Abstract
A recently described family of TGN (trans-Golgi network) proteins, all of which contain a GRIP domain targeting sequence, has been proposed to play a role in membrane transport. On the basis of the high content of heptad repeats, GRIP domain proteins are predicted to contain extensive coiled-coil regions that have the potential to mediate protein–protein interactions. Four mammalian GRIP domain proteins have been identified which are targeted to the TGN through their GRIP domains, namely p230, golgin-97, GCC88 and GCC185. In the present study, we have investigated the ability of the four mammalian GRIP domain proteins to interact. Using a combination of immunoprecipitation experiments of epitope-tagged GRIP domain proteins, cross-linking experiments and yeast two-hybrid interactions, we have established that the GRIP proteins can self-associate to form homodimers exclusively. Two-hybrid analysis indicated that the N- and C-terminal fragments of GCC88 can interact with themselves but not with each other, suggesting that the GRIP domain proteins form parallel coiled-coil dimers. Analysis of purified recombinant golgin-97 by CD spectroscopy indicated a 67% α-helical structure, consistent with a high content of coiled-coil sequences. These results support a model for GRIP domain proteins as extended rod-like homodimeric molecules. The formation of homodimers, but not heterodimers, indicates that each of the four mammalian TGN golgins has the potential to function independently.
Keywords: dimer, golgin, GRIP domain protein, homodimer, trans-Golgi network, yeast two-hybrid
Abbreviations: DTSSP, dithiobis(sulphosuccinimidyl propionate); TGN, trans-Golgi network
INTRODUCTION
A large number of long coiled-coil proteins have been identified on intracellular organelles, in particular the Golgi and endosomes [1]. On the basis of their high content of coiled coils, these molecules are predicted to adopt a rod-like or extended fibrous structure. Members of the family of coiled-coil proteins specifically located on the Golgi (termed golgins), such as p115 and GM130, have been shown to be important as tethering molecules and in the biogenesis of membranes of the Golgi stack [1–5]. A number of golgins specifically associated with the TGN (trans-Golgi network) have recently been identified [6,7] based on the presence of a modestly conserved, 45-residue Golgi targeting sequence located at the C-terminus, called the GRIP domain [8–10].
The importance of the TGN golgins is highlighted by the finding that these GRIP domain proteins are conserved throughout evolution, as functional GRIP targeting sequences have been identified in yeast, plants and protozoan parasites [10–12]. Alteration in the levels of TGN golgins has been shown to result in abnormalities in TGN structure and/or a block in membrane transport pathways, indicating that they may play a role similar to the golgins of the Golgi stack, as either matrix components and/or in vesicular tethering [7,13,14].
There are four mammalian GRIP domain proteins which have been identified and all are localized to the TGN of HeLa cells [7]. The mammalian TGN golgins include a 230 kDa peripheral membrane protein (p230, also known as golgin-245), shown to be specifically associated with buds/vesicles of TGN membranes [15–18], golgin-97 [19], GGC88 and GGC185 (GCC stands for Golgi-localized coiled-coil protein) [7]. Like other members of the golgin family, the sequence of GRIP domain proteins indicates a high percentage (75–85%) of α-helical heptad repeats, sequences typical of coiled coils. The prediction of coiled-coil domains suggests that the TGN golgins probably exist as dimers or oligomers. An important question is whether such dimers/oligomers are homo-oligomers or whether different members of the GRIP family can associate to form hetero-oligomers. This is a relevant issue in defining whether the TGN golgins can function independent of each other or whether their activities are closely linked. In the present study, using a combination of approaches, we have investigated the interaction between different members of the TGN golgin family and have analysed the structural properties of one family member.
EXPERIMENTAL
Antibodies
Epitope-tagged proteins were detected with the monoclonal antibody anti-Myc (9E10) [20] and monoclonal antibody anti-FLAG (M2) (Sigma, Castle Hill, NSW, Australia). Horseradish peroxidase-conjugated rabbit anti-mouse Ig and porcine anti-rabbit Ig were obtained from Dako (Carpinteria, CA, U.S.A.). Rabbit antibodies to GCC88 have been described in [7]. Sheep anti-rabbit Ig-FITC, sheep anti-mouse Ig-FITC and sheep anti-human IgG-FITC were purchased from Silenus Laboratories (Melbourne, VIC, Australia).
Cell culture and transfection
HeLa and COS cells were maintained as monolayers in Dulbecco's modified Eagle's medium, supplemented with 10% (v/v) foetal calf serum, 2 mM glutamine and 100 μg/ml penicillin/streptomycin, in a humidified 37 °C incubator with 10% CO2. Transient transfections of cells were performed using FuGENE™ transfection reagent (Roche, Castle Hill, NSW, Australia) as described previously [6].
Mammalian expression vectors
pCIneoGCC88, Myc-tagged GCC185, Myc-tagged GCC88 and Myc-tagged p230 have been described previously [7,9,21]. To generate full-length golgin-97 with a triple Myc tag at the N-terminus, the golgin-97 cDNA was subcloned into the vector pCMU-MYC. Constructs encoding the full-length GCC88, p230, golgin-97 and GCC185 with a triple FLAG epitope at the N-terminus were generated by cloning into pCMU-FLAG vector [22].
