Abstract
In γ-irradiation, •OH is directly generated from water and causes DNA damage leading to carcinogenesis. Exposure of proteins to γ-irradiation, in the presence of oxygen, gives high yields of hydroperoxides. To clarify whether these hydroperoxides, particularly those formed on DNA-binding histone proteins, participate in γ-irradiation-induced carcinogenesis, experiments using 32P-labelled DNA fragments obtained from human cancer-related genes were undertaken. Histone protein-hydroperoxides induced significant DNA damage in the presence of Cu(I). Histone H1- and H3-hydroperoxides showed stronger DNA damage compared with histone H2A- and H4-hydroperoxides at 0.7 μM. Histone H1-hydroperoxides caused Cu(I)-dependent DNA damage predominantly at guanine residues, especially at 5′-GGC-3′, 5′-GGA-3′, 5′-GGT-3′ and single G bases. In contrast, histone H3-hydroperoxides/Cu(I) induced DNA damage at 5′-G in GG sequences; this sequence specificity is identical with that generated by 2,2′-azobis (2-amidinopropane) dihydrochloride, which is known to produce peroxyl radicals (RO2•). The difference in site specificity of DNA damage induced by histone H1- and H3-hydroperoxides may arise from their amino acid composition or their mode of binding to DNA. The histone H1-hydroperoxides/Cu(I) system also induced 8-oxo-7,8-dihydro-2′-deoxyguanosine formation in calf thymus DNA. It is concluded that histone protein-hydroperoxides can induce guanine-specific DNA damage, which may contribute to γ-irradiation-induced carcinogenesis.
Keywords: carcinogenesis; 8-oxo-7,8-dihydro-2′-deoxyguanosine; DNA damage; guanine; histone; hydroperoxide
Abbreviations: 8-oxodG, 8-oxo-7,8-dihydro-2′-deoxyguanosine; AAPH, 2,2′-azobis (2-amidinopropane) dihydrochloride; DTPA, diethylenetriamine-N,N,N′,N″,N″-penta-acetic acid; ECD, electrochemical detector; Fpg, Escherichia coli formamidopyrimidine-DNA glycosylase
INTRODUCTION
Histone proteins are localized inside the nuclei and play a key role in the ordering of DNA and packaging of the nucleosome into chromatin. The binding of histones to DNA and its organization into higher order chromatin structures significantly protects DNA against hydroxyl radical-induced DNA strand breaks [1]; this probably arises from the preferential oxidation of these proteins. Thus histones can be considered as sacrificial targets that protect DNA against oxidative damage [2]. Histone proteins can suppress lymphoid and solid tumours in p53-deficient mice [3,4].
On the other hand, oxidative damage to nuclear proteins may not be entirely benign. Oxidative stress in the nucleus may result in the modification of histone amino acids, formation of protein–protein cross-links and DNA–protein cross-links [5,6]. It was reported that proteins were the most abundant target within cells for radicals such as •OH [7]. Oxidized proteins have been shown to oxidize antioxidants and reducing agents such as ascorbate and glutathione [8,9]. Heavily oxidized proteins generally show decreased susceptibility to proteolytic attack by most proteinases [10,11]. Accumulation of oxidized proteins has been associated with the development of a number of diseases, including diabetes, atherosclerosis and neurodegenerative conditions [10,12,13], raising the possibility that modified histone proteins may contribute to DNA damage, for example, by base oxidation.
In γ-irradiation, •OH is directly generated from water and causes DNA damage, which leads to carcinogenesis. Exposure of amino acids, peptides and proteins to •OH and other radicals in the presence of O2 gives high yields of hydroperoxides [8]. We have shown that γ-irradiation induces the formation of hydroperoxides on histone H1 and other histone proteins and amino acids in the presence of O2 [14]. Decomposition of such amino acid-, peptide- and protein-hydroperoxides in the presence of exogenous catalysts such as light, heat or transition metal ions, results in the formation of further reactive radicals [15]. Protein- and amino acid-derived radicals have been previously reported to cause DNA double-strand breaks [15,16] and DNA–protein cross-links [16,17]. Histone protein-hydroperoxides caused the formation of 8-oxodG (8-oxo-7,8-dihydro-2′-deoxyguanosine, also known as 8-hydroxy-2′-deoxyguanosine), the most frequently used marker for oxidative damage to DNA [14]. Therefore DNA damage induced by radicals generated from histone protein-hydroperoxides may contribute to carcinogenesis, as an indirect effect of γ-irradiation.
To clarify the role of histone protein-hydroperoxides in γ-irradiation-induced DNA damage and carcinogenesis, we have investigated the sequence specificity of DNA damage by histone protein-hydroperoxides in the presence of copper ions in comparison with peroxyl radicals (RO2•) generated from the thermolabile compound AAPH [2,2′-azobis (2-amidinopropane) dihydrochloride]. We have also quantified the formation of 8-oxodG in calf thymus DNA treated with histone protein-hydroperoxides.
