Abstract
Ribosome biogenesis in Saccharomyces cerevisiae occurs primarily in a specialized nuclear compartment termed the nucleolus within which the rRNA genes are transcribed by RNA polymerase I into a large 35 S rRNA precursor. The ensuing association/dissociation and catalytic activity of numerous trans-acting protein factors, RNAs and ribosomal proteins ultimately leads to the maturation of the precursor rRNAs into 25, 5.8 and 18 S rRNAs and the formation of mature cytoplasmic 40 and 60 S ribosomal subunits. Although many components involved in ribosome biogenesis have been identified, our understanding of this essential cellular process remains limited. In the present study we demonstrate a crucial role for the previously uncharacterized nucleolar protein Nop53p (Ypl146p) in ribosome biogenesis. Specifically, Nop53p appears to be most important for biogenesis of the 60 S subunit. It physically interacts with rRNA processing factors, notably Cbf5p and Nop2p, and co-fractionates specifically with pre-60 S particles on sucrose gradients. Deletion or mutations within NOP53 cause significant growth defects and display significant 60 S subunit deficiencies, an imbalance in the 40 S:60 S ratio, as revealed by polysome profiling, and defects in progression beyond the 27 S stage of 25 S rRNA maturation during 60 S biogenesis.
Keywords: biogenesis, Nop53p, proteome, rRNA, SDS/PAGE, 60 S ribosome
Abbreviations: GFP, green fluorescent protein; ITS, internally transcribed spacer; ORF, open reading frame; pA, Protein A; RFP, red fluorescent protein; snoRNAs, small nucleolar RNAs; WT, wild-type
INTRODUCTION
Ribosome biogenesis in eukaryotes is a complex, tightly regulated and highly dynamic process that is orchestrated over a broad spatial and temporal context. For example, ribosomal proteins are synthesized in the cytoplasm and must be imported into the nucleus before assembling with rRNAs, which are spliced and modified in conjunction with the assembly. This assembly occurs primarily within the specialized nucleolar compartment of the nucleus, and demands the co-ordinated function of over 170 trans-acting protein factors and as many as 66 or more snoRNAs (small nucleolar RNAs), which serve to guide enzymatic modifications along the rRNA during ribosome biogenesis [1] (see also http://www.bio.umass.edu/biochem/rna-sequence/Yeast_snoRNA_Database/mastertable.html and http://rna.wustl.edu/snoRNAdb/Sc/Sc-snos-bysno.html). In most cases, snoRNAs must also assemble with protein counterparts and undergo maturation before they are transported to the nucleolus. Before completion, precursors of the 60 and 40 S ribosomal subunits are exported to the cytoplasm, where they undergo the final stages of maturation and perform their role in translation. Co-ordination of these events is crucial to the cell, and accordingly, defects at almost any stage lead to profound effects on cell viability and/or fitness [1].
Ribosome biogenesis is evidently intimately linked with nucleocytoplasmic transport and cells appear to exploit the transport process and machinery to create checkpoints ensuring fidelity in the maturation process. Macromolecular transport between the nucleus and cytoplasm is active and selective, and occurs through nuclear pore complexes embedded in the nuclear envelope. Most often, macromolecular transport is accomplished by carrier proteins, collectively called karyopherins, which bind to their cargoes, the nuclear pore complex and other regulators of transport [2,3].
Owing primarily to its ease of genetic and experimental manipulation, the budding yeast Saccharomyces cerevisiae has provided many fundamental insights into eukaryotic ribosome biogenesis. The rRNA is transcribed by two distinct polymerases. RNA polymerase I produces a large 35 S rRNA precursor, which is processed into three mature rRNAs: 18, 5.8, and 25 S [4,5], although RNA polymerase III synthesizes the 5 S RNA. Mature 60 S ribosomal subunits contain the 5, 5.8 and 25 S rRNAs, whereas the small 40 S subunit contains the 18 S rRNA. The ribosome biogenesis pathway is initiated by the transcription of a precursor 35 S rRNA, which is assembled with early ribosomal and assembly proteins. The pathway then bifurcates on endonucleolytic cleavage of the precursor rRNA yielding precursors of the small (40 S) and large (60 S) ribosomal subunits. These major steps were initially detected by sucrose gradient sedimentation analysis in the early 1970s as 90, 66 and 43 S preribosomes [6]; but, over the ensuing three decades, rRNA analyses have revealed additional specific intermediates. Recently, the advent of proteomics-based approaches have also contributed to our molecular inventory and coupling these results with data from experiments assessing the maturation of rRNA has enabled the ordering of complex ribonucleoprotein intermediates of the pathway [7–12]. The majority of trans-acting factors identified through various approaches do not appear to participate directly in rRNA processing but are thought to be involved in integrating and regulating assembly events [4,13]. The complexity revealed by such studies suggests the potential for hundreds of intermediates. Notably, current models derived from proteomic data propose a number of previously undetected intermediates in the 60 S pathway (pre-60 S early 1 (E1), early 2 (E2), middle (M) and late (L)) [4]. This type of analysis will allow more precise assessment of the numerous assembly steps that will ultimately lead to a detailed molecular understanding of the complete ribosome biogenesis program.
In spite of the large inventory of proteins detected in association with the assembling ribosomal subunits, much remains to be learned about their specific functions. In the present study, we characterize one such protein, which we term Nop53p. We demonstrate that this previously uncharacterized nucleolar protein is required for efficient 60 S ribosome biogenesis, specifically for progression beyond the 27 S stage of 25 S rRNA maturation.
EXPERIMENTAL
Yeast strains and microbiological techniques
The S. cerevisiae strains used in the present study were derivatives of the diploid DF5 strain [14]. All yeast genetic manipulations were performed according to established procedures [15]. Deletion of the NOP53 ORF (open reading frame) was accomplished by integrative transformation of DF5 cells with a PCR-synthesized HIS5 marker containing short flanking homology to the upstream and downstream regions of the NOP53 ORF, generated by using the following oligonucleotides: NOP53-5′, CGTTAATGAATCATTTATTTAAAAATGGCTCCAACTAATCTAACCAAGAAACCATCTCAATAACCTGACATAATCCAATTCATC, and NOP53-3′, CTAGAGATATCGACATGGGAATGCCCTCTCAAACTAGAAAAGGGCAGAGAACGAAAAGAAGCTGACGGTATCGATAAGCTT. Transformants were selected on SC (synthetic complete) plates lacking histidine (SC–His plates), sporulated and tetrads were dissected to obtain a haploid Δnop53 strain (MATα ura3-52 his3Δ200 trp1-1 leu2-3,112 lys2-801 nop53::HIS5).
