Abstract
Glucose repression is a global transcriptional regulatory mechanism commonly observed in micro-organisms for the repression of enzymes that are not essential for glucose metabolism. In Saccharomyces cerevisiae, Mig1p, a homologue of Wilms' tumour protein, is a global repressor protein dedicated to glucose repression. Mig1p represses genes either by binding directly to the upstream repression sequence of structural genes or by indirectly repressing a transcriptional activator, such as Gal4p. In addition, some genes are repressed by both of the above mechanisms. This raises a fundamental question regarding the physiological relevance of the varied mechanisms of repression that exist involving Mig1p. We address this issue by comparing two well-known glucose-repression systems, that is, SUC2 and GAL gene expression systems, which encompass all the above three mechanisms. We demonstrate using steady-state analysis that these mechanisms lead to a hierarchical glucose repression profile of different family of genes. This switch over from one carbon source to another is well-calibrated as a function of glucose concentration through this hierarchical transcriptional response. The mechanisms prevailing in this repression system can achieve amplification and sensitivity, as observed in the well-characterized MAPK (mitogen-activated protein kinase) cascade system, albeit through a different structure. A critical feature of repression predicted by our steady-state model for the mutant strain of S. cerevisiae lacking Gal80p agrees well with the data reported here as well as that available in the literature.
Keywords: glucose repression, Mig1p, mitogen-activated protein kinase (MAPK), transcriptional activator, transcriptional repressor, yeast
Abbreviations: MAPK, mitogen-activated protein kinase; UAS, upstream activation sequence; URS, upstream repression sequence
INTRODUCTION
Micro-organisms, in their natural habitat, encounter growth media consisting of fermentable and non-fermentable carbon sources, of which cells prefer glucose to the exclusion of others [1–4]. Micro-organisms have evolved a hierarchical utilization system for various sugars, which allows them to switch from one carbon source to the other. This hierarchy appears to have evolved in the order of pathways that consume increasingly more cellular energy for the synthesis of metabolic machinery. Accordingly, as the cells grow, they reorient their metabolism through a genetic regulatory mechanism to utilize different sugars as dictated by them. This orderly expression of the sugar-utilization system is established through a hierarchical transcriptional regulation. Furthermore, the derepression of a family of genes that are under the negative control of glucose occurs as a smooth function of glucose concentration. How is this hierarchy in sugar utilization established at the transcriptional level?
In Saccharomyces cerevisiae, glucose exhaustion leads to differential expression of a family of genes, which is under negative regulation of glucose repression [5–9]. For example, invertase is induced simply by glucose removal and does not require a positive signal [10,11]. At another level, certain genes are not expressed merely upon glucose exhaustion, but also require a positive signal (usually through a transcriptional activator) for induction [9]. For example, the GAL/MEL regulon of yeast is one such well-established system. A common feature of genes that are repressed by glucose is the presence of a GC-rich URS (upstream repression sequence) for Mig1p binding [5,12–14].
Mig1p is a constitutively expressed [15] global repressor protein whose activity is regulated through a phosphorylation–dephosphorylation cycle [9,16–18]. In the presence of glucose, it is believed that Snf1 kinase (a homologue of ADP/AMP-activated protein kinase in humans) is inactivated through a mechanism that is not clearly understood [16,17]. Under these conditions, Mig1p is predominantly in the dephosphorylated state, and translocates into the nucleus [17,19] to repress the genes by binding to the URS of various genes.
In S. cerevisiae, three different mechanisms can be distinguished for glucose repression through Mig1p. Expression is repressed directly by binding of Mig1p to the URS of genes such as SUC2 and GAL4 [5,12,20]. In GAL/MEL regulon, Mig1p represses the structural (such as MEL1, GAL1 and GAL7) and regulatory (such as GAL3 and GAL80) genes indirectly through a transcriptional activator (that is Gal4p). In this case, Mig1p represses only the expression of the activator and thus represses the structural genes indirectly (like in GAL7) [12]. In addition, a set of structural genes (GAL1 and MEL1), as well as a regulatory gene (GAL3), has URS for Mig1p binding [5,9,12,20] as well as a UAS (upstream activation sequence) for the transcriptional activator [21]. Intuitively, by repressing the genes through a common activator, such as Gal4p, the cell achieves the repression in a co-ordinated fashion, instead of repressing each gene through an independent URS. However, this reason alone does not explain why only a few genes are repressed through an activator.
