZIP1 in pCI-neo still decreased luciferase activities by approx. three times in CHSE-214 cells when compared with control cells transfected with pCI-neo (Figure 4B). These results suggest that DrZIP1, FrZIP2 and FrECaC proteins have significant zinc-binding abilities that can decrease intracellular weakly bound Zn2+ level when CHSE-214 cells are exposed to excessive zinc, and that DrZIP1 protein displays much higher affinity to bind zinc compared with FrZIP2 and FrECaC.
Figure 4. Effects of DrZIP1 expression on luciferase activities in CHSE-214 cells.
(A) Luciferase activities in CHSE-214 cells that were co-transfected with zinc-reporter plasmid, pMT-793, and one among pIRES2EGFP-DrZIP1, pIRES2EGFP-FrZIP2, pIRES2EGFP-FrECaC and pIRES2EGFP, and subsequently treated with 30 μM ZnSO4 for 24 h. (B) Luciferase activities in CHSE-214 cells, that were co-transfected with pMT-793 and either pCI-neo-DrZIP1 or pCI-neo, and subsequently treated with 30 μM ZnSO4 for 24 h. Values are means±S.E.M. (n=3). Significant differences between experimental groups (P<0.05) were determined using Mann–Whitney U test.
Functional characterization of DrZIP1 expressed in CHSE-214 cells
To examine whether the expression of DrZIP1 can directly stimulate zinc uptake, we generated stable transfectant CHSE-214 cells overexpressing DrZIP1, hereafter referred to as CHSE-214-DrZIP1. The stable overexpression of DrZIP1 in CHSE-214 cells was demonstrated on the basis of the examination of EGFP proteins using an inverted fluorescence microscope. As we failed to generate the vector-only stable transfectant CHSE-214 cell line, the untransfected CHSE-214 cells, referred to as CHSE-214 cells, were thus used as the control in the assays of 65Zn2+ accumulation. CHSE-214 cells may share the same properties of zinc uptake as vector-only stable transfectant CHSE-214 cells as it has been shown that transfection with GFP alone has no effects on zinc uptake activity in human K562 erythroleukaemia cells [39].
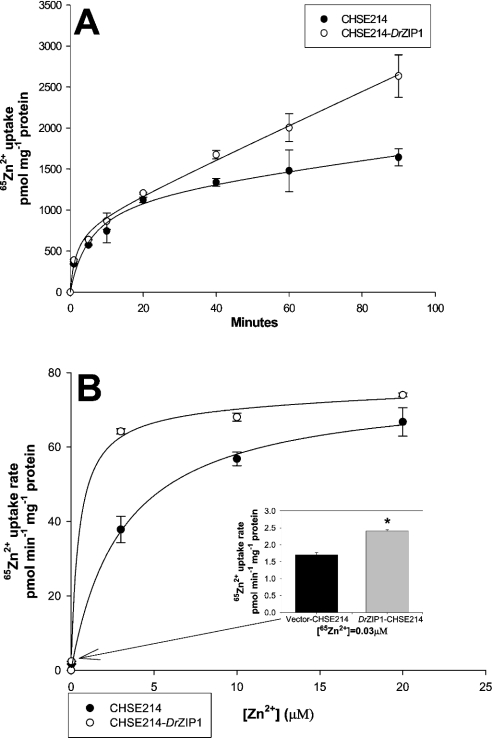
Examination of 65Zn2+ accumulation in CHSE-214 cells indicated the presence of endogenous zinc uptake systems. The endogenous zinc uptake is a saturable process in a time- and concentration-dependent fashion (Figures 5A and 5B). When assayed over a range of zinc concentrations (0.03–20 μM), this endogenous system showed Michaelis–Menten kinetics with an apparent Michaelis constant Km of 3 μM zinc and a Jmax (the maximum flux of substrate in an enzymatic assay) of 76 pmol Zn2+·min−1·(mg of protein)−1 (Figure 5B).