Immunofluorescence
Cells were processed for immunofluorescence as described previously [21] and examined by confocal microscopy using a Bio-Rad MRC-1024 imaging system.
Immunoblotting
Cell extracts were diluted in 50 mM Tris/HCl (pH 6.8) containing 100 mM dithiothreitol, 2% (w/v) SDS and 10% (v/v) glycerol and subjected to SDS/PAGE. Proteins were then transferred on to PVDF membranes, and the membrane was dried. Antibodies were diluted in PBS containing 5% (w/v) skim milk powder and incubated on the membrane for 1 h, followed by three 10 min washes with 0.05% Tween 20/PBS. Membranes were then incubated with peroxidase-conjugated anti-mouse and bound antibodies detected by enhanced chemiluminescence (Lumi-Light; Roche) as described previously [18].
Cross-linking
Subconfluent cultures of HeLa cells were either untransfected or transfected with pCIneo-GCC88 24 h before harvesting. The cells were washed with PBS and harvested using a cell scraper, then lysed with 1% Igepal CA-630 (Sigma) in PBS on ice for 30 min. The extract was centrifuged at 700 g for 10 min, and the supernatant was incubated with 0.05–1.0 mM 3,3′-DTSSP [3,3′-dithiobis(sulphosuccinimidyl propionate); Pierce Biotechnology, Rockford, IL, U.S.A.] for 30 min at room temperature (25 °C). The reaction was stopped by the addition of Tris/HCl (pH 7) to a final concentration of 50 mM and incubation for 15 min. Cross-linked extracts were analysed by non-reducing SDS/PAGE and immunoblotting.
Co-immunoprecipitation of mammalian GRIP proteins
HeLa cells were co-transfected with pairs of Myc and FLAG constructs. After 24–48 h, monolayers were washed with PBS, harvested and resuspended in lysis buffer (1% Igepal in PBS) containing a protease inhibitor cocktail (Roche Diagnostics, Mannheim, Germany) for 30 min at 4 °C. The extract was clarified by centrifuging at 13000 g for 10 min. Anti-Myc monoclonal antibodies were then added to the supernatant at a final concentration of 2–4 μg/ml, and the mixture was gently rotated for 2 h at 4 °C. Protein G–Sepharose beads (Amersham Biosciences, Uppsala, Sweden) were added to a concentration of 4% (v/v) and the mixtures rotated for 30 min. The Protein G–Sepharose beads were collected by centrifuging at 2000 g for 5 min and washed four times with lysis buffer. Bound proteins were eluted by boiling the beads in SDS/PAGE loading buffer and analysed by immunoblotting with either anti-Myc or anti-FLAG antibodies.
Yeast two-hybrid analysis
pHybLex-GCC88, pHybLex-GCC88aa1-330, pHybLex-GCC88aa331-775, pGAD-GCC88 and pGAD-GCC88aa441-775 were obtained by subcloning the full-length [7] and partial GCC88 cDNAs, isolated from full-length GCC88 cDNA by digestion with restriction enzymes, into the polylinker sites of pHybLex/Zeo (LexA-binding domain; Invitrogen) and pGAD GH (GAL4 activation domain; Clontech, Palo Alto, CA, U.S.A.) respectively. The pHybLex/Zeo and pGAD GH constructs were co-transformed in to the Saccharomyces cerevisiae reporter stain L40 [23] using a lithium acetate method [24]. Transformants were selected on synthetic medium lacking leucine and containing 300 μg/ml zeocin. HIS3 reporter gene activation was detected by analysing growth on a medium lacking histidine and leucine and containing zeocin and 7.5 mM 3-aminotriazole. Constructs were initially analysed to ensure absence of autoactivation of HIS3 reporter gene. LacZ reporter gene activation was assessed using the liquid β-galactosidase assay [25]
Expression and purification of His-tagged golgin-97
Full-length golgin-97 was cloned into the pET28a expression vector (Clontech), which contains a His tag and T7 epitope at the N-terminus. Recombinant protein was expressed in Escherichia coli BL21(DES) strain grown in Luria–Bertani broth containing kanamycin and chloramphenicol. Overnight cultures were diluted in Luria–Bertani broth and kanamycin and incubated for 1 h at 30 °C before recombinant protein expression was induced with 0.1 mM isopropyl β-D-thiogalactoside by incubation at 30 °C for 3–4 h, with shaking. Bacteria were lysed by freeze–thawing followed by sonication, and cell lysates prepared by the addition of 1% sarkosyl in PBS for 30 min at 4 °C. His-tagged golgin-97 was purified by cobalt affinity chromatography. Weakly bound or non-specific proteins were eluted from the TALON cobalt affinity column with PBS containing 10 mM imidazole, before eluting His-tagged proteins with PBS containing 200 mM imidazole. Eluted samples were dialysed against PBS and protein concentration was determined by measuring absorbance A at 280 nm (mass absorption coefficient ε=0.335 mg·ml−1·cm−1).