The NOP53-GFP (green fluorescent protein) strain (MATa ura3-52 his3Δ200 trp1-1 leu2-3,112 lys2-801 NOP53-GFP-spHIS5+Nop1p–RFP) was generated by appending the coding sequence of Aequoria victoria GFP at the 3′ end of NOP53 resulting in the in-frame fusion of GFP to the C-terminus of Nop53p as described previously [16]. The NOP53-A strain (MATα ura3-52 his3Δ200 trp1-1 leu2-3,112 lys2-801 NOP53-pA-spHIS5), which expresses Staphylococcus aureus pA (Protein A) fused to the C-terminus of Nop53p, was described previously [17]. The nop53-FL and nop53-ΔC strains are isogenic to the nop53Δ strain except that they harbour plasmids pBH1 and pBH2 respectively.
Plasmids
pBH1 and pBH2 were constructed using two PCR products corresponding to both full-length NOP53 gene (pBH1) and a mutant version lacking nucleotides 1201–1368 encoding the C-terminal amino acids 401–456, which contains a heptad repeat of the leucine zipper motif (amino acids 402–434) and a pBH2 (NOP53-ΔC). PCR products were generated using the following oligonucleotides: YPL146C-5′ Xho1, CGGCTCGAGCTTCCTTTAATTATTGAT; YPL146C-3′Sac1, GCCGAGCTCGCCAACACAAATATGAAC; YPL146C amino acids 401-3′Sac1 CGGGAGCTCTTATTCGTCAGAAAATTTAAT. PCR products were digested with SacI and XhoI and then ligated into these restriction sites of pRS316. pGAL-NOP53 was obtained by subcloning a PCR product corresponding to full-length YPL146c into the BamHI–SalI sites of pYEUra3 (Clonetech, Palo Alto CA, U.S.A.). The PCR product was obtained by using the following oligonucleotides: YPL146C-5′ BamHI, AAAGGATCCATGGCTCCAACTAATCTAACCAAGAAACCA; YPL146C-3′ SalI, AAACAGCTGGATAAACTTCAGGAATACACAGGTGAAAAG. The Nop1p–RFP plasmid [containing Nop1p fused to RFP (red fluorescent protein)] was kindly provided by E. Hurt (Biochemie-Zentrum Heidelberg, Heidelberg, Germany).
Microscopy
Cells expressing GFP-tagged proteins were grown in selective media and the GFP chimaeras were visualized directly by fluorescence microscopy using a confocal microscope (LSM 510 NLO; Carl Zeiss, Cologne, Germany). Further processing of images was performed using Adobe® Photoshop® 6.0 (Adobe Systems, Seattle, WA, U.S.A.).
SDS/PAGE and Western-blot analysis
For Western-blot analysis, proteins were resolved on 7–15% (w/v) SDS/polyacrylamide gels and transferred on to nitrocellulose membranes by standard methods [18,19]. The following antibodies were used as primary antibodies: affinity-purified rabbit anti-mouse IgG (Cappel, Organon Teknika, West Chester, PA, U.S.A.) for detection of pA, mouse monoclonal anti-Tcm1p (rpL3) (kindly provided by J. L. Woolford, Department of Biological Sciences, The Mellon College of Science, Carnegie Mellon University, Pittsburgh, PA, U.S.A.). Horseradish peroxidase-conjugated anti-mouse or anti-rabbit Ig (Amersham Biosciences, Piscataway, NJ, U.S.A.) were used as secondary antibodies, and immunoreactive bands were visualized using SuperSignal West Pico Chemiluminescent Substrate (Pierce, Rockford, IL, U.S.A.).
Isolation of Nop53p–pA interacting proteins
Nop53p–pA and associated proteins were isolated from isolated nuclei by affinity purification. Nuclei were isolated from 4 litres of mid-logarithmically growing yeast as described previously [20–22]. Nuclei were pelleted, resuspended in 10 ml of 50 mM Tris/HCl (pH 8.0), 250 mM NaCl, 0.1 mM MgCl2, 4 μg/ml Pepstatin A and 180 μg/ml PMSF and treated with 20 μg/ml DNase I on ice for 15 min before passage through a prechilled French pressure cell (3 times at 1000 Ψ). The resulting lysate was clarified by centrifugation (27000 g for 15 min) and Nop53p–pA was isolated using IgG-Sepharose beads (Pharmacia, Piscataway, NJ, U.S.A.) essentially as described in [20,23]. Beads were washed ten times with 1 ml of wash buffer [50 mM Tris/HCl (pH 8.0), 150 mM NaCl, 0.1 mM MgCl2, 4 μg/ml Pepstatin A and 180 μg/ml PMSF]. Bound proteins were eluted with wash buffer containing increasing concentrations of MgCl2 (0.05–4 M). Proteins in the eluted fractions were precipitated with trichloroacetic acid, resolved by SDS/PAGE and visualized by Coomassie Blue staining. Bands of interest were excised from the gel, subjected to in-gel digestion, and identified by microcapillary HPLC tandem MS, and comparison of peptide tandem mass spectra to sequences in a non-redundant protein database as described previously [20,24].
Northern blot and primer extension analysis
RNA was isolated by the hot-phenol technique [18]. The analysis of rRNA by Northern hybridization was performed as described previously [25]. The following primers were used: 5′ETS-A0, GGTCTCTCTGCTGCCGG; 5′18 S, CATGGCTTAATCTTTGAGAC; D-A2, GCTCTCATGCTCTTGCCA; A2-A3, ATGAAAACTCCACAGTG; A3-B1L, CCAGTTACGAAAATTCTTG; 5′5.8 S, CGCTGCGTTCTTCATCGATG; C2-C1, GTTCGCCTAGACGCTCTC; 5′25 S, CTCCGCTTATTGATATGC.
Primer extension was performed using Promega AMV reverse transcriptase primer extension system (Promega, Madison, WI, U.S.A.) using the 5′18 S, 5′5.8 S and 5′25 S primers described above.
Pulse–chase analysis
Metabolic labelling was performed as described previously [26]. nop53Δ and WT (wild-type) strains harbouring the pRS316 plasmid were grown in 8 ml of CSM-Ura medium to an A600 0.4. The cells were concentrated to 4 ml cultures and labelled by the addition of 200 μCi of [3H]uracil (Perkin Elmer, Boston, MA, U.S.A.). After 1 min, an excess of unlabelled uracil (1.2 mg) was added and 1 ml aliquots were removed after continued incubation for 1, 5, 15 or 30 min. Total RNA was isolated from each sample using hot phenol as described in [27] and 50000 c.p.m. of labelled RNA derived from each strain was resolved by electrophoresis on a 1.3% (v/v) agarose gel. Labelled species were detected by exposure of the dried gel to Kodak X-Omat AR film (Eastman Kodak Company, Rochester, NY, U.S.A.).