EXPERIMENTAL
Materials
Restriction enzymes (ApaI, AvaI, EcoRI, BssHII, XbaI and HindIII) and alkaline phosphatase from calf intestine were purchased from Boehringer Mannheim (Mannheim, Germany). Restriction enzyme (BamHI) and T4 polynucleotide kinase were obtained from New England Biolabs (Beverly, MA, U.S.A.). [γ-32P]ATP (222 TBq/mmol) was obtained from Amersham Biosciences (Little Chalfont, Bucks., U.K.) and DTPA (diethylenetriamine-N,N,N′,N″,N″-penta-acetic acid) from Dojin Chemicals (Kumamoto, Japan). Acrylamide, bisacrylamide and AAPH were obtained from Wako Pure Chemical Industries (Osaka, Japan). Ethanol and CuCl were obtained from Nacalai Tesque (Kyoto, Japan). Nuclease P1 (400 units/mg) was purchased from Yamasa Shoyu (Chiba, Japan), Fpg (Escherichia coli formamidopyrimidine-DNA glycosylase) from Trevigen (Gaithersburg, MD, U.S.A.) and histone H1 (subgroup f1-lysine-rich fraction, calf thymus) and other histone proteins from Sigma (St. Louis, MO, U.S.A.).
Preparation of 32P-5′-end-labelled DNA fragments
DNA fragments were obtained from the human p53 tumour suppressor gene (EMBL Data library, accession number X54156) and the human c-Ha-ras-1 proto-oncogene [19]. The 5′-endlabelled 650 bp fragment (HindIII*13972–EcoRI* 14621) and 470 bp fragment (HindIII*13038–EcoRI*13507) were obtained by dephosphorylation using calf intestine phosphatase and rephosphorylation using [γ-32P]ATP and T4 polynucleotide kinase (* indicates 32P labelling). The 650 bp fragment was further digested with ApaI to obtain a singly labelled 443 bp fragment (ApaI 14179–EcoRI*14621) and 211 bp fragment (HindIII*13972–ApaI 14182) as described previously [20]. A DNA fragment was prepared from plasmid pbcNI, which carries a 6.6 kb BamHI chromosomal DNA segment containing the c-Ha-ras-1 proto-oncogene [21,22]. The 435 bp fragment (AvaI*2247–AvaI*2681) was further digested with PstI to obtain a singly labelled 337 bp fragment (PstI 2345–AvaI*2681) and the 602 bp fragment (AvaI*1645–AvaI*2246) was digested with XbaI to obtain a singly labelled 261 bp fragment (AvaI*1645–XbaI 1905) by the method described previously [21,22]. Nucleotide numbering starts with the BamHI site [19].
Formation of protein hydroperoxides by γ-irradiation
Solutions of histone proteins (∼1 mg/ml) in water were continuously oxygenated during γ-irradiation using a 60Co source at a dose rate of approx. 60–65 Gy/min to total doses of up to 2000 Gy. Immediately after irradiation, each sample was treated with catalase (0.25–0.5 mg total) for 10 min at room temperature (21 °C) to remove H2O2 formed during irradiation. It has been shown previously that this treatment does not affect the hydroperoxide yields [14]. The samples were then aliquoted and snap-frozen on solid CO2 to minimize loss of the hydroperoxides. Hydroperoxide concentrations were determined by iodometric [23] or modified FOX (no added sorbitol) assays [24,25], using H2O2 as standards; these two assays have been previously shown to give similar values for these peroxides [12]. The values quoted are given as H2O2 equivalents and correspond to 0.5–3.5 mol of peroxide/mol of protein.
Analysis of DNA damage induced by histone protein-hydroperoxides in the presence of Cu(I)
Standard reaction mixtures in a microtube (1.5 ml Eppendorf) contained various concentrations of histone protein-hydroperoxides, CuCl, 32P-labelled DNA fragment and 20 μM in DNA bases of calf thymus DNA in 200 μl of 10 mM sodium phosphate buffer (pH 7.4) containing 5 μM DTPA. The concentration dependence of hydroperoxide was determined by adding different concentrations of γ-irradiated histone protein. The concentration of 32P-labelled DNA fragment added was several orders of magnitude lower than that of the calf thymus DNA fragment. Therefore the total concentration of DNA fragments is approx. 20 μM in DNA bases. DTPA was used to chelate contaminating metal ions in the sodium phosphate buffer. CuCl solutions were made up just before use. After incubation at 37 °C for 1 h, the DNA fragments were treated with 10 units of Fpg protein in the reaction buffer [10 mM Hepes/KOH (pH 7.4), 100 mM KCl, 10 mM EDTA and 0.1 mg/ml BSA] at 37 °C for 2 h. Fpg protein catalyses the excision of 8-oxodG as well as 2,6-diamino-4-hydroxy-5-formamidopyrimidine residues [26–28].