While genetic and biochemical analysis have elucidated the molecular mechanisms of transcriptional repression through Mig1p, the need for such diverse features of repression mechanisms has not been clearly understood. It is not clear as to why, of the 37 known Mig1p-binding genes, only SUC2 has two URSs for Mig1p binding [5,9,20]. To obtain an in-depth understanding of these subtle features of repression, it is necessary to address the significance of direct binding of Mig1p to the URS and the indirect effect of Mig1p through the recruitment of a positive transcriptional activator in the repression mechanism. In other words, is the presence of the activator useful only for the induction mechanism or does it also play a crucial mechanistic role in repression? To address the above issues, we chose to analyse the repression profile of SUC2 and the GAL system, which provide contrasting features in the Mig1p-dependent glucose repression mechanism. An analysis based on steady-state modelling of the Mig1p-dependent repression clearly reveals that a transcriptional hierarchy can be established solely through the various mechanisms that exist for glucose repression without sacrificing amplification and sensitivity.
THEORY
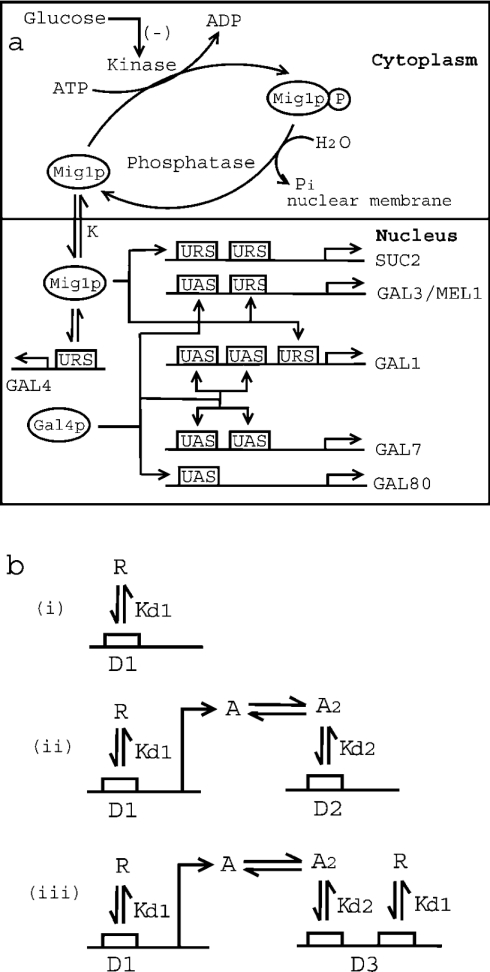
Figure 1(a) shows a schematic diagram of glucose repression of the SUC2 and GAL genes of S. cerevisiae. A steady-state model was developed to quantify the glucose repression. The steady-state model accounts for the phosphorylation/dephosphorylation cycle of Mig1p in the cytoplasm, and subsequent translocation of unphosphorylated Mig1p into the nucleus [9,16–18]. The phosphorylation of Mig1p by Snf1p kinase [17–19] through the monocyclic cascade was modelled by the procedure described by Goldbeter and Koshland [22,23]. Glucose inhibition of Snf1p kinase to phosphorylate Mig1p was quantified by a Michaelis–Menten-type relationship. The active Mig1p, once inside the nucleus, interacts with the URS of GAL4 to repress synthesis of Gal4p, the transcriptional activator of GAL genes. The model also accounts for Mig1p binding to the URS of 37 genes, including GAL4, three genes of the GAL family (GAL1, GAL3 and MEL1) and two URS sites for SUC2 [5,9,12].
Figure 1. Mechanism of glucose repression in S. cerevisiae.