To monitor the effects of Nop53p depletion, nop53Δ cells harbouring pGAL-NOP53, pregrown in CSM-Ura medium with 2% (w/v) galactose as a carbon source, were incubated for an additional 14 h in CSM-Ura medium containing either 2% (w/v) glucose or 2% galactose as the carbon source. Cells were concentrated, labelled and RNA was isolated as described above.
Polysome analysis
For polysome preparations, 250 ml yeast cultures were grown to an A600 0.6, treated with 100 μg/ml cycloheximide, harvested and disrupted by glass-bead lysis as described previously [28,29]. After the removal of cellular debris by sequential centrifugation at 2000, 5000 and 12000 g, 200 A260 units of each cell lysate were loaded on to a 36 ml linear (7–42%) sucrose gradient and subjected to centrifugation for 6–8 h at 141000 g in a Beckman SW28 rotor at 4 °C. Sucrose gradient fractions were analysed by continuous monitoring at 254 nm with a UV monitor (LKB UV-M II; LKB-Pharmacia, Uppsala, Sweden).
RESULTS
Deletion of NOP53 results in a severe growth defect
The protein encoded by the ORF YPL146c was initially identified by proteomic analysis of a highly enriched subcellular fraction of yeast NPCs (nuclear pore complexes) [17]. In that screen for potential nucleoporins, Ypl146p was localized primarily to a portion of the nucleus, which was reminiscent of a nucleolar signal, suggesting that Ypl146p was not a nucleoporin, but was probably a contaminant of the NPC fraction. On the basis of further characterization of Ypl146p, including its localization, function and predicted molecular mass, we have named Ypl146p as Nop53p.
Analysis of the Nop53p sequence indicated that the protein is well-conserved from yeast to humans. Nop53p was subjected to structural prediction analysis using Robetta [30] that predicted the presence of a large central coiled-coil domain that matched several proteins containing this signature structure of myosin heavy chain. Interestingly, the C-terminal region (amino acid residues approx. 380–450) did not conform to this family, but contained the highest primary sequence similarity to a putative human orthologue (glioma tumour suppressor candidate region 2; GTSCR2). Modelling of this domain predicted a leucine zipper motif (amino acid residues 402–434), which we targeted during characterization (see below).
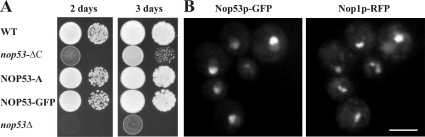
To examine the function of Nop53p in vivo, we created a null mutant by targeted gene disruption. In contrast to a previous finding from a yeast high-throughput study [31], the resulting haploid null mutants were viable, exhibiting growth at all temperatures tested. However, the growth rate of the null mutant was severely impaired, indicating that although not essential for cell survival on rich medium, Nop53p is critical for normal growth and its loss renders cells extremely unfit (Figure 1A). Indeed, log-phase nop53Δ cells growing in liquid-rich medium exhibited an 8-fold reduction in doubling time relative to WT cells (12 h for nop53Δ cells compared with 90 min for WT cells). This generalized growth defect observed in the nop53Δ strain was completely rescued after the introduction of a plasmid expressing WT NOP53 under the control of its endogenous promoter (results not shown). Moreover, C-terminally tagged protein A and GFP versions of Nop53p chimaeras, integrated into the NOP53 locus, also grew at WT rates (Figure 1) indicating functional complementation by the tagged versions of Nop53p used in the present study. To create a strain with a less severe growth defect, we also deleted the C-terminal region of Nop53p, which contains the predicted leucine zipper and is the region of highest identity with the human protein, GTSCR2. Indeed, deletion of the C-terminal 56 amino acids of Nop53p (nop53-ΔC) yielded a phenotype intermediate between that of the WT and Δnop53 strains in terms of growth (Figure 1A) and other aspects of Nop53p function (see below).
Figure 1. Nop53p is a non-essential protein that localizes to the nucleolus.
(A) Cells lacking the NOP53 ORF (nop53Δ) are viable but exhibit a strongly decreased growth rate compared with WT cells on rich media. Strains expressing genomically encoded GFP or pA chimaeras of Nop53p (NOP53–GFP, NOP53-A) show growth rates similar to those of WT cells, indicating that these fusion proteins are functional. The nop53-ΔC strain, expressing a truncation mutant of NOP53 lacking the C-terminal 56 amino acids, also shows a growth defect, but it is less dramatic compared with that of the NOP53 knockout. (B) Double labelling fluorescence micrographs of live cells expressing Nop53p–GFP and the nucleolar marker Nop1p–RFP. Note the co-localization of Nop53p–GFP with the Nop1p–RFP. Scale bar, 5 μm.
To monitor the localization of Nop53p in live cells, the Nop53p–GFP was observed by fluorescence microscopy. In agreement with our preliminary analysis [17] Nop53p–GFP was localized predominantly to the nucleolus as detected by co-localization with the nucleolar protein Nop1p–RFP. However, it should be noted that Nop53p–GFP was not exclusively nucleolar. By comparison to Nop1p–RFP, Nop53p–GFP showed significant nucleoplasmic signal suggesting that the protein may perform part of its function outside the confines of the nucleolus.
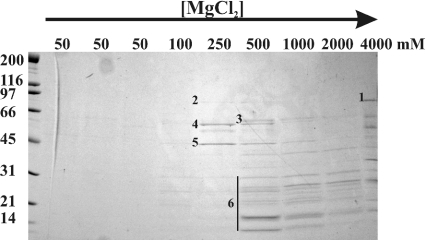
To gain insight about the function of Nop53p, we attempted to identify proteins with which it physically interacts. To do this, nuclei were isolated from strains producing a Nop53–protein A (Nop53p–pA) chimaera as the only version of Nop53p in the cell. Nuclei were lysed, the extract was incubated with IgG-Sepharose, and associated proteins were eluted from the resin with a gradient of MgCl2. Proteins from each fraction were separated by SDS/PAGE and the proteins were identified by tandem MS of excised gel slices (Figure 2) [20]. In addition to Nop53p, four prominent bands above the 31 kDa protein marker were identified: Cbf5p, Nop2p, Fpr3p and Fpr4p. All four proteins have been localized to the nucleolus and have been implicated in ribosomal biogenesis. Fpr3p and Fpr4p are members of the FKBP (FK506-binding protein) family, and are predicted to have peptidyl-prolyl cis-trans isomerase activity [32,33]. Cbf5p is an essential protein, and is proposed to function in snoRNA-guided uridine to pseudouridine conversions in rRNA [34,35]. Nop2p is also essential, is implicated in 60 S ribosomal subunit maturation, and is proposed to have RNA m(5)C methyltransferase activity [36,37]. In addition to these four ribosome biogenesis factors, MS also identified several ribosomal proteins that migrated below the 31 kDa protein marker. Together, these results suggest that Nop53p associates with assembling ribosomal precursors.