The preferred cleavage sites were determined by direct comparison of the positions of the oligonucleotides with those produced by chemical reactions of the Maxam–Gilbert procedure [29] using a DNA-sequencing system (LKB 2010 Macrophor; Amersham Biosciences). A laser densitometer (Personal Densitometer SI, Amersham Biosciences) was used for the measurement of the relative amounts of oligonucleotides from treated DNA fragments.
Analysis of 8-oxodG formation in calf thymus DNA induced by histone H1-hydroperoxides in the presence of Cu(I)
8-OxodG formation was quantified by the method of Kasai et al. [30] with some modifications. Calf thymus DNA fragments (100 μM in DNA bases) were incubated with histone H1-hydroperoxides and CuCl or with AAPH at 37 °C for 1 h. After ethanol precipitation, DNA was digested with nuclease P1 and calf intestine phosphatase and analysed by HPLC with an ECD (electrochemical detector) [31].
RESULTS
Damage to 32P-labelled DNA fragments induced by histone H1-hydroperoxides in the presence of Cu(I)
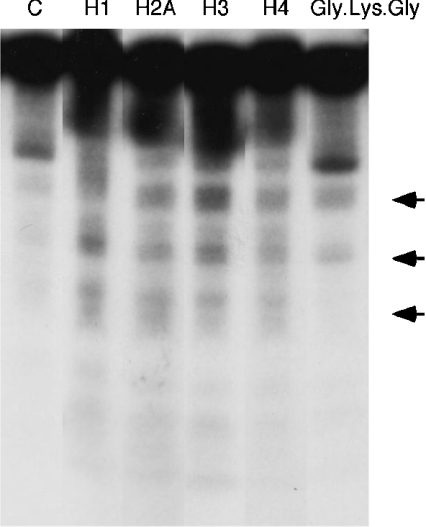
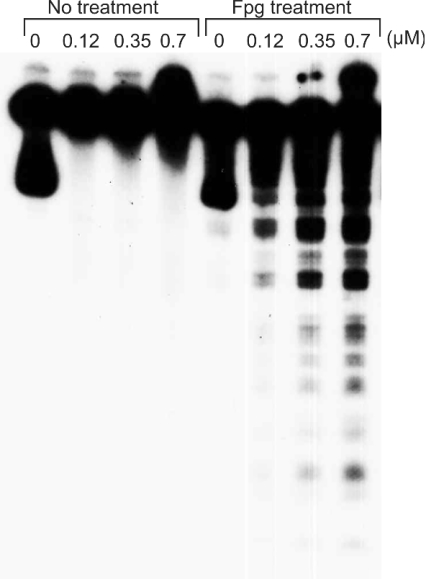
We compared the intensity of DNA damage induced by four histone protein-hydroperoxides: H1, H2A, H3 and H4 in the presence of Cu(I). Histone H1- and H3-hydroperoxides showed stronger DNA damage compared with histone H2A- and H4-hydroperoxides at 0.7 μM (Figure 1). We also examined DNA damage induced by a small peptide hydroperoxide; Gly-Lys-Gly-hydroperoxides caused comparatively weak DNA damage in the presence of Cu(I). Neither these peptide- and histone protein-hydroperoxides (results not shown) nor 20 μM Cu(I) alone (Figures 1 and 2) caused significant DNA damage under the conditions employed. Mixtures of non-irradiated histone proteins and Cu(I) caused little or no DNA damage (results not shown). Figure 2 shows that DNA damage increased with increasing concentrations of histone H1-hydroperoxides in the presence of Cu(I). Histone H1-hydroperoxides and Cu(I) caused DNA damage with Fpg treatment, although no DNA cleavage was observed without Fpg treatment.
Figure 1. Autoradiogram of 32P-labelled DNA fragments incubated with various histone protein-hydroperoxides in the presence of Cu(I).
The reaction mixtures contained the 32P-5′-end-labelled 337 bp DNA fragment, calf thymus DNA (20 μM in DNA bases), 0.7 μM of histone protein-hydroperoxides (histone H1, H2A, H3 and H4) and 20 μM CuCl in 200 μl of 10 mM sodium phosphate buffer (pH 7.4) containing 5 μM DTPA. After incubation at 37 °C for 60 min, DNA fragments were treated with 10 units of Fpg protein at 37 °C for 120 min. The DNA fragments were electrophoresed on an 8% (w/v) polyacrylamide/8 M urea gel (12 cm×16 cm), and autoradiograms were obtained by exposing an X-ray film to the gel. Oligonucleotides generated by hydroperoxide-induced DNA damage are indicated by arrows. The bands above these oligonucleotides are single- and double-strand intact DNA fragments.
Figure 2. Effect of Fpg treatment on DNA damage induced by histone H1-hydroperoxides in the presence of Cu(I).