CD spectroscopy
CD spectra of pure golgin-97 (0.42 mg/ml) dissolved in PBS (pH 7.4) were recorded on an Aviv 62DS CD spectrophotometer between the wavelengths 205 and 250 nm at a temperature of 20 °C. Data were collected using a step size of 0.5 nm and a slit bandwidth of 1.5 nm in a 1 mm path-length quartz cuvette. Signal averaging time was 2.0 s and ellipticities were reported as Δε in units of M−1·cm−1. CD spectra were fitted using a database comprising 43 soluble proteins (IBasis 4) employing the programs CDSSTR, CONTINLL and SELCON3 [26], which are included in the CDPro package available at http://lamar.colostate.edu/~sreeram/CDPro/.
RESULTS
The four mammalian GRIP domain proteins can interact to form homo-oligomers, but not hetero-oligomers
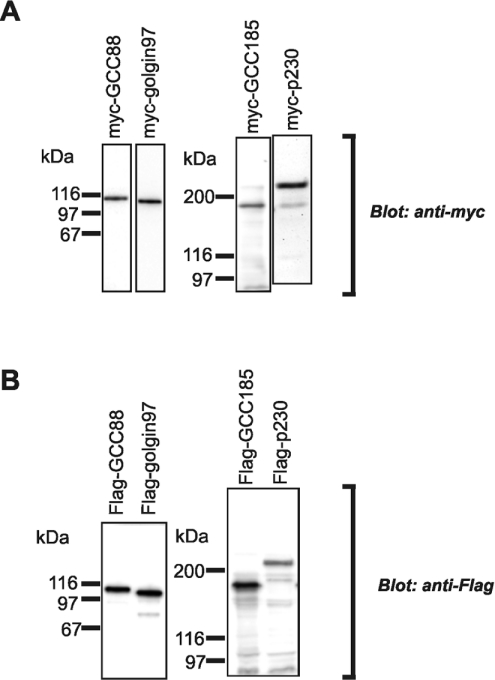
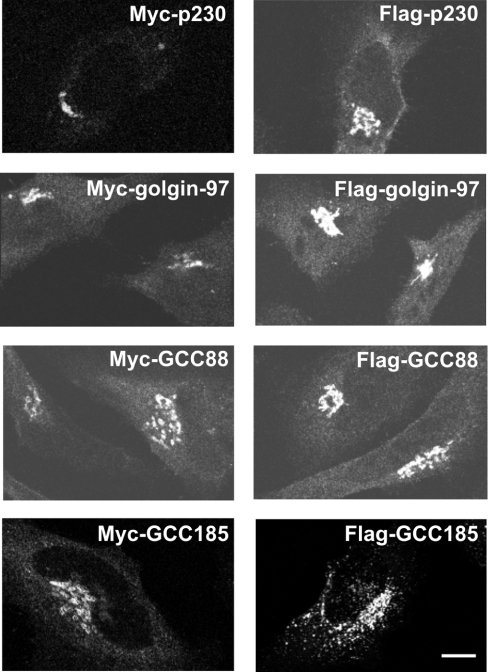
To determine whether the four mammalian GRIP proteins interact to form oligomers, a series of co-immunoprecipitation experiments was performed using epitope-tagged constructs. Myc- and FLAG-tagged full-length constructs were generated for each of the four TGN golgins as described in the Experimental section. Immunoblotting of HeLa cells transfected with each construct showed that each of the four Myc-tagged and four FLAG-tagged golgins were the expected size, with only low levels of degradation detected (Figure 1). Each of the Myc- and FLAG-tagged TGN golgins localized predominantly to the juxtanuclear Golgi region of transfected HeLa cells, as detected by immunofluorescence (Figure 2), suggesting that the proteins were folding correctly.
Figure 1. Expression of full-length Myc- and FLAG-tagged GRIP domain proteins.
HeLa cells were transfected with Myc- or FLAG-tagged GCC88, golgin-97, GCC185 or p230, as indicated. Extracts of transfected cells were analysed by immunoblotting with (A) monoclonal anti-Myc antibodies and (B) monoclonal anti-FLAG antibodies, as indicated, using a chemiluminescence detection system.
Figure 2. Localization of Myc- and FLAG-tagged GRIP domain proteins in transfected HeLa cells.
HeLa cells were transfected with constructs encoding either Myc- or FLAG-tagged GRIP domain proteins, as indicated, fixed, permeabilized and stained with anti-Myc or anti-FLAG monoclonal antibody followed by FITC goat anti-mouse Ig. Scale bar, 10 μm.