Figure 2. Nop53p physically interacts with rRNA processing factors involved in pre-60 S ribosomal subunit biogenesis.
Nop53p was immunopurified from a nuclear fraction prepared from cells expressing an Nop53p–pA chimaera. Proteins bound to Nop53p–pA were eluted with MgCl2 at increasing concentrations and analysed by SDS/PAGE followed by Coomassie Blue staining. The MgCl2 concentrations are noted above each lane. Molecular mass standards are shown at the left. Proteins corresponding to the numbered bands were identified by tandem MS of excised gel slices. Bands 1–6 were identified as Nop53p, Nop2p, Cbf5p, Fpr3p, Fpr4p and ribosomal proteins, respectively. Proteins that migrated below the 31 kDa marker were excised together and contained numerous 40 and 60 S ribosomal subunit proteins.
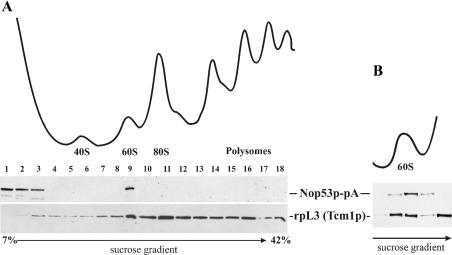
To further investigate this possibility, we examined whether Nop53p was directly associated with ribosomes. For these experiments, whole cell lysates were obtained from cells expressing Nop53p–pA and separated into ribosomal subunits, mature 80 S ribosomes and polysomes by sucrose-density-gradient centrifugation. The gradient was fractionated and simultaneously monitored by recording the A254 (Figure 3A). Proteins in each fraction were analysed by Western blotting using rabbit IgG to detect Nop53p–pA and the monoclonal antibody, TCM1, directed against the 60 S subunit protein rpL3p. Nop53p–pA was detected in the load fraction and associated with unassembled 60 S subunits, but was absent from fractions containing 40 S subunits, 90 S precursors, 80 S mature ribosomes and polysomes. Moreover, on further resolution of the 60 S region of the sucrose gradient (Figure 3B), it became apparent that Nop53p associates only with a subset of free Tcm1p-reactive subunits. These results suggest that Nop53p associates with immature 60 S subunits after processing of 90 S precursors to precursors of the small and large subunits, but is not retained on its further maturation and assembly into functional 80 S ribosomes. These results are consistent with the mass-spectrometric identification of Nop53p (Ypl146p) among several proteins associated with the 60 S preribosomal particle [7,10,11].
Figure 3. Nop53p co-fractionates with 60 S ribosomal subunits.
(A) Yeast lysates from cells expressing Nop53p–pA were fractionated on a linear 5–42% sucrose gradient and monitored by A254 to identify ribosomal fractions (upper trace). Peak fractions are labelled. Fractions derived from the gradient were analysed by Western blotting (lower panel) using monoclonal antibodies against rpL3 (Tcm1p) to detect 60 S-containing fractions and rabbit IgG to detect Nop53p–pA. Note that Nop53p–pA co-fractionates with the 60 S ribosomal subunit peak but not with the 40 S subunits, assembled 80 S ribosomes or polysomes. (B) Further resolution of the 60 S region of the gradient. The relevant region of a sucrose gradient similar to that in (A) is shown, but subjected to longer centrifugation. Note that the peaks of rpL3 (Tcm1p) and Nop53p–pA do not precisely coincide.
Nop53p is involved in 60 S preribosome biogenesis and rRNA processing
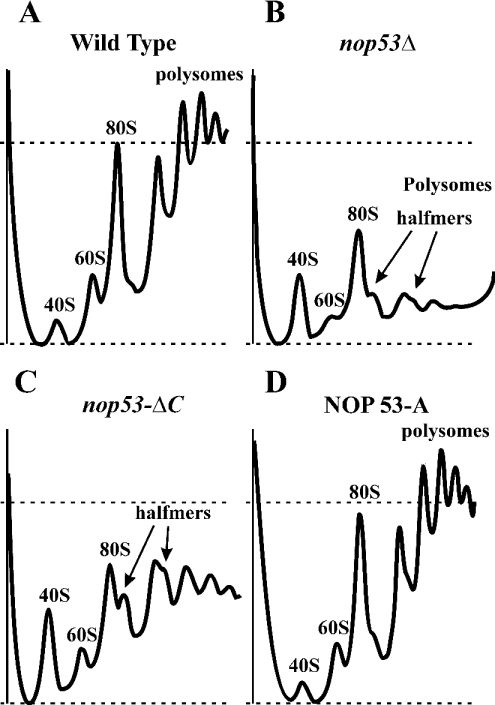
To investigate a potential role of Nop53p in ribosome biogenesis, ribosome profiles were obtained from WT and nop53Δ strains. nop53Δ cells showed a significant paucity of assembled 60 S subunits, revealed by a significant decrease in the mature 60 S:40 S subunit ratio (results not shown). This was also reflected in the polysome profiles (Figure 4). WT cells displayed a typical distribution of 40, 60 and 80 S particles and polysomes. By comparison, the profiles from nop53Δ cells suggested significant defects in 60 S biogenesis. The 60 S:40 S ratio was much lower than that found in WT cells, and the abundance of 80 S ribosomes and polysomes was significantly decreased. In addition, shoulders on the 80 S and polysome peaks were evident in the profiles. This feature is indicative of halfmers, or stalled 40 S subunits that accumulate due to a lack of 60 S subunits available to complete ribosome assembly. Profiles from nop53ΔC cells exhibited similar features including an imbalance in the 40 S:60 S ratio, decreased ribosomes and polysomes and the presence of halfmers, albeit not as significantly as exhibited by cells completely lacking Nop53p. Importantly, the profiles were normal in haploid cells containing Nop53p–pA (Figure 4D) and Nop53p–GFP (results not shown) chimaeras as the sole versions of Nop53p, or in nop53Δ cells expressing NOP53 from a plasmid (results not shown).