Reaction mixtures contained the 32P-5′-end-labelled 337 bp fragment, calf thymus DNA (20 μM in DNA bases), the indicated concentrations of histone H1-hydroperoxides and 20 μM Cu(I) in 200 μl of 10 mM sodium phosphate buffer (pH 7.4) containing 5 μM DTPA. The mixture was incubated at 37 °C for 60 min. The DNA fragments were then treated with Fpg and electrophoresed on an 8% polyacrylamide/8 M urea gel (12 cm×16 cm), and autoradiograms were obtained by exposing an X-ray film to the gel.
Site specificity of Cu(I)-mediated DNA damage induced by histone H1-hydroperoxides
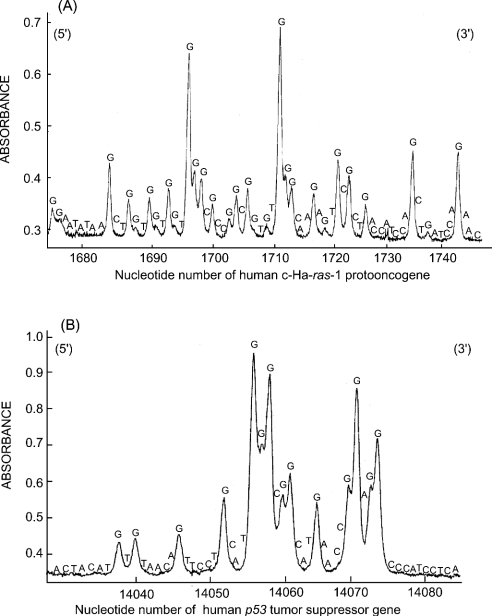
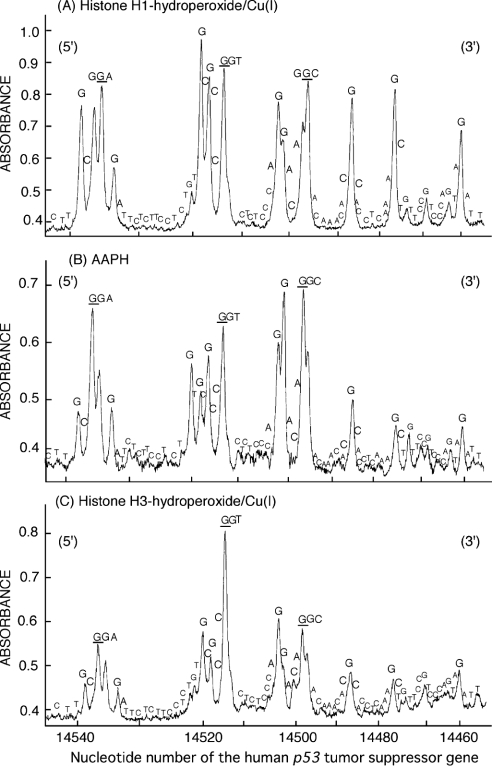
Figure 3 shows the site specificity of DNA damage induced by histone H1-hydroperoxides in the presence of Cu(I). Figure 4 shows a comparison of the site specificity of DNA damage induced by histone protein-hydroperoxides/Cu(I) and AAPH. AAPH has often been used as a hydrophilic radical initiator at ambient temperatures, with the rate of radical generation virtually constant for the first few hours at 37 °C in neutral water [32]. Histone H1-hydroperoxides in the presence of Cu(I) induced DNA damage predominantly at guanine in 5′-GGC-3′, 5′-GGA-3′, 5′-GGT-3′ sequences (damaged bases are underlined) and single G bases (Figures 3 and 4A), whereas histone H3-hydroperoxides induced DNA damage mainly at 5′-G in GG sequences and, to a lesser extent, at single G bases (Figure 4C). The site specificity of DNA damage induced by AAPH, a known source of RO2•, was similar to that observed with histone H3-hydroperoxides (Figure 4B).
Figure 3. Site specificity of DNA damage induced by histone H1-hydroperoxides in the presence of Cu(I).
The 32P-5′-end-labelled 261 bp fragment (A) and 211 bp fragment (B) were exposed to 0.7 μM histone H1-hydroperoxides, 20 μM Cu(I) and calf thymus DNA (20 μM in DNA bases) in 200 μl of 10 mM sodium phosphate buffer (pH 7.4) containing 5 μM DTPA. After incubation at 37 °C for 60 min, DNA fragments were treated with 10 units of Fpg protein at 37 °C for 120 min. The DNA fragments were electrophoresed on an 8% polyacrylamide/8 M urea gel using a DNA-sequencing system and visualized by autoradiography. The relative amounts of oligonucleotide were measured by scanning the autoradiogram with a laser densitometer. Horizontal axis: the nucleotide number of the human c-Ha-ras-1 proto-oncogene (A) and p53 tumour suppressor gene (B).