Figure 5. Functional expression of DrZIP1 in CHSE-214 cells.
(A) Zinc accumulation was assayed by incubating CHSE-214-DrZIP1 and CHSE-214 cells in uptake buffer containing 0.5 μM ZnCl2 plus 0.033 μCi 65Zn2+ at 20 °C for either 1, 5, 10, 20, 40, 60 or 90 min. (B) Zinc accumulation rate was determined by incubating cells in the uptake buffer containing a range of substrate concentrations at 20 °C for 40 min. Values are means±S.E.M. (n=3). Significant differences between experimental groups (*P<0.05) were determined using Mann–Whitney U test.
Similarly, 65Zn2+ accumulation in CHSE-214-DrZIP1 cells is also a saturable process in a time- and concentration-dependent way. 65Zn2+ accumulation was significantly increased in CHSE-214-DrZIP1 cells that accumulated approx. 1.6-fold higher zinc compared with CHSE-214 cells over 90 min (Figure 5A). Zinc accumulation in CHSE-214-DrZIP1 cells was also a concentration-dependent and saturable process with an apparent Km of 0.5 μM zinc and a Jmax of 75 pmol Zn2+·min−1·(mg of protein)−1 (Figure 5B). In comparison with wild-type CHSE-214 cells, expression of DrZIP1 markedly stimulated zinc uptake (1.5-fold) in CHSE-214-DrZIP1 cells at low zinc concentration in the uptake buffer (<2.5 μM), whereas this stimulation decreased at higher zinc concentration (>2.5 μM). Furthermore, the Km of zinc accumulation in CHSE-214-DrZIP1 cells is approx. six times lower than the endogenous level in CHSE-214 cells, whereas the Jmax of zinc accumulation in both cells is similar, suggesting that expression of DrZIP1 increased the affinity of CHSE-214 cells for zinc 6-fold. Thus when expressed in CHSE-214 cells, DrZIP1 can function as a zinc uptake transporter possibly with high affinity for zinc.
Functions of DrZIP1 expressed in X. laevis oocytes
For cross-examination of the zinc-transporting activity of DrZIP1, a classical robust expression system, X. laevis oocytes, was employed to express DrZIP1 followed by 65Zn2+ influx assays.
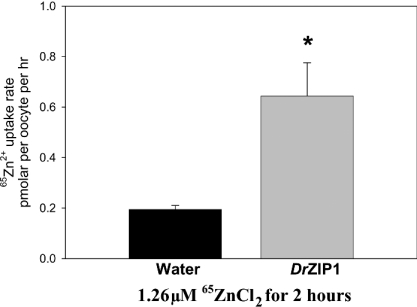
As shown in Figure 6, DrZIP1-injected oocytes accumulated approx. three times higher 65Zn2+ compared with water-injected ones after being incubated in a flux buffer containing 1.26 μM 65Zn2+ at 18 °C for 2 h. Thus DrZIP1 functioned as a zinc uptake transporter when expressed in X. laevis oocytes.
Figure 6. Functional characterization of DrZIP1 in X. laevis ooctyes.
A total of 12 oocytes injected with either water or DrZIP1 mRNA were incubated in influx buffer containing 1.26 μM 65ZnCl2 (6 μCi·ml−1) at 18 °C for 2 h. Values are expressed as means±S.E.M. (n=12). Significant differences between experimental groups (*P<0.05) were determined with Student's t test.