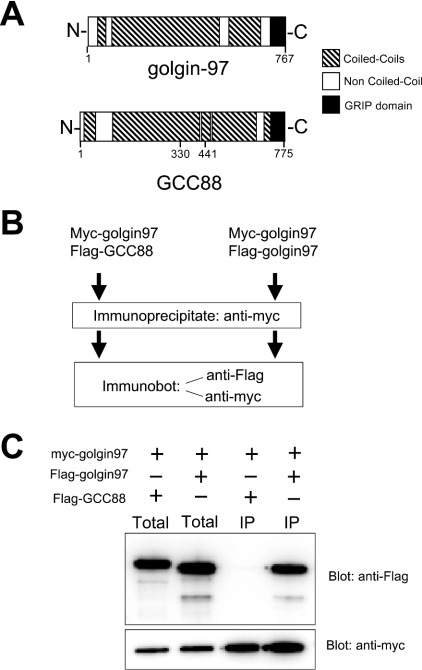
Details of the domain structure of untagged golgin-97 and GCC88 are shown in Figure 3(A) to illustrate the extensive coiledcoil regions and the C-terminal GRIP domain. The strategy for the co-immunoprecipitation experiments is outlined in Figure 3(B). HeLa cells were transfected with pairs of Myc- and FLAG-tagged constructs. The transfected cell lysates were then immunoprecipitated with anti-Myc antibodies and the precipitates were immunoblotted with anti-Myc or anti-FLAG. To ensure that the anti-Myc antibody did not cross-react with any of the FLAG-tagged constructs, HeLa cells were initially transfected with FLAG–GCC88, FLAG–golgin-97, FLAG–GCC185 or FLAG–p230, and immunoprecipitation was performed using anti-Myc antibodies. Immunoblotting of the total cellular extracts with anti-FLAG showed that abundant levels of the FLAG-tagged proteins had been produced, yet no FLAG-tagged proteins were detected in the anti-Myc immunoprecipitate, indicating that there was no cross-reactivity between the anti-Myc antibody and the FLAG-tagged proteins (results not shown).
Figure 3. Co-immunoprecipitation of epitope-tagged GRIP domain proteins.
(A) Strategy for determining interactions between GRIP domain proteins. HeLa cells were co-transfected with pairs of Myc- and FLAG-tagged constructs and extracts of transfected cells were immunoprecipitated with anti-Myc antibody. The immunoprecipitate was then blotted with anti-FLAG and anti-Myc antibodies. (B) Co-immunoprecipitation of Myc–golgin-97 with either FLAG–golgin-97 or FLAG–GCC88. (C) HeLa cells were co-transfected with pairs of Myc- and FLAG-tagged constructs, as indicated, and extracts of transfected cells were analysed directly (Total) or immunoprecipitated with anti-Myc antibody (IP). Samples were blotted with anti-FLAG or anti-Myc antibodies, as indicated.
In the example shown in Figure 3, HeLa cells were transfected with Myc–golgin-97 together with either FLAG–golgin-97 or FLAG–GCC88. Immunoblotting of the total lysates revealed that all recombinant proteins were produced in abundant levels. FLAG–golgin-97, but not FLAG–GCC88, was detected in the anti-Myc immunoprecipitate. This result indicates that golgin-97 can self-interact, but cannot interact with GCC88.
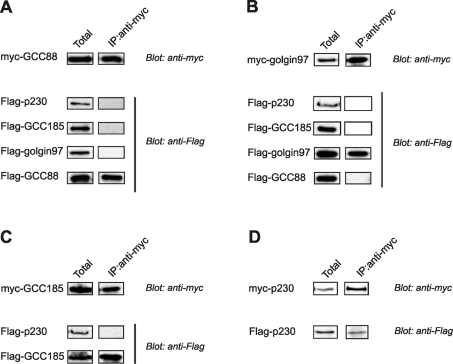
This approach was extended to analyse interactions between all possible pairings of the four GRIP domain proteins, as shown in Figure 4. Each of the panels of Figure 4 shows the interaction of one of the Myc-tagged golgins with the FLAG-tagged proteins. In each case, and as expected, the Myc-tagged golgin was detected in the blot of the anti-Myc immunoprecipitate. Of the four FLAG-tagged constructs that were co-transfected with Myc–GCC88, only FLAG–GCC88 was detected in the immunoprecipitate (Figure 4A). Similarly, of the four constructs co-transfected with Myc–golgin-97, only FLAG–golgin-97 was present in the immunoprecipitate (Figure 4B). FLAG–p230 and FLAG–GCC185 were co-transfected with Myc–GCC185, and only FLAG–GCC185 was present in the immunoprecipitate (Figure 4C). Finally, Myc–p230 co-precipitated FLAG–p230 (Figure 4D). Collectively, these results reveal that the four GRIP domain proteins are able to self-associate as homo-oligomers, and that this interaction is specific as no hetero-oligomers were observed between any of the pairs tested.
Figure 4. The GRIP domain proteins form homo-oligomers.
HeLa cell monolayers were co-transfected with (A) Myc–GCC88, (B) Myc–golgin-97, (C) Myc–GCC185, (D) Myc–p230 and FLAG-tagged full-length GRIP domain proteins, as indicated. The cells were harvested and complexes containing the Myc-tagged proteins were immunoprecipitated with anti-Myc antibodies. Samples of total extract (Total) and immunoprecipitate (IP) were analysed by immunoblotting with monoclonal anti-Myc or anti-FLAG antibodies, as indicated, followed by horseradish peroxidase-conjugated anti-mouse Ig antibodies. Immunoblots were developed by a chemiluminescence detection system. Loadings represent approx. 2% of input and 20% of immunoprecipitate.