Figure 4. Deletion of NOP53 results in a 60 S subunit assembly defect.
Polysome profiles for WT (A), Δnop53 (B), nop53-ΔC (C) and NOP53 genomically tagged with protein A (NOP53-A) (D) were obtained by fractionation of cell lysates on 7–42% sucrose gradients and monitoring the optical density at 254 nm. The peaks corresponding to 40, 60, 80 S and polysomes are indicated. Loads were normalized by A260 before fractionation; thus, peak heights are sensitive indicators of the levels of each subunit. The dashed lines indicate the abundance of 80 S ribosomes in the WT strain. The nop53 mutant strains (B and C) exhibit deficit of 60 S particles as indicated by the decrease in free 60 and 80 S ribosomes and polysomes, and by the appearance of halfmers. The polysome profile of the NOP53-A strain (D) is indistinguishable from that of the WT strain.
As the biogenesis of ribosomes is inextricably linked to the complex processing of rRNA, we evaluated rRNA processing through pulse–chase, Northern hybridization and primer extension analyses (Figures 5 and 6) in the presence or absence of Nop53p. The pulse–chase labelling with [3H]uracil showed that strains lacking Nop53p accumulated pre-rRNA at the 35 and 27 S stages (Figure 5A), indicative of a 27 to 25 S processing defect (Figure 3A). Similar results were obtained by depleting Nop53p by controlling its expression using a GAL1 promoter and repressing the promoter by shifting cells to glucose as the sole carbon source. In these experiments, there was no evidence of a significant perturbation of the 18 S biogenesis pathway. In agreement with the ribosome and polysome analyses, such an rRNA processing defect is expected to lead to the observed 60 S abnormalities [38]. This is similar to the previously observed defects on loss of Nop2p [39], which we detect in association with Nop53p.
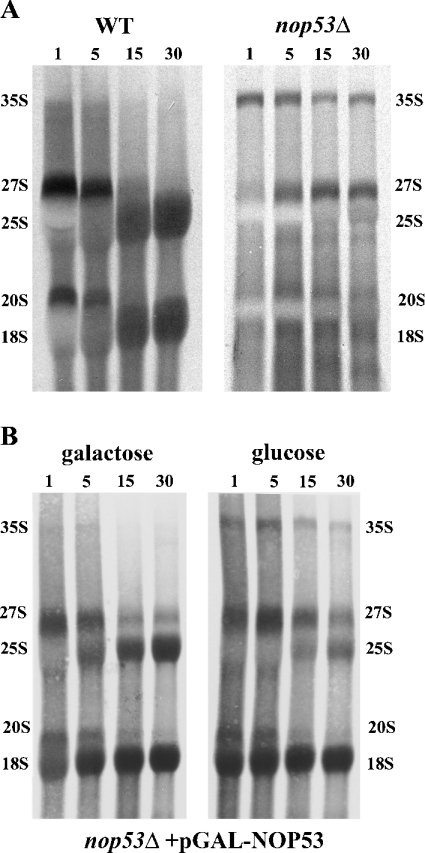
Figure 5. Pulse–chase analysis of RNA from Δnop53 cells (A) or from cells depleted of Nop53p (B).
(A) RNA from WT and Δnop53 cells was metabolically labelled with [3H]uracil for 1 min followed by a chase with excess of unlabelled uracil for 1, 5, 15 and 30 min (noted above each lane). For each lane, RNA corresponding to 50000 c.p.m. of the starting material was separated on a 1.3% (v/v) formaldehyde-agarose gel before autoradiography. (B) Δnop53 cells expressing pGAL-NOP53 were grown in either galactose or glucose for 14 h before pulse–chase analysis. The major rRNA species are marked. Cells lacking Nop53p exhibit defects in 27 to 25 S processing as indicated by the absence of labelled 25 S rRNA and accumulation of the larger 35 and 27 S precursor species.
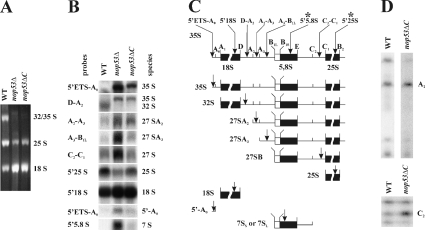
Figure 6. Northern hybridization and primer extension analysis reveal defects in rRNA processing in Δnop53 and nop53-ΔC strains.
(A) Total RNA isolated from WT and Δnop53 and nop53-ΔC cells was separated on a 1.4% formaldehyde/agarose gel and visualized by ethidium bromide staining. To normalize the 18 S signal between WT and nop53 strains, 2.5- and 2-fold more A260 units were loaded from Δnop53 and nop53-ΔC cells, respectively, relative to WT cells. Note that the intensities of the 25 S and 32 S bands are less in nop53 cells. (B) Northern-blot analysis of rRNA shows a specific defect in the 60 S subunit processing in nop53 strains. An identical gel to that shown in (A) was analysed by Northern blotting using oligonucleotide probes used to detect specific rRNA species. The probes are listed in the Experimental section. (C) The position of each probe and the rRNA species designed to detect are shown in the diagram of probe positions and pre-rRNA processing intermediates. Note the accumulation of the steady-state levels of the 35, 27 and 7 S species in nop53 cells and a decrease in 32 and 25 S levels indicating defects in 60 S processing. (D) Primer extension analyses were performed on the WT and nop53-ΔC strains. Analysis through ITS1 was performed using a 5′ to 5,8 S primer (marked with * in C), which hybridizes within mature 5.8 S rRNA, although analysis through ITS2 was performed using a 5′ 25 S primer (marked with * in C), that hybridizes within the mature 25 S rRNA. Note that the ITS1 and ITS2 regions of the 35 S rRNA transcript are not efficiently processed in nop53 cells as evidenced by the increased A2 and C2 signals.
Northern blotting using probes designed to identify different intermediates in the processing pathway and to reveal rRNA processing defects was also performed. Consistent with the pulse–chase results and polysome profiles, the nop53 strains were unable to produce normal amounts of mature 25 S rRNA. An analysis of the intermediates in nop53 cells revealed an accumulation of 35 and 27 S precursors, with a corresponding decrease in 32 and 25 S species (Figure 6B). This pattern is typical of mutants with defects early in the processing of rRNA (sites A0, A1, A2 and A3) or in the processing of the ITS2 (internally transcribed spacer 2) at sites C1 and C2. nop53 cells also accumulated 5′-A0 fragments along with 7 S precursors, suggesting that exosome function is compromised in the absence of Nop53p [40,41]. As predicted by results from polysome fractionations (Figure 4B) and pulse–chase analyses (Figure 5B), the biosynthesis of the 40 S subunit was relatively normal and we detected no accumulation of the 23 and 20 S precursors of the 18 S rRNA (Figure 6B; results not shown).