Figure 4. Comparison of the site specificity of DNA cleavage induced by histone H1-hydroperoxides and AAPH.
Reaction mixtures containing the 32P-5′-end-labelled 443 bp DNA fragment, calf thymus DNA (20 μM in DNA bases), 0.7 μM histone H1-hydroperoxides and 20 μM Cu(I) (A), 1 mM AAPH (B) or 0.7 μM histone H3-hydroperoxides and 20 μM Cu(I) (C) in 200 μl of 10 mM sodium phosphate buffer (pH 7.4) containing 5 μM DTPA were incubated at 37 °C for 60 min. After Fpg treatment, the DNA fragments were analysed by the method described in Figure 3.
Formation of 8-oxodG in calf thymus DNA by histone H1-hydroperoxides/Cu(I) or AAPH
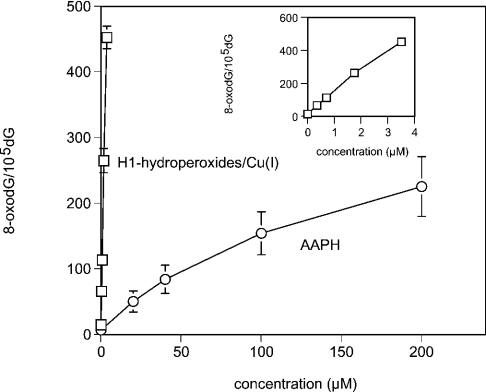
8-OxodG formation was quantified in calf thymus DNA treated with histone H1-hydroperoxides/Cu(I), or AAPH, by using HPLC ECD. The amount of 8-oxodG increased with increasing concentrations of histone H1-hydroperoxides or AAPH, with histone H1-hydroperoxides/Cu(I) giving higher yields of this oxidized base when compared with AAPH (Figure 5).
Figure 5. Formation of 8-oxodG in calf thymus DNA induced by histone H1-hydroperoxides in the presence of Cu(I).
Calf thymus DNA (100 μM in DNA bases) was incubated with the indicated concentrations of histone H1-hydroperoxides plus 20 μM Cu(I) or AAPH. After ethanol precipitation, DNA was subjected to enzyme digestion and analysed by HPLC ECD. The inset shows 8-oxodG formation by H1 using a different scale for the concentration.
DISCUSSION
The present study has demonstrated that histone protein-hydroperoxides have an ability to cause guanine-specific damage to DNA in the presence of Cu(I). Cu(I)-mediated DNA damage is of particular relevance since this metal ion is known to be present in the cell nucleus and to be bound to histone proteins [33,34]. Histone protein-hydroperoxides in the presence of Cu(I) caused DNA damage detected after Fpg treatment, although no DNA damage was observed without Fpg treatment. This result indicates that histone protein-hydroperoxides plus Cu(I) caused base modification, such as 8-oxodG formation, without significant yields of DNA strand breaks under the conditions employed. This is in contrast with results of a previous study [15] that detected strand breaks with much higher peroxide concentrations. Histone H1-hydroperoxides/Cu(I) also induced significant 8-oxodG formation in calf thymus DNA in line with previous studies using histone protein-hydroperoxides/Cu(II) and Ti(III) systems [12,14]. The formation of this oxidized base has been reported to lead to G→T transversion through DNA misreplication [35–37]. To clarify the reactive species responsible for this damage, we compared the site specificity of DNA damage induced by histone protein-hydroperoxides/Cu(I) and AAPH, which has been shown to produce RO2• in a stoichiometric manner [32,38,39]. Histone H1-hydroperoxides/Cu(I) induced DNA cleavage most frequently at guanine residues, especially at the 5′-GGC-3′, 5′-GGA-3′ and 5′-GGT-3′ sequences (damaged bases are shown underlined). In addition, histone H1-hydroperoxides/Cu(I) induced DNA damage at single G bases to a similar extent. In contrast, AAPH and histone H3-hydroperoxides/Cu(I) gave DNA cleavage predominantly at the 5′-G in GG sequences and less extensive DNA damage at single G bases.
The binding of histones to DNA arises from electrostatic interactions between the highly positively charged histone tails and the negative charges present on the phosphate groups of the DNA backbone [40]. The manner in which histones bind to DNA is dependent on their amino acid sequence, and composition, and therefore appears to play a key role in determining both the site specificity and extent of DNA damage. Basic amino acids, such as lysine and arginine residues, are probably involved in these electrostatic interactions with DNA. Histone H1 contains a much higher content of lysine when compared with histone H3. The smaller extent of damage observed with Gly-Lys-Gly peroxide when compared with identical concentrations of histone peroxides, both of which are expected to contain similar peroxide groups on lysine residues, suggests that the extent of damage is determined by other factors in addition to the peroxide concentration. This observation is consistent with the electrostatic binding of the hydroperoxide-containing materials to DNA, which is probably much stronger with the histones than Gly-Lys-Gly, being a key process in determining the extent of DNA damage. Lysine is known to give rise to high concentrations of hydroperoxides on oxidation by free radicals [41]. Therefore lysinehydroperoxides may play an important role in site-specific DNA damage. However, hydroperoxides can also be formed at other amino acids, such as isoleucine, leucine, proline, glutamic acid and valine [41]. The possibility that hydroperoxides formed at these amino acids participate in site-specific DNA damage cannot be excluded, although these amino acids are at a lower abundance compared with lysine residues, particularly in histone H1. A difference in the population of the hydroperoxides between histones H1 and H3, and the chemistry of the derived radicals, may contribute to the different specificity of damage detected with these two proteins.