GCC88 from HeLa cells is a dimer
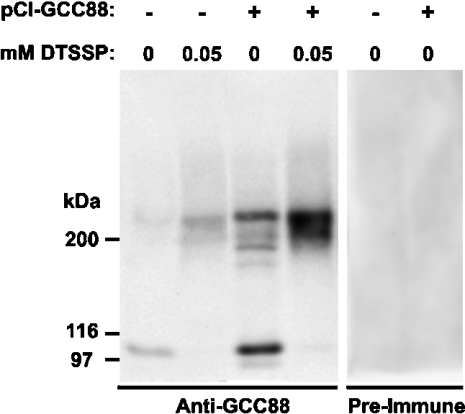
To investigate the size of the oligomers, we analysed the behaviour of one of the TGN golgins, namely GCC88, in more detail. Endogenous GCC88 of HeLa cells was detected by immunoblotting with anti-GCC88 antibodies as a monomer of 88 kDa (apparent molecular mass 105 kDa; Figure 5). In addition, we observed a weakly stained band of approx. 210 kDa with the anti-GCC88 serum, but it was absent from the preimmune serum. This approx. 210 kDa band may represent an SDS-resistant dimer of GCC88. To investigate further the ability of GCC88 to form dimers, HeLa cell extracts were treated with the chemical cross-linker, DTSSP, before analysis by immunoblotting with anti-GCC88 antibodies. After cross-linking with 0.05 mM DTSSP, the 105 kDa monomeric band of GCC88 was no longer detected and a broad band of approx. 200–230 kDa was very prominent, consistent with the formation of covalently cross-linked GCC88 dimers (Figure 5). A similar experiment was also performed using HeLa cells transfected with recombinant GCC88 to increase the level of expression of this TGN golgin. In untreated transfected cell extracts, the 105 kDa monomeric band of GCC88 was detected together with the approx. 210 kDa band as well as additional weakly stained bands around 200 kDa. In transfected cell extracts treated with 0.05 mM DTSSP, GCC88 migrated exclusively with an apparent molecular mass of approx. 200–210 kDa (Figure 5). The broad nature of this band may reflect the behaviour of the dimer in SDS/PAGE under non-reducing conditions. No components were detected in either untransfected or cell extracts transfected with preimmune serum (Figure 5). Therefore GCC88 can form a dimer.
Figure 5. Cross-linking of cytosolic GCC88.
Untransfected HeLa cells, or pCI-GCC88 transfected HeLa cells, were extracted and extracts treated with the indicated concentrations (mM) of the cross-linker DTSSP and analysed by non-reducing SDS/PAGE followed by immunoblotting with rabbit anti-GCC88 antibodies or preimmune serum as indicated, using a chemiluminescence detection system.
GCC88 forms a parallel coiled-coil dimer
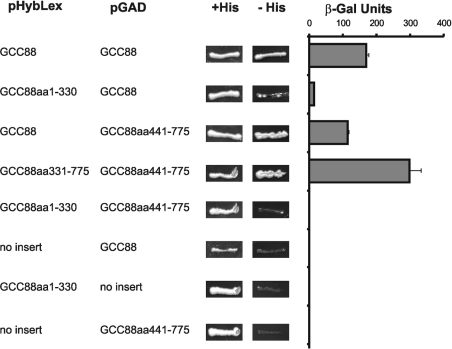
On the basis of the above, GCC88 probably forms a coiled-coil homodimer. Coiled-coil dimers may be orientated in a parallel (head-to-head) or anti-parallel (head-to-tail) fashion [27]. To distinguish between these possibilities, we utilized the yeast two-hybrid assay to analyse interactions between full-length GCC88 and a number of N- and C-terminal truncation mutants. A number of GCC88 constructs were cloned into the bait vector pHybLex, which contains the LexA DNA-binding domain, and the prey vector pGAD-GH, which contains the GAL4 activation domain. The reporter yeast strain L40 was transformed with pairs of constructs, and grown on selective medium. Interactions between GCC88 fusion proteins were assessed by HIS3 and LacZ reporter gene activation, measured by the ability of transformed yeast to grow in a medium lacking histidine and by assaying for β-galactosidase activity.
Strong interaction was observed between the full-length GCC88 constructs, LexA-GCC88 and GAD-GCC88, confirming that GCC88 forms homodimers (Figure 6), and consistent with the cross-linking and immunoprecipitation experiments. The N-terminal construct LexA-GCC88aa1-330 (refer also to Figure 3A for GCC88 constructs) interacted, albeit weakly, with the full-length GCC88 construct GAD-GCC88, and the C-terminal construct GCC88aa441-775 strongly interacted with the full-length construct LexA-GCC88, indicating that these truncated constructs probably contain sufficient number of coiled-coil domains to dimerize. Next, interactions between these truncated constructs were examined to determine the orientation of GCC88 dimer. The C-terminal construct, LexA-GCC88aa331-775 showed strong interaction with the C-terminal construct GAD-GCC88aa441-775, while the N-terminal construct LexA-GCC88aa1-330 did not interact with the C-terminal construct. This indicates that GCC88 is able to form parallel dimers but not anti-parallel dimers. Control transformations with empty vectors in combination with each of the constructs tested confirmed that the interactions observed were specific.