Primer extension studies were also performed to specifically detect the accumulation of uncleaved precursors in the internally transcribed spacers ITS1 and ITS2. Because sufficient high-quality RNA could not be isolated from Δnop53 cells, nop53-ΔC cells were used for this analysis. In these cells primer extension through ITS1 and ITS2 revealed an accumulation of the products cleaved at sites A2 and C2 (Figure 6C). These sites are present in the 27SA2 and 26 S species and are often abundant in 25 S deficient mutants [38]; thus, Nop53p affects events at this stage of ribosome biogenesis as well as earlier events. In the aggregate, these results suggest that Nop53p is required for efficient 60 S biogenesis and that this manifests primarily by an accumulation of 27 S processing intermediates.
DISCUSSION
Ribosome biogenesis is a temporally and spatially regulated assembly process involving the co-ordinated activity of numerous protein and nucleic acid cis- and trans-acting factors. In the present study, we have investigated the role of S. cerevisiae Nop53p, a highly conserved eukaryotic ribosomal biogenesis factor. Nop53p is primarily localized to the nucleolus and associates with assembling 60 S ribosomal subunits after cleavage of the 90 S precursor. Nop53p sediments with approx. 60 S particles, immunopurifies with other ribosome biogenesis factors and loss of Nop53p results in a dramatic decrease in the abundance of mature 60 S ribosome subunits, an accumulation of halfmers, but relatively little change in 40 S subunit levels. Because Nop53p was absent from the cytoplasm by fluorescence microscopy and from 80 S ribosome or polysome fractions, it appears that Nop53p associates early with the assembling 60 S subunit after the cleavage of the 35 S rRNA/90 S preribosome complex and departs the assembling 60 S complex before the formation of the mature 80 S ribosome.
Large-scale interaction studies based on affinity purifications followed by MS have detected Nop53p (Ypl146p), and numerous other proteins, in association with several pre-60 S biogenesis factors including Nsa3p, Nug1p, Rix1p, Sda1p and Arx1p, Rpl24p, and Nog1p [7,10,11]. In addition, Nop53p was absent from fractions associated with 90 or 40 S preribosomal components [9,12]. The detection of Nop53p with numerous different 60 S biogenesis factors suggests that Nop53p may be associated with a large multicomponent complex, or may associate with several different complexes during the biogenesis of 60 S subunits. Our results suggest that one such complex contains Cbf5p, Nop2p and Fkb3p and Fkb4p. Each of them was detected in association with Nop53p isolated by affinity purification. These four proteins also appear to be among the most tightly associated with Nop53p as they were resistant to extensive washing of the Nop53p–pA complex with an increasing concentration of MgCl2. Taken together, the abundance of interaction results from the present study and others clearly places Nop53p in the 60 S biogenesis pathway.
In agreement with this role for Nop53p, pulse–chase and Northern blotting revealed that the most significant defect in nop53 cells was in 27 S processing to the mature 25 S rRNA species. The concomitant accumulation of 35 S species is common to 27 S processing defects and was also observed in nop53 cells. Moreover, primer extension analysis also showed defects in processing of 27SA2 to 25 S. Cells were inefficient in processing both the A2 and C2 intermediates, although there was no obvious accumulation of A0 or A1, which are processed quickly, before the formation of the 27 S intermediate. This is similar to the situation with Nop2p, which we found to be tightly associated with Nop53p. In particular, the paucity of 60 S subunits and the accumulation of 35 and 27 S rRNA species are similar phenotypes to those observed on the loss of functional Nop2p [36,39]. Indeed, Nop2p is absolutely required for A0 processing as well as for efficient A1 and A2 processing [42]. Although these steps were the most sensitive to Nop53p (or Nop2p) loss, nop53 mutants had an overall decrease in the levels of rRNA, suggesting general defects as well as stage-specific defects. This could result from the specific involvement of Nop53p in many different events of 60 S processing, and/or the loss of additional proteins (interactors of Nop53p, such as Nop2p and Cbf5p). For example, both Nop2p and Cbf5p are involved in snRNP guiding and base-specific rRNA modifications (pseudouridine formation and 2′-O-ribose methylation, respectively). Lack of pseudouridines or 2′-O-methyl modifications of both rRNA and snoRNA can slow down or prevent multiple rRNA processing steps leading to general rRNA and ribosome defects [43,44]. rRNA undermodification has also been shown to decrease the efficiency of Rat1p exonuclease activity, perhaps by a quality control mechanism [43]. The 5′ to 3′ exonuclease activity of Rat1p is responsible for A3>B1 trimming (27SA3>27SB) and C2>C1 trimming (25.5 S>25 S) [45]. The accumulation of intermediate cleavage products C1 and possibly A2 (precursors to 5.8 S/25 S), revealed by primer extension, may be an indirect result of Nop53p loss leading to inefficient Rat1p function. Interestingly, similar C2-C1 processing delays were observed in nop2 mutants [36], further supporting the contention that Nop53p is important for Nop2p function.
It is interesting to note that although Nop53p interacts with Cbf5p, the most dramatic phenotypes of nop53 cells are not the same as those resulting from the loss of functional Cbf5p. Cbf5p is proposed to be the pseudouridine synthase and a component of the box H-ACA class of snoRNPs [35]. Accordingly, depletion of Cbf5p leads to defects in both 60 and 40 S biogenesis. In contrast, depletion of Nop53p had very little effect on 18 S rRNA biogenesis. Thus the lack of Nop53p did not appear to dramatically alter Cbf5p function, but, like Nop2p, had primary effects on 25 S rRNA and 60 S subunit biogenesis. We do note, however, that complete absence of Nop53p did lead to phenotypes that were not restricted to the 60 S pathway. For example, to compare rRNA from WT and Δnop53 mutants, more than twice the number of Δnop53 cells were required to get similar amounts of 18 S rRNA, suggesting that the defects associated with nop53 also (eventually) affected 40 S biogenesis. Thus, the observed 60 S defect appears to dominate the phenotype and might mask a less significant role in Cbf5p function and 40 S biogenesis.