Histone H3-hydroperoxides/Cu(I) showed similar site specificity to AAPH, suggesting that DNA damage is mainly attributed to RO2•. These results are supported by our previous studies showing that Cu(I)-catalysed decomposition of benzoyl peroxide into benzoyloxyl radicals caused DNA damage specifically at the 5′-G of GG and GGG sequences [42]. Results of the present study also suggest, however, that free radicals other than RO2• are involved in DNA damage induced by histone H1-hydroperoxides/Cu(I), since this protein hydroperoxide induced more extensive DNA damage at single G residues when compared with histone H3 hydroperoxide and AAPH. It has been previously reported that one-electron reduction of amino acid-, peptide- and protein-hydroperoxides by an Fe(II)–EDTA complex occurs through a pseudo-Fenton reaction, leading to the formation of alkoxyl radicals (RO•) [43,44]. Since RO• are more powerful oxidants compared with RO2• (reduction potentials of approx. 1.6 and 1.0 V respectively [45]), it is expected that RO• will be less site-specific compared with RO2• in DNA damage. We have also previously reported that hydroperoxide-derived alkoxyl radicals undergo rapid intramolecular rearrangement and fragmentation reactions to give carbon-centred radicals (C•) [43,44]. Therefore there remains the possibility that DNA damage by histone H1-hydroperoxides involves alkoxyl radicals and carbon-centred radicals (C•), in addition to RO2•.
In conclusion, it has been shown that formation of hydroperoxides on histone proteins and their subsequent decomposition to radicals can give rise to DNA damage specifically at guanine bases. Since these proteins are localized inside the nuclei and play a key role in the ordering of DNA and packaging of the nucleosome into chromatin, these hydroperoxides and the radicals derived from them are probably formed in proximity to the DNA. This protein-hydroperoxide-induced oxidative DNA damage may contribute to carcinogenesis through a delayed effect of γ-irradiation, in addition to the rapid effects induced by •OH radical formation.
Acknowledgments
This work was partially supported by a Grant-in-aid for Scientific Research from the Ministry of Education, Science and Culture of Japan and by the Australian Research Council.
References
- 1.Enright H. U., Miller W. J., Hebbel R. P. Nucleosomal histone protein protects DNA from iron-mediated damage. Nucleic Acids Res. 1992;20:3341–3346. doi: 10.1093/nar/20.13.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ljungman M., Hanawalt P. C. Efficient protection against oxidative DNA damage in chromatin. Mol. Carcinog. 1992;5:264–269. doi: 10.1002/mc.2940050406. [DOI] [PubMed] [Google Scholar]
- 3.Bassing C. H., Suh H., Ferguson D. O., Chua K. F., Manis J., Eckersdorff M., Gleason M., Bronson R., Lee C., Alt F. W. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell (Cambridge, Mass.) 2003;114:359–370. doi: 10.1016/s0092-8674(03)00566-x. [DOI] [PubMed] [Google Scholar]
- 4.Celeste A., Difilippantonio S., Difilippantonio M. J., Fernandez-Capetillo O., Pilch D. R., Sedelnikova O. A., Eckhaus M., Ried T., Bonner W. M., Nussenzweig A. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell (Cambridge, Mass.) 2003;114:371–383. doi: 10.1016/s0092-8674(03)00567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ullrich O., Grune T. Proteasomal degradation of oxidatively damaged endogenous histones in K562 human leukemic cells. Free Radical Biol. Med. 2001;31:887–893. doi: 10.1016/s0891-5849(01)00672-4. [DOI] [PubMed] [Google Scholar]
- 6.Altman S. A., Zastawny T. H., Randers-Eichhorn L., Cacciuttolo M. A., Akman S. A., Dizdaroglu M., Rao G. Formation of DNA-protein cross-links in cultured mammalian cells upon treatment with iron ions. Free Radical Biol. Med. 1995;19:897–902. doi: 10.1016/0891-5849(95)00095-f. [DOI] [PubMed] [Google Scholar]