Expression distribution of DrZIP1 and FrZIP1 mRNA in various tissues
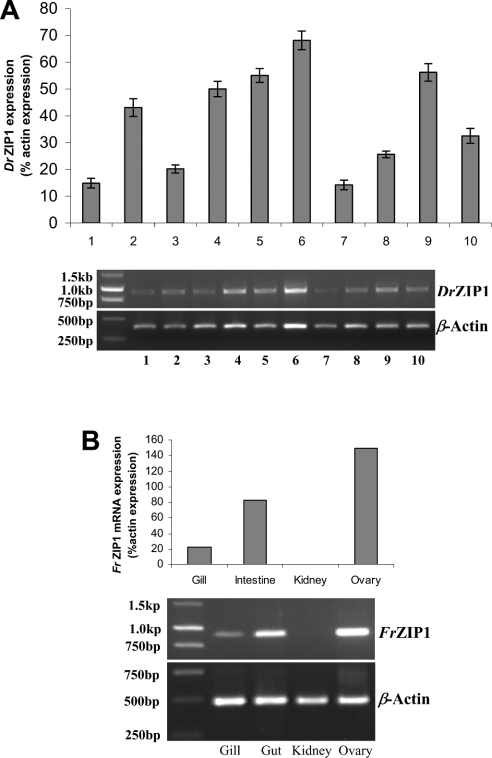
DrZIP1 mRNA is ubiquitously expressed in all tissues of ten individual zebrafish herein examined. The highest level of DrZIP1 mRNA was detected in the ovary (Figure 7A), whereas substantial DrZIP1 mRNA levels were also detected in the heart, eye, kidney and brain (Figure 7A). There were relatively modest levels of DrZIP1 mRNA in the intestine and the lowest levels of DrZIP1 mRNA were found in gill and skin (Figure 7A).
Figure 7. Tissue distribution of DrZIP1 and FrZIP1 mRNA in zebrafish and pufferfish.
(A) DrZIP1 mRNA levels, zebrafish gill (1), brain (2), intestine (3), kidney (4), eye (5), ovary (6), skin (7), fin (8), heart (9), and muscle (10) were determined by RT–PCR for 30 cycles. Upper panel: relative amount of DrZIP1 mRNA to β-actin mRNA in various tissues; and lower panel: expression pattern of DrZIP1 and β-actin mRNA in agarose gels. Values are means±S.E.M. (n=10), significant difference (P<0.05) between groups was tested with ANOVA followed by post-hoc test. (B) Levels of FrZIP1 mRNA in pufferfish gill, intestine, kidney and ovary were determined by RT–PCR for 30 cycles. Upper panel: relative amount of FrZIP1 mRNA to β-actin mRNA in various tissues; and lower panel: expression pattern of FrZIP1 and β-actin mRNA in agarose gels.
Similarly, in pufferfish, the highest level of FrZIP1 mRNA was detected in the ovary and the lowest in the gill (Figure 7B). As in the zebrafish, the FrZIP1 mRNA level was higher in the intestine compared with the gill (Figure 7B). However, extremely low levels of FrZIP1 mRNA seem to be present in the kidney (Figure 7B).
DISCUSSION
Zinc is both a vital metal nutrient and a problematic toxicant to aquatic biota. To understand the interplay between nutrition and toxicity, it will be important to determine the mechanisms that regulate zinc uptake into the body and organs. In yeast, plants and mammals, ZIP members (SLC39A) have been identified to play prominent roles in cellular zinc uptake [20]. However, it remains to be studied which proteins are functionally responsible for cellular zinc uptake in fish, although fish ZIP homologues have been predicted to exist in fish to perform similar roles [19]. In the present study, to understand the molecular mechanism of zinc uptake in fish cells, we cloned a novel ZIP homologue from zebrafish gill, DrZIP1. Orthologues of DrZIP1 were also identified in both seawater and freshwater pufferfish (Ta. rubripes and Te. nigroviridis) genomes, as Takifugu ZIP1 (FrZIP1) shares a high amino acid identity (73%) with DrZIP1 throughout the protein, especially in the predicted TM domains. Thus fish ZIP1 may be well conserved and functionally important among fish species.