Figure 6. Dimerization of GCC88 constructs analysed by yeast two-hybrid interactions.
The indicated GCC88 constructs were generated as fusions with the DNA-binding protein LexA in pHybLex/Zeo and the GAL4 activation domain in pGAD GH. GCC88 refers to the full-length 775-amino-acid sequence, and the amino acid numbers denote truncated constructs. The pHybLex/Zeo and pGAD constructs were co-transformed into yeast reporter strain L40. Transformants were initially selected on medium lacking leucine and containing zeocin. The interaction between LexA and GAL4 fusions was assessed by HIS3 reporter gene activation as measured by the ability of transformants to grow on medium lacking histamine and by LacZ reporter gene activation using the liquid β-galactosidase assay. β-Galactosidase activity is given in Miller units. Assays were performed in triplicate.
Analysis of golgin-97 constructs by two-hybrid analysis also demonstrated, as expected, an interaction between the full-length golgin-97 constructs, LexA-golgin-97 and GAD-golgin-97, but not between LexA-golgin-97 and GAD-GCC88, again indicating the specific formation of homodimers (results not shown).
Analysis of purified His6-tagged golgin-97 by CD
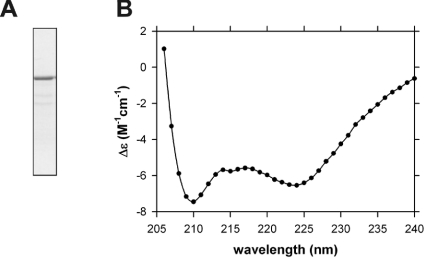
The high content of heptad repeats of the TGN golgins predicts extensive coiled-coil regions. To analyse directly the α-helical content of GRIP domain proteins, recombinant full-length proteins were generated for structural analysis. The two smaller TGN golgins, namely GCC88 and golgin-97, were expressed in bacteria as His6-tagged proteins; however, only His6-tagged golgin-97 was expressed at a high level to allow purification at sufficient material purity for analysis (Figure 7A).
Figure 7. CD spectrum of purified His6-tagged golgin-97.
(A) Purified His6-tagged golgin-97 analysed by SDS/PAGE and stained with Coomassie Blue. (B) CD spectrum of purified His6-tagged golgin-97 (0.42 mg/ml). The change in ellipticity, Δε (M−1·cm−1), is plotted as a function of wavelength (nm) for golgin-97 (0.42 mg/ml) dissolved in PBS (open symbols). The data are superimposed with the non-linear best-fit using the program CDSSTR [26], yielding 67% α-helix, 12% β-strand, 9% turn and 12% unordered structure (—) with an r.m.s.d. of 0.277.
To determine the degree of secondary structure in a sample of pure recombinant His-tagged golgin-97, CD spectroscopy was conducted in aqueous solution under physiological conditions. The data plotted as Δε versus wavelength are shown in Figure 7(B). The CD spectrum of golgin-97 reveals double minima at approx. 210 and 222 nm (Figure 7B), which is characteristic of the α-helical structure. This assertion is confirmed by non-linear least-squares analysis using the program CDSSTR (1), which yielded a best-fit to 67% α-helical structure with an r.m.s.d. (root mean-square deviation) of 0.277 (Figure 7B, solid line). Similar analyses were obtained using the programs CONTINLL and SELCON3 [26]. The CD data are consistent with the predominantly coiled-coil structure predicted from the golgin-97 protein sequence.
DISCUSSION
GRIP domain proteins have emerged as important components of the TGN involved in maintaining the highly complex structure of the TGN and in the regulation of the membrane traffic [28]. Mammalian cells express four TGN golgins; however, it is not known if each TGN golgin has a specific function or if their functions are redundant. In defining the function of the mammalian GRIP proteins, it is important to ascertain whether they exist as homo-oligomers or whether different members of the GRIP family can associate to form hetero-oligomers. In the present study, we have demonstrated that the TGN golgins can self-interact to form parallel homodimers. First, GCC88 can be cross-linked yielding a product of approx. twice the size of the monomer. Secondly, immunoprecipitation experiments using epitope-tagged GRIP domain proteins demonstrated that all four TGN golgins can self-interact, but not interact with any of the other three members. Thirdly, analysis of yeast two-hybrid interactions has established that the GRIP proteins can self-interact to form parallel coiled-coil homodimers; and fourthly, analysis of purified recombinant full-length golgin-97 confirmed the high content of α-helical structure. The formation of homodimers, but not heterodimers, indicates that each TGN golgin has the potential to function independently from the other members of the GRIP family.