In addition, nop53 mutants exhibit an accumulation of 7 S and 5′-A0 species, which are indicative of exosome defects [46–48]. However, because the exosome-dependent modifications take place late in the rRNA processing pathway, the accumulation of these fragments may also be attributed to the quality control mechanism, rather than to a specific Nop53p function. Moreover, we have observed such defects associated with Rat1p and its interacting partner, Rai1p, neither of which have been directly implicated in exosome function [20]. Similar results have been observed for Dob1/Mtr4p and Cgr1p [40,49].
Taken together, our results demonstrate that Nop53p plays an integral role in 60 S subunit biogenesis, through its association with key players such as Nop2p and (to a lesser extent) Cbf5p. On the basis of its association with Nop2p and other factors (Cbf5p, Fpr3p, Fpr4p and numerous ribosomal proteins), we prefer a model in which Nop53p plays more of a structural function than an enzymatic one of site-specific modification; however, an enzymatic activity for Nop53p cannot be ruled out at this point. Understanding its molecular role in the context of the maturing 60 S subunit will demand a more comprehensive analysis of the complex molecular events of ribosome biogenesis.
Acknowledgments
We thank M. Marelli and other members of Aitchison Laboratory (ISB, Seattle, WA, U.S.A.) for helpful discussions. This work was supported by operating grants and salary support from the Canadian Institutes for Health Research and Alberta Heritage Foundation for Medical Research to J.D.A. and R.W.W. and operating support from the Institute for Systems Biology and Merck (Whitehouse Station, NJ, U.S.A.) to J.D.A. R.W.W. is an AHFMR and CIHR Scientist. Y.S. is a recipient of a University of Alberta CHIA Fellowship for Foreign Students.
References
- 1.Fromont-Racine M., Senger B., Saveanu C., Fasiolo F. Ribosome assembly in eukaryotes. Gene. 2003;313:17–42. doi: 10.1016/s0378-1119(03)00629-2. [DOI] [PubMed] [Google Scholar]
- 2.Rout M. P., Aitchison J. D. Pore relations: nuclear pore complexes and nucleocytoplasmic exchange. Essays Biochem. 2000;36:75–88. doi: 10.1042/bse0360075. [DOI] [PubMed] [Google Scholar]
- 3.Rout M. P., Aitchison J. D. The nuclear pore complex as a transport machine. J. Biol. Chem. 2001;276:16593–16596. doi: 10.1074/jbc.R100015200. [DOI] [PubMed] [Google Scholar]
- 4.Fatica A., Tollervey D. Making ribosomes. Curr. Opin. Cell Biol. 2002;14:313–318. doi: 10.1016/s0955-0674(02)00336-8. [DOI] [PubMed] [Google Scholar]
- 5.Kressler D., Linder P., de La Cruz J. Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:7897–7912. doi: 10.1128/mcb.19.12.7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Warner J. R., Kumar A., Udem S. A., Wu R. S. Ribosomal proteins and the assembly of ribosomes in eukaryotes. Biochem. J. 1972;129:29–30. doi: 10.1042/bj1290029p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bassler J., Grandi P., Gadal O., Lessmann T., Petfalski E., Tollervey D., Lechner J., Hurt E. Identification of a 60 S preribosomal particle that is closely linked to nuclear export. Mol. Cell. 2001;8:517–529. doi: 10.1016/s1097-2765(01)00342-2. [DOI] [PubMed] [Google Scholar]
- 8.Harnpicharnchai P., Jakovljevic J., Horsey E., Miles T., Roman J., Rout M., Meagher D., Imai B., Guo Y., Brame C. J., et al. Composition and functional characterization of yeast 66 S ribosome assembly intermediates. Mol. Cell. 2001;8:505–515. doi: 10.1016/s1097-2765(01)00344-6. [DOI] [PubMed] [Google Scholar]
- 9.Grandi P., Rybin V., Bassler J., Petfalski E., Strauss D., Marzioch M., Schafer T., Kuster B., Tschochner H., Tollervey D., et al. 90 S pre-ribosomes include the 35 S pre-rRNA, the U3 snoRNP, and 40 S subunit processing factors but predominantly lack 60 S synthesis factors. Mol. Cell. 2002;10:105–115. doi: 10.1016/s1097-2765(02)00579-8. [DOI] [PubMed] [Google Scholar]
- 10.Nissan T. A., Bassler J., Petfalski E., Tollervey D., Hurt E. 60 S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 2002;21:5539–5547. doi: 10.1093/emboj/cdf547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saveanu C., Namane A., Gleizes P. E., Lebreton A., Rousselle J. C., Noaillac-Depeyre J., Gas N., Jacquier A., Fromont-Racine M. Sequential protein association with nascent 60 S ribosomal particles. Mol. Cell. Biol. 2003;23:4449–4460. doi: 10.1128/MCB.23.13.4449-4460.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schafer T., Strauss D., Petfalski E., Tollervey D., Hurt E. The path from nucleolar 90 S to cytoplasmic 40 S pre-ribosomes. EMBO J. 2003;22:1370–1380. doi: 10.1093/emboj/cdg121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lalev A. I., Nazar R. N. A chaperone for ribosome maturation. J. Biol. Chem. 2001;276:16655–16659. doi: 10.1074/jbc.M101157200. [DOI] [PubMed] [Google Scholar]
- 14.Aitchison J. D., Rout M. P., Marelli M., Blobel G., Wozniak R. W. Two novel related yeast nucleoporins Nup170p and Nup157p: complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J. Cell Biol. 1995;131:1133–1148. doi: 10.1083/jcb.131.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guthrie C., Fink G. R. Guide to yeast genetics and molecular biology. Methods Enzymol. 1991;194:1–863. [PubMed] [Google Scholar]
- 16.Dilworth D. J., Suprapto A., Padovan J. C., Chait B. T., Wozniak R. W., Rout M. P., Aitchison J. D. Nup2p dynamically associates with the distal regions of the yeast nuclear pore complex. J. Cell Biol. 2001;153:1465–1478. doi: 10.1083/jcb.153.7.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rout M. P., Aitchison J. D., Suprapto A., Hjertaas K., Zhao Y., Chait B. T. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ausubel F. M. New York: John Wiley & Sons; 1994. Current Protocols in Molecular Biology. [Google Scholar]
- 19.Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J. Biol. Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- 20.Sydorskyy Y., Dilworth D. J., Yi E. C., Goodlett D. R., Wozniak R. W., Aitchison J. D. Intersection of the Kap123p-mediated nuclear import and ribosome export pathways. Mol. Cell. Biol. 2003;23:2042–2054. doi: 10.1128/MCB.23.6.2042-2054.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strambio-de-Castillia C., Blobel G., Rout M. P. Isolation and characterization of nuclear envelopes from the yeast Saccharomyces. J. Cell Biol. 1995;131:19–31. doi: 10.1083/jcb.131.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rout M. P., Kilmartin J. V. Components of the yeast spindle and spindle pole body. J. Cell Biol. 1990;111:1913–1927. doi: 10.1083/jcb.111.5.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aitchison J. D., Blobel G., Rout M. P. Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science. 1996;274:624–627. doi: 10.1126/science.274.5287.624. [DOI] [PubMed] [Google Scholar]