DrZIP1 belongs to the ZIP family and is a zinc uptake transporter in fish based on its structural and functional properties. Similar to the ZIP members in yeast, plants and mammals, DrZIP1 contains all the typical conserved structural features of the ZIP family, including eight TM domains, a potential cytoplasmic metal binding domain between TM III and IV and functionally important histidine residues in TM IV and TM V. Moreover, DrZIP1 protein also shares a significant amino acid similarity with the known members of the ZIP family, especially mammalian ZIP proteins such as human SLC39A1 and SLC39A2. As both human SLC39A1 and SLC39A2 have been functionally characterized as zinc uptake transporters when expressed in human erythroleukaemia K562 cells [31,39], DrZIP1 may play a similar role in cellular zinc transport in fish. Furthermore, functional expression analysis suggested that DrZIP1 protein can be expressed as a membrane protein in cultured cells and identified to be located at the plasma membrane in CHSE-214 cells, a fish embryonic cell line. The plasma membrane localization of DrZIP1 protein provided the functional basis at cellular level for its role in cellular zinc uptake. The role of DrZIP1 as a zinc uptake transporter is subsequently confirmed by the experimental facts that expression of DrZIP1 stimulated zinc uptake in both CHSE-214 cells and X. laevis oocytes.
Binding zinc, at least temporarily, should be an essential structural property of zinc transporters to facilitate the transport of zinc across cell membrane and this property may therefore influence the affinity of zinc transporters for zinc. Like the known zinc uptake transporters, FrZIP2 and FrECaC [16,17], DrZIP1 behaved like a zinc-binding protein by decreasing the intracellular weakly bound zinc level when being overexpressed in zinc-treated CHSE-214 cells. However, in comparison with FrZIP2 and FrECaC proteins, DrZIP1 exhibited much higher binding affinity for zinc (decreasing luciferase activities eight and six times when compared with FrZIP2 and FrECaC respectively), suggesting that it may represent a zinc uptake transporter with much higher affinity for zinc in fish. Consistently, the function of DrZIP1 as a high-affinity zinc importer is also supported by the observation that stable expression of DrZIP1 increased the endogenous affinity of CHSE-214 cells for zinc approx. six times by decreasing the Km from 3 to 0.5 μM.
DrZIP1 markedly stimulated zinc uptake of CHSE-214 cells at lower zinc levels but no difference in zinc influx between DrZIP1 and mock transfected cells was observed at higher extracellular zinc concentrations. Thus DrZIP1 enhanced zinc affinity of CHSE-214 cells but had no effect on the endogenous maximal capacity of zinc uptake. A possible reason for this feature of DrZIP1 is the post-translational modification of its zinc uptake activity in response to increased external zinc levels, just like the yeast high-affinity zinc uptake transporter, Zrt1. In yeast Saccharomyces cerevisae, after zinc treatment, Zrt1 protein can be quickly ubiquitinated, recruited into the endocytic pathway and removed from the cell surface [40,41]. Without this inactivation, total cellular zinc levels could increase from less than 100 μM to nearly 100 mM within 1 h of treating a zinc-limiting yeast cell with zinc [42]. Similarly, mouse ZIP4 protein expressed in human embryonic kidney 293 cells is post-translationally inactivated in response to prompt zinc treatment [43]. The proposed post-translational inactivation of DrZIP1 protein after zinc treatment is supported by the existence of two predicted PKC phosphorylation sites in DrZIP1 protein, including a serine residue between TM III and IV. In yeast, the phosphorylation of a serine residue between TM III and IV of Zrt1 protein in response to zinc is proposed to be the pathway of the ubiquitinated inactivation of Zrt1 protein [42]. However, it remains to be demonstrated how the functions of DrZIP1 are post-translationally regulated.