(a) Schematic representation of Mig1p-mediated glucose repression of SUC2 and GAL genes. Kinase (Snf1) phosphorylates Mig1p to Mig1p-P (phosphorylated product), while phosphatase dephosphorylates Mig1p. Glucose inhibits the activity of Snf1 kinase. Unphosphorylated Mig1p is translocated to the nucleus with a distribution coefficient of K. Furthermore, Mig1p in the nucleus binds to the URS of SUC2, GAL4, GAL1, GAL3 and MEL1, with a dissociation constant Kd1. It may be noted that SUC2 has two binding sites for Mig1p, while GAL4, GAL1, GAL3 and MEL1 have only one binding site. Also, the transcriptional activator, Gal4p, binds to the UAS of GAL3/MEL1 and GAL80 (with one binding site for Gal4p), and GAL1 and GAL7 (with two binding sites for Gal4p). (b) Schematic representation of protein–protein and protein–DNA interactions for three different mechanisms for gene repression. Mechanism of repression in which (i) repressor protein ‘R’ binds to the URS of gene ‘D1’ with a dissociation constant of Kd1, (ii) repressor protein ‘R’ binds to the URS of transcriptional activator gene ‘D1’. The product of gene ‘D1’ is transcriptional activator protein ‘A’, which dimerizes with dissociation constant Kd1 before binding to the UAS of gene ‘D2’ with dissociation constant Kd2. (iii) Both mechanisms (i) and (ii) described above are operational.
The expression of GAL genes to galactose is based on a steadystate model described by Verma et al. [24]. The model accounts for the dimerization of Gal4p and Gal80p, binding of the Gal4p dimer to the UAS of GAL genes and interaction between Gal80p with Gal4p to repress the genes. In the presence of galactose, Gal3p is activated and binds further to Gal80p in the cytoplasm. This initiates shuttling of Gal80p from the nucleus to the cytoplasm and relieves repression of the GAL system. The activation of Gal3p was linked to the galactose concentration through a typical Michaelis–Menten expression. The fractional transcriptional expression of GAL genes with one binding site (f1) and two binding sites (f2) are defined as:
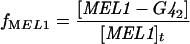 |
1 |
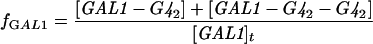 |
2 |
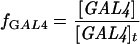 |
3 |
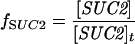 |
4 |
This means that fMEL1 and fGAL1 are the ratio of mRNA that is transcribed to a given input stimulus to the maximum mRNA that can be transcribed by the system for MEL1 and GAL1 with one and two binding sites respectively. Furthermore, fGAL4 and fSUC2 are the ratio that is transcribed and is defined as the ratio of free to total gene concentrations for GAL4 and SUC2 respectively. It should be noted that the binding sites present in different GAL promoters have intrinsic differences in affinities for Gal4p [25,26]. In our model, we have not considered different binding affinities of Gal4p for promoters of MEL1 (a gene with one Gal4p-binding site) and GAL1 (a gene with two Gal4p-binding sites).
All protein–protein and protein–DNA interactions were assumed to be at equilibrium. The translocation of Mig1p [27] and shuttling of Gal80p [28] were quantified based on a distribution coefficient which was defined as the ratio of concentration in the nucleus to that in the cytoplasm [24]. Molar balances were invoked on all total component concentrations, that is on Gal4p, Gal80p, Gal3p, Mig1p and operator site concentrations of various genes. The translational response was related to the transcriptional response through power-law formalism and was quantified by a co-response coefficient [24,29,30]. The detailed model equations accounting for both glucose repression and galactose activation are documented in the supplementary information available at http://www.BiochemJ.org/bj/388/bj3880843add.htm. The model equations consisting of non-linear algebraic equations were solved simultaneously using the fsolve function in MATLAB (The MathWorks, Natick, MA, U.S.A.).
MATERIALS AND METHODS
Yeast strains
S. cerevisiae strain Sc 285 with genotype MATa ura3-52 leu2-3 2-112gal80 [31] was used for measuring α-galactosidase expression (the protein expressed from MEL1 having one Gal4p-binding site and one Mig1p-binding site), whereas strain YM 3544 with genotype MATa ura3-52 his3-200 ade2-101 trp1-901 CANr met2 gal80-538, LEU2::GAL1-lacG lys2-801::GAL4 gal4-CAT-URA3 [32] was used to measure β-galactosidase expression (the protein expressed from the promoter of GAL1 having two Gal4p-binding sites and one Mig1p-binding site).