The Golgi membrane binding of each of the four mammalian GRIP domain proteins is G-protein-dependent [7,28]. Recruitment of p230 and golgin-97 to the TGN is mediated through an interaction with Arl1, a member of the ARF/Arl small G-protein family. The structure of the Arl1-GTP complexed with the GRIP domain of p230/golgin-245 has recently been solved by crystallography by two groups [29,30]. These studies provided insight into the structural basis for the GTPase-dependent recruitment of the GRIP domain to TGN membranes. Of particular relevance to the present study was the finding that the isolated GRIP domain forms a homodimer that interacts with two Arl1-GTP molecules, a result consistent with our findings on the behaviour of the full-length molecule. Furthermore, the structure of the Arl1–GRIP domain complex predicts that the coiled-coil regions of these TGN golgins are orientated to face away from the lipid bilayer. Dimerization of the GRIP domain is critical for Golgi targeting, since mutations which disrupt the formation of GRIP domain dimers also result in loss of Golgi targeting [29,30]. On the basis of structural requirements for the interaction of Arl1-GTP and the p230 GRIP domain, those authors proposed that the full-length GRIP domain proteins form parallel homodimers. In the present study, we have confirmed this prediction and directly demonstrated that the full-length TGN golgins form parallel homodimers. The structure of the TGN golgins would predict that recruitment to TGN involves two C-termini in close proximity to the TGN membranes, with the long homodimeric coiled-coil domain orientated away from the membrane.
Given the high content of the α-helix, TGN golgins probably form extended rod-like molecules. For example, with a 67% α-helical content and assuming a rod-like shape, golgin-97 is predicted to be 75 nm in length. As the C-terminal GRIP domain interacts with membranes, the N-terminus of the molecule has the potential to form contacts between membranes or other structures at some distance from the GRIP attachment site. The N-terminal domains of the different mammalian TGN golgins do not show any sequence similarity and, therefore, may have distinct properties. Given the formation of homodimers, it is now important to define the nature of the interactive partners with the individual members of the TGN golgin family.
Acknowledgments
This work was supported by funding from the Australian Research Council (to P.A.G.). M.R.L. is supported by an Australian Postgraduate Award. We thank Dr M. Fitzler (Departments of Medicine and Medical Biochemistry, University of Calgary, Calgary, AB, Canada) for the golgin-97 cDNA.
References
- 1.Gillingham A. K., Munro S. Long coiled-coil proteins and membrane traffic. Biochim. Biophys. Acta. 2003;1641:71–85. doi: 10.1016/s0167-4889(03)00088-0. [DOI] [PubMed] [Google Scholar]
- 2.Seemann J., Jokitalo E., Pypaert M., Warren G. Matrix proteins can generate the higher order architecture of the Golgi apparatus. Nature (London) 2000;407:1022–1026. doi: 10.1038/35039538. [DOI] [PubMed] [Google Scholar]
- 3.Short B., Preisinger C., Korner R., Kopajtich R., Byron O., Barr F. A. A GRASP55-rab2 effector complex linking Golgi structure to membrane traffic. J. Cell Biol. 2001;155:877–883. doi: 10.1083/jcb.200108079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pfeffer S. R. Constructing a Golgi complex. J. Cell Biol. 2001;155:873–875. doi: 10.1083/jcb.200109095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barr F. A., Short B. Golgins in the structure and dynamics of the Golgi apparatus. Curr. Opin. Cell Biol. 2003;15:405–413. doi: 10.1016/s0955-0674(03)00054-1. [DOI] [PubMed] [Google Scholar]
- 6.Brown D. L., Heimann K., Lock J., Kjer-Nielsen L., van Vliet C., Stow J. L., Gleeson P. A. The GRIP domain is a specific targeting sequence for a population of trans-Golgi network derived tubulo-vesicular carriers. Traffic. 2001;2:336–344. doi: 10.1034/j.1600-0854.2001.002005336.x. [DOI] [PubMed] [Google Scholar]
- 7.Luke M. R., Kjer-Nielsen L., Brown D. L., Stow J. L., Gleeson P. A. GRIP domain-mediated targeting of two new coiled-coil proteins, GCC88 and GCC185, to subcompartments of the trans-Golgi network. J. Biol. Chem. 2003;278:4216–4226. doi: 10.1074/jbc.M210387200. [DOI] [PubMed] [Google Scholar]
- 8.Barr F. A. A novel Rab6-interacting domain defines a family of Golgi-targeted coiled-coil proteins. Curr. Biol. 1999;9:381–384. doi: 10.1016/s0960-9822(99)80167-5. [DOI] [PubMed] [Google Scholar]
- 9.Kjer-Nielsen L., Teasdale R. D., van Vliet C., Gleeson P. A. A novel Golgi-localisation domain shared by a class of coiled-coil peripheral membrane proteins. Curr. Biol. 1999;9:385–388. doi: 10.1016/s0960-9822(99)80168-7. [DOI] [PubMed] [Google Scholar]