The ubiquitous expression of DrZIP1 mRNA, coupled with its function as a zinc importer, suggests that DrZIP1 may be a major constitutive zinc uptake transporter in many cells of the zebrafish. The highest level of ZIP1 mRNA exists in the ovary of both zebrafish and pufferfish, which is consistent with the observation that high levels of zinc are accumulated in fish ovary during oocyte development [44,45]. Moreover, the levels of the main zinc-binding proteins, MT, began to increase at the onset of vitellogenesis, and levels peaked (approx. 7-fold of basic levels) when spawning occurred [46,47]. Substantial levels of DrZIP1 mRNA also exist in heart, eye, kidney and brain of zebrafish. Eye and brain are known to have a high-concentration of zinc [6,48] and kidney is an important organ to reabsorb zinc from the urine in freshwater fish since there is an extremely low urinary excretion rate for zinc [49]. In contrast, we found no FrZIP1 mRNA in pufferfish kidney. To counteract the passive diffusion of salts from seawater, marine teleosts such as the pufferfish kidney excrete divalent ions [50]. There appears to be no evidence for renal zinc reabsorption in marine fish. In both fish species, ZIP1 mRNA is expressed at higher levels in the intestine when compared with the levels in gill. During standard conditions of divalent ions, the intestine is thought to act as the major pathway for zinc uptake and the gill to supplement absorption when dietary zinc levels become limited [19]. With decreasing dietary zinc levels, the gill becomes increasingly important, especially when waterborne zinc levels are increased [51]. Thus it can be speculated that ZIP1 mRNA in fish gill could be up-regulated when dietary zinc becomes limited and availability of waterborne zinc is increased.
Acknowledgments
This work was supported by K. C. Wong Education Foundation, a Chinese Schorlarship to A.Q. and an U.S. EPA grant (R 826104-01-1) to C.H. The authors thank Dr A. T. Mckie, King's College London, for providing facilities and valuable advice for the Xenopus laevis oocytes expression experiments.
References
- 1.Vallee B. L., Auld D. S. Zinc coordination, function, and structure of zinc enzymes and other proteins. Biochemistry. 1990;29:5647–5659. doi: 10.1021/bi00476a001. [DOI] [PubMed] [Google Scholar]
- 2.Wu F. Y., Wu C. W. Zinc in DNA replication and transcription. Annu. Rev. Nutr. 1987;7:251–272. doi: 10.1146/annurev.nu.07.070187.001343. [DOI] [PubMed] [Google Scholar]
- 3.MacDiarmid C. W., Gaither L. A., Eide D. J. Zinc transporters that regulate vacuolar zinc storage in Saccharomyces cerevisiae. EMBO J. 2000;19:2845–2855. doi: 10.1093/emboj/19.12.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koh J. Y., Suh S. W., Gwag B. J., He Y. Y., Hsu C. Y., Choi D. W. The role of zinc in selective neuronal death after transient global cerebral ischemia. Science. 1996;272:1013–1016. doi: 10.1126/science.272.5264.1013. [DOI] [PubMed] [Google Scholar]
- 5.Frederickson C. J., Bush A. I. Synaptically released zinc: physiological functions and pathological effects. Biometals. 2001;14:353–366. doi: 10.1023/a:1012934207456. [DOI] [PubMed] [Google Scholar]
- 6.Hogstrand C., Wood C. M. The physiology and toxicology of zinc in fish. In: Taylor E. W, editor. Toxicology of Aquatic Pollution. Cambridge: Cambridge University Press; 1996. pp. 61–84. [Google Scholar]