Medium for the pre-culture and inoculum size
The inoculum was prepared in a cotton-stoppered 500 ml shake flask containing 100 ml of medium of the composition 0.025 g/l adenine, 5.0 g/l yeast extract, 10.0 g/l peptone and 30.0 g/l glycerol. The pH of the medium was adjusted to 5.5 with 1 M HCl. A loopful of culture from a fresh slant was inoculated, and cells were grown at 240 rev./min on a rotary shaker at 30 °C for 12–16 h, until the cell concentration had reached a D600 of 1.0–1.5. After this point, the bioreactor was inoculated with 10% cell mass with a D600 of 1.
Cultivation conditions
S. cerevisiae strains were grown in a batch bioreactor until the biomass reached a D600 of approx. 0.5–0.75 in a medium of composition 0.025 g/l adenine, 5.0 g/l yeast extract, 10.0 g/l peptone and 30.0 g/l glycerol. The bioreactor was operated in a fed-batch mode by maintaining different average steady-state glucose concentrations (±10%). The aerobic bioreactor (Vaspan Industries, Mumbai, India) used was a stirred tank of 2 litre capacity (1 litre working volume) having two Rushton turbines. The dissolved oxygen and pH data were obtained on-line using two probes (Bela Instruments, Mumbai, India) interfaced with a PC, at an airflow rate of 1.5 litres/min and agitation at 300 rev./min at 30 °C. The glucose concentration in the reactor was maintained by continuous feeding of a standard glucose solution using calibrated peristaltic pumps (Watson Marlow 101U) through a feedback-control mechanism. Different average glucose concentrations (with a set point for each) were maintained by altering the feed rate and the concentration of standard glucose solution. α-Galactosidase and β-galactosidase were measured as a function of time for different average steady-state glucose concentrations. The data obtained from these experiments were tabulated as a steady-state fractional protein expressed at different steady-state glucose concentrations.
Analyses
Samples were taken aseptically at regular intervals to measure the expressions of α- and β-galactosidase. The α-galactosidase activity was measured in strain Sc 285 as described in [33]. The activity of β-galactosidase was measured as described by Rose and Botstein [34]. For glucose estimation, samples were immediately filtered through a 0.45-μm-pore-size cellulose acetate filter (Sartorius AG, Göttingen, Germany), and determined further by the orthotolidine reagent (Sigma) method as described in the manufacturer's catalogue.
All of the experiments were conducted in triplicate, and the deviation in the protein expression data was within acceptable limits (not more than 9%). The fluctuations in glucose concentrations from their average values in a fed-batch reactor were within acceptable limits (variation of 10–15%).
RESULTS
Comparison of transcriptional response of one-step and two-step repression (through a transcriptional activator)
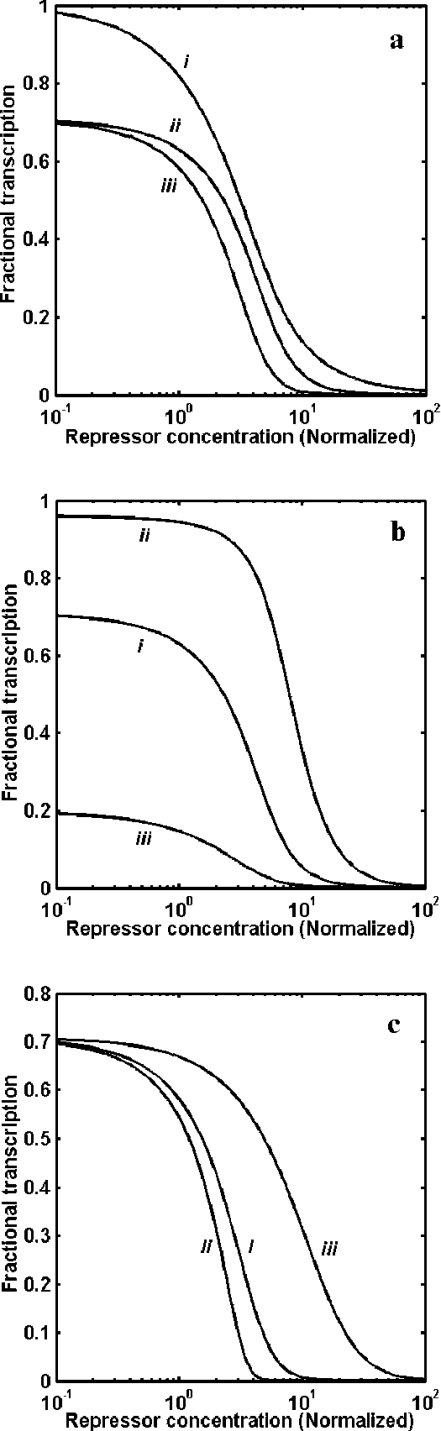