- 7.Du J., Gebicki J. M. Proteins are major initial cell targets of hydroxyl free radicals. Int. J. Biochem. Cell Biol. 2004;36:2334–2343. doi: 10.1016/j.biocel.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 8.Simpson J. A., Narita S., Gieseg S., Gebicki S., Gebicki J. M., Dean R. T. Long-lived reactive species on free-radical-damaged proteins. Biochem. J. 1992;282:621–624. doi: 10.1042/bj2820621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu S., Gebicki S., Jessup W., Gebicki J. M., Dean R. T. Biological fate of amino acid, peptide and protein hydroperoxides. Biochem. J. 1995;311:821–827. doi: 10.1042/bj3110821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dean R. T., Fu S., Stocker R., Davies M. J. Biochemistry and pathology of radical-mediated protein oxidation. Biochem. J. 1997;324:1–18. doi: 10.1042/bj3240001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davies K. J., Lin S. W., Pacifici R. E. Protein damage and degradation by oxygen radicals. IV. Degradation of denatured protein. J. Biol. Chem. 1987;262:9914–9920. [PubMed] [Google Scholar]
- 12.Luxford C., Dean R. T., Davies M. J. Radicals derived from histone hydroperoxides damage nucleobases in RNA and DNA. Chem. Res. Toxicol. 2000;13:665–672. doi: 10.1021/tx000053u. [DOI] [PubMed] [Google Scholar]
- 13.Berlett B. S., Stadtman E. R. Protein oxidation in aging, disease, and oxidative stress. J. Biol. Chem. 1997;272:20313–20316. doi: 10.1074/jbc.272.33.20313. [DOI] [PubMed] [Google Scholar]
- 14.Luxford C., Morin B., Dean R. T., Davies M. J. Histone H1- and other protein- and amino acid-hydroperoxides can give rise to free radicals which oxidize DNA. Biochem. J. 1999;344:125–134. [PMC free article] [PubMed] [Google Scholar]
- 15.Luxford C., Dean R. T., Davies M. J. Induction of DNA damage by oxidised amino acids and proteins. Biogerontology. 2002;3:95–102. doi: 10.1023/a:1015228001561. [DOI] [PubMed] [Google Scholar]
- 16.Distel L., Distel B., Schüsler H. Formation of DNA double-strand breaks and DNA-protein crosslinks by irradiation of DNA in the presence of a protein. Radiat. Phys. Chem. 2002;65:141–149. [Google Scholar]
- 17.Gebicki S., Gebicki J. M. Crosslinking of DNA and proteins induced by protein hydroperoxides. Biochem. J. 1999;338:629–636. [PMC free article] [PubMed] [Google Scholar]
- 18. Reference deleted.
- 19.Capon D. J., Chen E. Y., Levinson A. D., Seeburg P. H., Goeddel D. V. Complete nucleotide sequences of the T24 human bladder carcinoma oncogene and its normal homologue. Nature (London) 1983;302:33–37. doi: 10.1038/302033a0. [DOI] [PubMed] [Google Scholar]
- 20.Yamashita N., Murata M., Inoue S., Hiraku Y., Yoshinaga T., Kawanishi S. Superoxide formation and DNA damage induced by a fragrant furanone in the presence of copper(II) Mutat. Res. 1998;397:191–201. doi: 10.1016/s0027-5107(97)00210-8. [DOI] [PubMed] [Google Scholar]
- 21.Kawanishi S., Yamamoto K. Mechanism of site-specific DNA damage induced by methylhydrazines in the presence of copper(II) or manganese(III) Biochemistry. 1991;30:3069–3075. doi: 10.1021/bi00226a013. [DOI] [PubMed] [Google Scholar]
- 22.Yamamoto K., Kawanishi S. Site-specific DNA damage induced by hydrazine in the presence of manganese and copper ions: the role of hydroxyl radical and hydrogen atom. J. Biol. Chem. 1991;266:1509–1515. [PubMed] [Google Scholar]
- 23.Jessup W., Dean R. T., Gebicki J. M. Iodometric determination of hydroperoxides in lipids and proteins. Methods Enzymol. 1994;233:289–303. [Google Scholar]
- 24.Wolff S. P. Ferrous ion oxidation in the presence of ferric ion indicator xylenol orange for measurement of hydroperoxides. Methods Enzymol. 1994;233:182–189. [Google Scholar]
- 25.Gay C., Collins J., Gebicki J. M. Hydroperoxide assay with the ferric-xylenol orange complex. Anal. Biochem. 1999;273:149–155. doi: 10.1006/abio.1999.4208. [DOI] [PubMed] [Google Scholar]
- 26.Bruner S. D., Norman D. P., Verdine G. L. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature (London) 2000;403:859–866. doi: 10.1038/35002510. [DOI] [PubMed] [Google Scholar]
- 27.David-Cordonnier M. H., Laval J., O'Neill P. Clustered DNA damage, influence on damage excision by XRS5 nuclear extracts and Escherichia coli Nth and Fpg proteins. J. Biol. Chem. 2000;275:11865–11873. doi: 10.1074/jbc.275.16.11865. [DOI] [PubMed] [Google Scholar]