- 7.King J. C., Shames D. M., Woodhouse L. R. Zinc homeostasis in humans. J. Nutr. 2000;130:1360S–1366S. doi: 10.1093/jn/130.5.1360S. [DOI] [PubMed] [Google Scholar]
- 8.Zia S., McDonald D. G. Role of the gills and gill chloride cells in metal uptake in the freshwater-adapted rainbow trout, Oncorhynchus mykiss. Can. J. Fish. Aquat. Sci. 1994;51:2482–2492. [Google Scholar]
- 9.McCormick S. D., Hasegawa S., Hirano T. Calcium-uptake in the skin of a fresh-water teleost. Proc. Natl. Acad. Sci. U.S.A. 1992;89:3635–3638. doi: 10.1073/pnas.89.8.3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spry D. J., Wood C. M. A kinetic method for the measurement of zinc influx in vivo in the rainbow trout, and the effects of waterborne calcium on flux rates. J. Exp. Biol. 1989;142:425–446. [Google Scholar]
- 11.Bentley P. J. Influx of zinc by channel catfish (Ictaurus punctatus): uptake from external environmental solutions. Comp. Biochem. Physiol. C. 1992;101:215–217. doi: 10.1016/0742-8413(92)90263-7. [DOI] [PubMed] [Google Scholar]
- 12.Hogstrand C., Verbost P. M., Bonga S. E. W., Wood C. M. Mechanisms of zinc uptake in gills of freshwater rainbow trout: Interplay with calcium transport. Am. J. Physiol. 1996;270:R1141–R1147. doi: 10.1152/ajpregu.1996.270.5.R1141. [DOI] [PubMed] [Google Scholar]
- 13.Rothbassell H. A., Clydesdale F. M. The influence of zinc, magnesium, and iron on calcium-uptake in brush-border membrane-vesicles. J. Am. Coll. Nutr. 1991;10:44–49. doi: 10.1080/07315724.1991.10718125. [DOI] [PubMed] [Google Scholar]
- 14.Gunshin H., Noguchi T., Naito H. Effect of calcium on the zinc uptake by brush-border membrane-vesicles isolated from the rat small-intestine. Agr. Biol. Chem. Tokyo. 1991;55:2813–2816. [Google Scholar]
- 15.Bertolo R. F., Brttger W. J., Atkinson S. A. Calcium competes with zinc for a channel mechanism on the brush border membrane of piglet intestine. J. Nutr. Biochem. 2001;12:66–72. doi: 10.1016/s0955-2863(00)00125-x. [DOI] [PubMed] [Google Scholar]
- 16.Qiu A. Ph.D. Thesis. U.K.: King's College London, University of London; 2004. Function and regulation of zinc transporters in model fish species. [Google Scholar]
- 17.Qiu A., Hogstrand C. Functional characterisation and genomic analysis of an epithelial calcium channel (ECaC) from pufferfish, Fugu rubripes. Gene. 2004;342:113–123. doi: 10.1016/j.gene.2004.07.041. [DOI] [PubMed] [Google Scholar]
- 18.Glover C. N., Hogstrand C. In vivo characterisation of intestinal zinc uptake in freshwater rainbow trout. J. Exp. Biol. 2002;205:141–150. doi: 10.1242/jeb.205.1.141. [DOI] [PubMed] [Google Scholar]
- 19.Bury N. R., Walker P. A., Glover C. N. Nutritive metal uptake in teleost fish. J. Exp. Biol. 2003;206:11–23. doi: 10.1242/jeb.00068. [DOI] [PubMed] [Google Scholar]
- 20.Gaither L. A., Eide D. J. Eukaryotic zinc transporters and their regulation. Biometals. 2001;14:251–270. doi: 10.1023/a:1012988914300. [DOI] [PubMed] [Google Scholar]
- 21.Lioumi M., Ferguson C. A., Sharpe P. T., Freeman T., Marenholz I., Mischke D., Heizmann C., Ragoussis J. Isolation and characterization of human and mouse ZIRTL, a member of the IRT1 family of transporters, mapping within the epidermal differentiation complex. Genomics. 1999;62:272–280. doi: 10.1006/geno.1999.5993. [DOI] [PubMed] [Google Scholar]
- 22.Eide D. J. The SLC39 family of metal ion transporters. Pflug. Arch. Eur. J. Phys. 2004;447:796–800. doi: 10.1007/s00424-003-1074-3. [DOI] [PubMed] [Google Scholar]