We carried out a theoretical analysis by comparing the repression of transcription of structural genes (genes encoding elements of the metabolic pathway) through three different mechanisms as illustrated in Figure 1(b): (i) repression through the binding of a repressor ‘R’ to the URS of a gene ‘D1’, (ii) repression of gene ‘D2’ through the binding of a repressor to the URS of a gene ‘D1’ encoding transcriptional activator ‘A’, and (iii) repression through the combination of mechanisms described in (i) and (ii). In the case of (ii) and (iii), the transcriptional activator ‘A’ dimerizes before binding to the UAS of the target gene. Figure 2(a) shows the transcriptional repression as a function of repressor ‘R’ concentration. As shown by our analysis, if repression occurs through a transcriptional activator alone [curve (ii) in Figure 2a], the output response is sensitive (with a Hill coefficient of 2.1), and the repressor concentration required to completely repress the system decreased, as compared with mechanism (i). In case of both mechanisms operating together, the amount of repressor concentration required to repress the system completely is reduced 10-fold (100 instead of 10), as compared with mechanism (i), and, furthermore, the response is also ultrasensitive [see curve (iii) in Figure 2a]. Owing to the limitation caused by the dimerization of the transcriptional activator ‘A’ in mechanism (ii), the expression is incomplete (only 70% of the maximum transcription) in the absence of the repressor. This implies that the amplification and sensitivity are obtained at the expense of maximum expression. Dimerization of transcriptional activator ‘A’ (see Figure 1b) is the cause for the ultrasensitive response, and, in the absence of dimerization, a similar high-sensitivity response cannot be achieved.
Figure 2. Repression of transcriptional response.
(a) Repression of transcription of genes by three different mechanisms as illustrated in Figure 1(b) at various repressor concentrations normalized with Kd2 (=0.2 nM). Response curve (i) represents repression by binding of ‘R’ to the URS of gene ‘D1’, (ii) represents repression of gene ‘D2’ through transcriptional activator ‘A’, and (iii) represents repression of the gene by both of the above mechanisms. (b) Indirect repression of transcription of gene ‘D2’ through transcriptional activator ‘A’ alone [mechanism (ii) in Figure 1b] at different values of Kd2. Curve (i) is for Kd2=0.2 nM, curve (ii) is for Kd2=0.02 nM, and curve (iii) is for Kd2=2 nM. (c) Repression of transcription of genes through transcriptional activator ‘A’ and repressor ‘R’ [mechanism (iii) in Figure 1b] at different values of Kd1. Curve (i) is for Kd1=0.2 nM, curve (ii) is for Kd1=0.02 nM, and curve (iii) is for Kd1=2 nM.
Our analysis indicates that the response is highly dependent on the dissociation constant for binding of repressor ‘R’ and dimerization constant of the transcriptional activator ‘A’. The dimerization constant mainly affects the sensitivity and extent of transcription (see Figure 2b), while the dissociation constant for the binding of the repressor affects both the amount of repressor required for complete repression and the sensitivity of the response (see Figure 2c). An increase in the dimerization constant of transcriptional activator reduces the maximum transcription in the absence of repressor, whereas an increase in the dissociation constant for binding of the repressor increases the amount of repressor required for complete repression and the sensitivity of the transcriptional response. Thus a unique combination of values of the two parameters can yield a distinct transcriptional response depending on the mechanisms employed. The above analysis provides a strong theoretical basis to infer that these mechanisms may have biological significance.