- 10.Munro S., Nichols B. J. The GRIP domain – a novel Golgi-targeting domain found in several coiled-coil proteins. Curr. Biol. 1999;9:377–380. doi: 10.1016/s0960-9822(99)80166-3. [DOI] [PubMed] [Google Scholar]
- 11.McConville M. J., Ilgoutz S. C., Teasdale R. D., Foth B. J., Matthews A., Mullin K. A., Gleeson P. A. Targeting of the GRIP domain to the trans-Golgi network is conserved from protists to animals. Eur. J. Cell Biol. 2002;81:485–495. doi: 10.1078/0171-9335-00268. [DOI] [PubMed] [Google Scholar]
- 12.Gilson P. R., Vergara C. E., Kjer-Nielsen L., Teasdale R. D., Gleeson P. A. Identification of a Golgi-localised GRIP domain protein in Arabidopsis thaliana. Planta. 2004;219:1050–1056. doi: 10.1007/s00425-004-1311-9. [DOI] [PubMed] [Google Scholar]
- 13.Lu L., Tai G., Hong W. Autoantigen Golgin-97, an effector of Arl1 GTPase, participates in traffic from the endosome to the TGN. Mol. Biol. Cell. 2004;15:4426–4443. doi: 10.1091/mbc.E03-12-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshino A., Bieler B. M., Harper D. C., Cowan D. A., Sutterwala S., Gay D. M., Cole N. B., McCaffery J. M., Marks M. S. A role for GRIP domain proteins and/or their ligands in structure and function of the trans Golgi network. J. Cell Sci. 2003;116:4441–4454. doi: 10.1242/jcs.00746. [DOI] [PubMed] [Google Scholar]
- 15.Kooy J., Toh B. H., Pettitt J. M., Erlich R., Gleeson P. A. Human autoantibodies as reagents to conserved Golgi components – characterization of a peripheral, 230-kDa compartment-specific Golgi protein. J. Biol. Chem. 1992;267:20255–20263. [PubMed] [Google Scholar]
- 16.Fritzler M. J., Lung C. C., Hamel J. C., Griffith K. J., Chan E. K. L. Molecular characterization of golgin-245, a novel Golgi complex protein containing a granin signature. J. Biol. Chem. 1995;270:31262–31268. doi: 10.1074/jbc.270.52.31262. [DOI] [PubMed] [Google Scholar]
- 17.Erlich R., Gleeson P. A., Campbell P., Dietzsch E., Toh B. H. Molecular characterization of trans-Golgi p230 – a human peripheral membrane protein encoded by a gene on chromosome 6p12-22 contains extensive coiled-coil alpha-helical domains and a granin motif. J. Biol. Chem. 1996;271:8328–8337. doi: 10.1074/jbc.271.14.8328. [DOI] [PubMed] [Google Scholar]
- 18.Gleeson P. A., Anderson T. J., Stow J. L., Griffiths G., Toh B. H., Matheson F. p230 is associated with vesicles budding from the trans-Golgi network. J. Cell Sci. 1996;109:2811–2821. doi: 10.1242/jcs.109.12.2811. [DOI] [PubMed] [Google Scholar]
- 19.Griffith K. J., Chan E. K. L., Lung C. C., Hamel J. C., Guo X. Y., Miyachi K., Fritzler M. J. Molecular cloning of a novel 97-Kd golgi complex autoantigen associated with Sjogrens-syndrome. Arthritis Rheum. 1997;40:1693–1702. doi: 10.1002/art.1780400920. [DOI] [PubMed] [Google Scholar]
- 20.Evan G. I., Lewis G. K., Ramsay G., Bishop J. M. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol. Cell. Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kjer-Nielsen L., van Vliet C., Erlich R., Toh B. H., Gleeson P. A. The Golgi-targeting sequence of the peripheral membrane protein p230. J. Cell Sci. 1999;112:1645–1654. doi: 10.1242/jcs.112.11.1645. [DOI] [PubMed] [Google Scholar]
- 22.Teasdale R. D., Loci D., Houghton F., Karlsson L., Gleeson P. A. A large family of endosome-localized proteins related to sorting nexin 1. Biochem. J. 2001;358:7–16. doi: 10.1042/0264-6021:3580007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vojtek A. B., Hollenberg S. M., Cooper J. A. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell (Cambridge, Mass.) 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 24.Gietz R. D., Schiestl R. H., Willems A. R., Woods R. A. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 25.Guarente L. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 1983;101:181–191. doi: 10.1016/0076-6879(83)01013-7. [DOI] [PubMed] [Google Scholar]
- 26.Sreerama N., Woody R. W. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 27.Kohn W. D., Mant C. T., Hodges R. S. Alpha-helical protein assembly motifs. J. Biol. Chem. 1997;272:2583–2586. doi: 10.1074/jbc.272.5.2583. [DOI] [PubMed] [Google Scholar]
- 28.Gleeson P. A., Lock J. G., Luke M. R., Stow J. L. Domains of the TGN: coats, tethers and G proteins. Traffic. 2004;5:315–326. doi: 10.1111/j.1398-9219.2004.00182.x. [DOI] [PubMed] [Google Scholar]
- 29.Panic B., Perisic O., Veprintsev D. B., Williams R. L., Munro S. Structural basis for Arl1-dependent targeting of homodimeric GRIP domains to the Golgi apparatus. Mol. Cell. 2003;12:863–874. doi: 10.1016/s1097-2765(03)00356-3. [DOI] [PubMed] [Google Scholar]
- 30.Wu M., Lu L., Hong W., Song H. Structural basis for recruitment of GRIP domain golgin-245 by small GTPase Arl1. Nat. Struct. Mol. Biol. 2004;11:1–10. doi: 10.1038/nsmb714. [DOI] [PubMed] [Google Scholar]