- 23.Taylor K. M., Nicholson R. I. The LZT proteins; the LIV-1 subfamily of zinc transporters. Biochim. Biophys. Acta Biomembranes. 2003;1611:16–30. doi: 10.1016/s0005-2736(03)00048-8. [DOI] [PubMed] [Google Scholar]
- 24.Taylor K. M., Morgan H. E., Johnson A., Hadley L. J., Nicholson R. I. Structure-function analysis of LIV-1, the breast cancer-associated protein that belongs to a new subfamily of zinc transporters. Biochem. J. 2003;375:51–59. doi: 10.1042/BJ20030478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamashita S., Miyagi C., Fukada T., Kagara N., Che Y.-S., Hirano T. Zinc transporter LIV1 controls epithelial–mesenchymal transition in zebrafish gastrula organizer. Nature (London) 2004;429:298–302. doi: 10.1038/nature02545. [DOI] [PubMed] [Google Scholar]
- 26.Aparicio S., Chapman J., Stupka E., Putnam N., Chia J.-M., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., et al. Whole-genome Shotgun assembly and analysis of the genome of Fugu rubripes. Science. 2002;297:1301–1310. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- 27.Dolphin C. T., Cullingford T. E., Shephard E. A., Smith R. L., Phillips I. R. Differential developmental and tissue-specific regulation of expression of the genes encoding three members of the flavin-containing monooxygenase family of man, FMO1, FM03 and FM04. Eur. J. Biochem. 1996;235:683–689. doi: 10.1111/j.1432-1033.1996.00683.x. [DOI] [PubMed] [Google Scholar]
- 28.Higgins D., Thompson J., Gibson T., Thompson J. D., Higgins D. G., Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moller S., Croning M. D. R., Apweiler R. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics. 2001;17:646–653. doi: 10.1093/bioinformatics/17.7.646. [DOI] [PubMed] [Google Scholar]
- 30.Kreegipuu A., Blom N., Brunak S. PhosphoBase, a database of phosphorylation sites: release 2.0. Nucleic Acids Res. 1999;27:237–239. doi: 10.1093/nar/27.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaither L. A., Eide D. J. Functional expression of the human hZIP2 zinc transporter. J. Biol. Chem. 2000;275:5560–5564. doi: 10.1074/jbc.275.8.5560. [DOI] [PubMed] [Google Scholar]
- 32.Eng B. H., Guerinot M. L., Eide D., Saier M. H. Sequence analyses and phylogenetic characterization of the ZIP family of metal ion transport proteins. J. Membr. Biol. 1998;166:1–7. doi: 10.1007/s002329900442. [DOI] [PubMed] [Google Scholar]
- 33.Berg J. M., Shi Y. G. The galvanisation of biology: a growing application for the roles of zinc. Science. 1996;271:1081–1085. doi: 10.1126/science.271.5252.1081. [DOI] [PubMed] [Google Scholar]
- 34.Cormack B. P., Valdivia R. H., Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 35.Olsson P. E., Kling P., Erkell L. J., Kille P. Structural and functional-analysis of the rainbow-trout (Oncorhyncus mykiss) metallothionein-A gene. Eur. J. Biochem. 1995;230:344–349. doi: 10.1111/j.1432-1033.1995.tb20569.x. [DOI] [PubMed] [Google Scholar]
- 36.Andrews G. K. Cellular zinc sensors: MTF-1 regulation of gene expression. Biometals. 2001;14:223–237. doi: 10.1023/a:1012932712483. [DOI] [PubMed] [Google Scholar]
- 37.Price-Haughey J., Bonham K., Gedamu L. Metallothionein gene-expression in fish cell-lines – its activation in embryonic-cells by 5-azacytidine. Biochim. Biophys. Acta. 1987;908:158–168. doi: 10.1016/0167-4781(87)90055-8. [DOI] [PubMed] [Google Scholar]
- 38.Mayer G. D., Leach A., Kling P., Olsson P. E., Hogstrand C. Activation of the rainbow trout metallothionein-A promoter by silver and zinc. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2003;134:181–188. doi: 10.1016/s1096-4959(02)00248-8. [DOI] [PubMed] [Google Scholar]