- 24.Eng J. K., McCormack A. L., Yates J. R., III An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 25.Iouk T. L., Aitchison J. D., Maguire S., Wozniak R. W. Rrb1p, a yeast nuclear WD-repeat protein involved in the regulation of ribosome biosynthesis. Mol. Cell. Biol. 2001;21:1260–1271. doi: 10.1128/MCB.21.4.1260-1271.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tollervey D., Lehtonen H., Carmo-Fonseca M., Hurt E. C. The small nucleolar RNP protein Nop1p (fibrillarin) is required for pre-rRNA processing in yeast. EMBO J. 1991;10:573–583. doi: 10.1002/j.1460-2075.1991.tb07984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmitt M. E., Brown T. A., Trumpower B. L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Foiani M., Cigan A. M., Paddon C. J., Harashima S., Hinnebusch A. G. GCD2, a translational repressor of the GCN4 gene, has a general function in the initiation of protein synthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 1991;11:3203–3216. doi: 10.1128/mcb.11.6.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kressler D., de la Cruz J., Rojo M., Linder P. Fal1p is an essential DEAD-box protein involved in 40 S-ribosomal-subunit biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 1997;17:7283–7294. doi: 10.1128/mcb.17.12.7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chivian D., Kim D. E., Malmstrom L., Bradley P., Robertson T., Murphy P., Strauss C. E., Bonneau R., Rohl C. A., Baker D. Automated prediction of CASP-5 structures using the Robetta server. Proteins. 2003;53:524–533. doi: 10.1002/prot.10529. [DOI] [PubMed] [Google Scholar]
- 31.Winzeler E. A., Shoemaker D. D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J. D., Bussey H., et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 32.Shan X., Xue Z., Melese T. Yeast NPI46 encodes a novel prolyl cis-trans isomerase that is located in the nucleolus. J. Cell Biol. 1994;126:853–862. doi: 10.1083/jcb.126.4.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dolinski K., Muir S., Cardenas M., Heitman J. All cyclophilins and FK506 binding proteins are, individually and collectively, dispensable for viability in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 1997;94:13093–13098. doi: 10.1073/pnas.94.24.13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cadwell C., Yoon H. J., Zebarjadian Y., Carbon J. The yeast nucleolar protein Cbf5p is involved in rRNA biosynthesis and interacts genetically with the RNA polymerase I transcription factor RRN3. Mol. Cell. Biol. 1997;17:6175–6183. doi: 10.1128/mcb.17.10.6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lafontaine D. L., Bousquet-Antonelli C., Henry Y., Caizergues-Ferrer M., Tollervey D. The box H+ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev. 1998;12:527–537. doi: 10.1101/gad.12.4.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hong B., Brockenbrough J. S., Wu P., Aris J. P. Nop2p is required for pre-rRNA processing and 60S ribosome subunit synthesis in yeast. Mol. Cell. Biol. 1997;17:378–388. doi: 10.1128/mcb.17.1.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.King M. Y., Redman K. L. RNA methyltransferases utilize two cysteine residues in the formation of 5-methylcytosine. Biochemistry. 2002;41:11218–11225. doi: 10.1021/bi026055q. [DOI] [PubMed] [Google Scholar]
- 38.Venema J., Tollervey D. Processing of pre-ribosomal RNA in Saccharomyces cerevisiae. Yeast. 1995;11:1629–1650. doi: 10.1002/yea.320111607. [DOI] [PubMed] [Google Scholar]
- 39.Hong B., Wu K., Brockenbrough J. S., Wu P., Aris J. P. Temperature sensitive nop2 alleles defective in synthesis of 25 S rRNA and large ribosomal subunits in Saccharomyces cerevisiae. Nucleic Acids Res. 2001;29:2927–2937. doi: 10.1093/nar/29.14.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de la Cruz J., Kressler D., Tollervey D., Linder P. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8 S rRNA in Saccharomyces cerevisiae. EMBO J. 1998;17:1128–1140. doi: 10.1093/emboj/17.4.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Allmang C., Mitchell P., Petfalski E., Tollervey D. Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res. 2000;28:1684–1691. doi: 10.1093/nar/28.8.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beltrame M., Henry Y., Tollervey D. Mutational analysis of an essential binding site for the U3 snoRNA in the 5′ external transcribed spacer of yeast pre-rRNA. Nucleic Acids Res. 1994;22:5139–5147. doi: 10.1093/nar/22.23.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Venema J., Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 44.Tollervey D., Lehtonen H., Jansen R., Kern H., Hurt E. C. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell (Cambridge, Mass.) 1993;72:443–457. doi: 10.1016/0092-8674(93)90120-f. [DOI] [PubMed] [Google Scholar]
- 45.Geerlings T. H., Vos J. C., Raue H. A. The final step in the formation of 25 S rRNA in Saccharomyces cerevisiae is performed by 5′→3′ exonucleases. RNA. 2000;6:1698–1703. doi: 10.1017/s1355838200001540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H. D., Zagorski J., Fournier M. J. Depletion of U14 small nuclear RNA (snR128) disrupts production of 18 S rRNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 1990;10:1145–1152. doi: 10.1128/mcb.10.3.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mitchell P., Petfalski E., Shevchenko A., Mann M., Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell (Cambridge, Mass.) 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 48.Butler J. S. The yin and yang of the exosome. Trends Cell Biol. 2002;12:90–96. doi: 10.1016/s0962-8924(01)02225-5. [DOI] [PubMed] [Google Scholar]
- 49.Moy T. I., Boettner D., Rhodes J. C., Silver P. A., Askew D. S. Identification of a role for Saccharomyces cerevisiae Cgr1p in pre-rRNA processing and 60 S ribosome subunit synthesis. Microbiology. 2002;148:1081–1090. doi: 10.1099/00221287-148-4-1081. [DOI] [PubMed] [Google Scholar]