- 28.Boiteux S., Gajewski E., Laval J., Dizdaroglu M. Substrate specificity of the Escherichia coli Fpg protein (formamidopyrimidine-DNA glycosylase): excision of purine lesions in DNA produced by ionizing radiation or photosensitization. Biochemistry. 1992;31:106–110. doi: 10.1021/bi00116a016. [DOI] [PubMed] [Google Scholar]
- 29.Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 30.Kasai H., Nishimura S., Kurosawa Y., Hayashi Y. Oral administration of the renal carcinogen, potassium bromate, specifically produces 8-hydroxydeoxyguanosine in rat target organ DNA. Carcinogenesis. 1987;8:1959–1961. doi: 10.1093/carcin/8.12.1959. [DOI] [PubMed] [Google Scholar]
- 31.Ito K., Inoue S., Yamamoto K., Kawanishi S. 8-Hydroxydeoxyguanosine formation at the 5′-site of 5′-GG-3′ sequences in double-stranded DNA by UV radiation with riboflavin. J. Biol. Chem. 1993;268:13221–13227. [PubMed] [Google Scholar]
- 32.Niki E. Free radical initiators as source of water- or lipid-soluble peroxyl radicals. Methods Enzymol. 1990;186:100–108. doi: 10.1016/0076-6879(90)86095-d. [DOI] [PubMed] [Google Scholar]
- 33.Chiu S. M., Xue L. Y., Friedman L. R., Oleinick N. L. Differential dependence on chromatin structure for copper and iron ion induction of DNA double-strand breaks. Biochemistry. 1995;34:2653–2661. doi: 10.1021/bi00008a032. [DOI] [PubMed] [Google Scholar]
- 34.Chiu S. M., Xue L. Y., Friedman L. R., Oleinick N. L. Copper ion-mediated sensitization of nuclear matrix attachment sites to ionizing radiation. Biochemistry. 1993;32:6214–6219. doi: 10.1021/bi00075a014. [DOI] [PubMed] [Google Scholar]
- 35.Proteggente A. R., England T. G., Rehman A., Rice-Evans C. A., Halliwell B. Gender differences in steady-state levels of oxidative damage to DNA in healthy individuals. Free Radical Res. 2002;36:157–162. doi: 10.1080/10715760290006475. [DOI] [PubMed] [Google Scholar]
- 36.Miller H., Prasad R., Wilson S. H., Johnson F., Grollman A. P. 8-OxodGTP incorporation by DNA polymerase beta is modified by active-site residue Asn279. Biochemistry. 2000;39:1029–1033. doi: 10.1021/bi991789x. [DOI] [PubMed] [Google Scholar]
- 37.Shibutani S., Takeshita M., Grollman A. P. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature (London) 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 38.Offer T., Samuni A. Nitroxides inhibit peroxyl radical-mediated DNA scission and enzyme inactivation. Free Radical Biol. Med. 2002;32:872–881. doi: 10.1016/s0891-5849(02)00750-5. [DOI] [PubMed] [Google Scholar]
- 39.Sakakibara H., Ashida H., Kanazawa K. A novel method using 8-hydroperoxy-2′-deoxyguanosine formation for evaluating antioxidative potency. Free Radical Res. 2002;36:307–316. doi: 10.1080/10715760290019336. [DOI] [PubMed] [Google Scholar]
- 40.Loyola A., Reinberg D. Histone deposition and chromatin assembly by RSF. Methods. 2003;31:96–103. doi: 10.1016/s1046-2023(03)00093-8. [DOI] [PubMed] [Google Scholar]
- 41.Gebicki S., Gebicki J. M. Formation of peroxides in amino acids and proteins exposed to oxygen free radicals. Biochem. J. 1993;289:743–749. doi: 10.1042/bj2890743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kawanishi S., Oikawa S., Murata M., Tsukitome H., Saito I. Site-specific oxidation at GG and GGG sequences in double-stranded DNA by benzoyl peroxide as a tumor promoter. Biochemistry. 1999;38:16733–16739. doi: 10.1021/bi990890z. [DOI] [PubMed] [Google Scholar]
- 43.Davies M. J. Protein and peptide alkoxyl radicals can give rise to C-terminal decarboxylation and backbone cleavage. Arch. Biochem. Biophys. 1996;336:163–172. doi: 10.1006/abbi.1996.0545. [DOI] [PubMed] [Google Scholar]
- 44.Davies M. J., Fu S., Dean R. T. Protein hydroperoxides can give rise to reactive free radicals. Biochem. J. 1995;305:643–649. doi: 10.1042/bj3050643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koppenol W. H. Oxyradical reactions: from bond-dissociation energies to reduction potentials. FEBS Lett. 1990;264:165–167. doi: 10.1016/0014-5793(90)80239-f. [DOI] [PubMed] [Google Scholar]