- 39.Gaither L. A., Eide D. J. The human ZIP1 transporter mediates zinc uptake in human K562 erythroleukemia cells. J. Biol. Chem. 2001;276:22258–22264. doi: 10.1074/jbc.M101772200. [DOI] [PubMed] [Google Scholar]
- 40.Gitan R. S., Luo H., Rodgers J., Broderius M., Eide D. Zinc-induced inactivation of the yeast ZRT1 zinc transporter occurs through endocytosis and vacuolar degradation. J. Biol. Chem. 1998;273:28617–28624. doi: 10.1074/jbc.273.44.28617. [DOI] [PubMed] [Google Scholar]
- 41.Gitan R. S., Eide D. J. Zinc-regulated ubiquitin conjugation signals endocytosis of the yeast ZRT1 zinc transporter. Biochem. J. 2000;346:329–336. [PMC free article] [PubMed] [Google Scholar]
- 42.Gitan R. S., Shababi M., Kramer M., Eide D. J. A cytosolic domain of the yeast Zrt1 zinc transporter is required for its post-translational inactivation in response to zinc and cadmium. J. Biol. Chem. 2003;278:39558–39564. doi: 10.1074/jbc.M302760200. [DOI] [PubMed] [Google Scholar]
- 43.Kim B. E., Wang F. D., Dufner-Beattie J., Andrews G. K., Eide D. J., Petris M. J. Zn2+-stimulated endocytosis of the mZIP4 zinc transporter regulates its location at the plasma membrane. J. Biol. Chem. 2003;279:4523–4530. doi: 10.1074/jbc.M310799200. [DOI] [PubMed] [Google Scholar]
- 44.Banks S. D., Thomas P., Baer K. N. Seasonal variations in hepatic and ovarian zinc concentrations during the annual reproductive cycle in female channel catfish (Ictalurus punctatus) Comp. Biochem. Physiol. C. 1999;124:65–72. doi: 10.1016/s0742-8413(99)00053-5. [DOI] [PubMed] [Google Scholar]
- 45.Thompson E. D., Mayer G. D., Balesaria S., Glover C. N., Walsh P. J., Hogstrand C. Physiology and endocrinology of zinc accumulation during the female squirrelfish reproductive cycle. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2003;134:819–828. doi: 10.1016/s1095-6433(03)00015-1. [DOI] [PubMed] [Google Scholar]
- 46.Olsson P. E., Haux C., Forlin L. Variations in hepatic metallothionen, zinc and copper levels during an annual reproductive-cycle in rainbow-trout, Salmo gairdneri. Fish Physiol. Biochem. 1987;3:39–47. doi: 10.1007/BF02183992. [DOI] [PubMed] [Google Scholar]
- 47.Olsson P. E., Zafarullah M., Foster R., Hamor T., Gedamu L. Developmental regulation of metallothionein messenger-RNA, zinc and copper levels in rainbow-trout, Salmo gairdneri. Eur. J. Biochem. 1990;193:229–235. doi: 10.1111/j.1432-1033.1990.tb19327.x. [DOI] [PubMed] [Google Scholar]
- 48.Sensi S. L., Canzoniero L. M. T., Yu S. P., Ying H. S., Koh J. Y., Kerchner G. A., Choi D. W. Measurement of intracellular free zinc in living cortical neurons: routes of entry. J. Neurosci. 1997;17:9554–9564. doi: 10.1523/JNEUROSCI.17-24-09554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spry D. J., Wood C. M. Ion flux rates, acid-base status, and blood gases in rainbow trout, Salmo gairdneri, exposed to toxic zinc in natural soft water. Can. J. Fish Aquat. Sci. 1985;42:1332–1341. [Google Scholar]
- 50.Nishimura H., Imai M. Control of renal-function in fresh-water and marine teleosts. Fed. Proc. Fed. Am. Soc. Exp. Biol. 1982;41:2355–2360. [PubMed] [Google Scholar]
- 51.Spry D. J., Hodson P. V., Wood C. M. Relative contributions of dietary and waterborne zinc in the rainbow-trout, Salmo gairdneri. Can. J. Fish. Aquat. Sci. 1988;45:32–41. [Google Scholar]