Comparison of repression through one and two binding sites for Mig1p
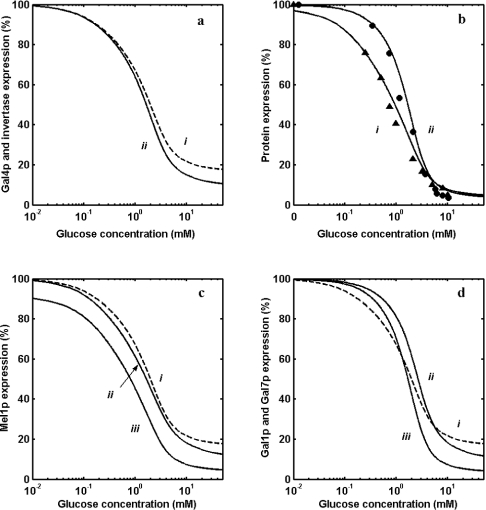
It is reported that out of the 37 genes known to have an URS for Mig1p binding, only SUC2 has two binding sites [5,9,20]. Figure 3(a) shows the comparison of repression of protein expression from genes with one and two binding sites for Mig1p (at a constitutively expressed total concentration of 3 nM) at different glucose concentrations. It is clear from the Figure that the repression is sensitive for SUC2 solely due to the number of binding sites as compared with genes having one binding site for Gal4p. The Gal4p expression falls to approx. 18% of the wild-type concentration at high glucose concentration with a Hill coefficient of 1.2 [Figure 3a (i)]. Invertase expression from SUC2 is repressed with a Hill coefficient of 1.3 and a half-saturation coefficient (K0.5) of 1.5 mM, implying that both sensitivity and amplification are achieved by having two binding sites for Mig1p.
Figure 3. Repression of GAL genes in a GAL80 mutant.
(a) Repression by glucose of protein expression by GAL4 (curve i) and SUC2 (curve ii). (b) Comparison of experimental data with model prediction of α-galactosidase expression from MEL1 and β-galactosidase expression from promoter of GAL1 having one and two binding sites for Gal4p respectively at different steady-state glucose concentrations. ▲ denotes α-galactosidase, and ● denotes β-galactosidase. Solid lines (i) and (ii) show model predictions. (c) Repression of expression from MEL1 (one binding site for Gal4p and Mig1p) through three different mechanisms. Curve (i) is for repression through Mig1p binding alone, curve (ii) is for repression through Gal4p alone, and curve (iii) is for dual repression through Mig1p and Gal4p. (d) Repression of expression from GAL1 (two binding sites for Gal4p and one binding site for Mig1p) through three different mechanisms. Curve (i) is for repression through Mig1p binding alone, curve (ii) is for repression through Gal4p alone, and curve (iii) is for dual repression through Mig1p and Gal4p.
Experimental results correlate with the theoretical analysis in a mutant strain lacking GAL80
In order to study the repression of MEL1 and GAL1 solely due to a decrease in the transcriptional activator (Gal4p), it was necessary to de-link the induction mechanism of a wild-type strain by deleting Gal80p. In such a mutant strain, the GAL genes are constitutive and repression can be studied only as a consequence of Mig1p-mediated repression. It should be noted that MEL1 with one and GAL1 with two binding sites for the transcriptional activator (Gal4p) have a single binding site for Mig1p (see Figure 1a). Therefore estimation of repression of MEL1 and GAL1 in such a strain should indicate the contribution of repression by different mechanisms. Expression from MEL1 and GAL1 was measured for a mutant strain lacking GAL80 at various glucose concentrations (Figure 3b). The repression of protein expression from genes with one (curve i) and two binding sites (curve ii) for Gal4p is shown in Figure 3(b). The model prediction matches well with the experimental data. The protein expression from genes with two binding sites is more sensitive than expression from gene with one binding site. Furthermore, the gene with two binding sites exhibits a higher expression than genes with one binding site, indicating that Gal4p is limiting. The Hill coefficients values for genes with one and two binding sites are 1.2 and 1.9, with K0.5 values of 1.0 and 1.6 mM respectively.
The above analysis allows us to delineate the degree of repression due to Mig1p binding to the structural gene and due to repression of transcriptional activator (only through Gal4p repression). Figure 3(c) shows the comparison of repression of MEL1 due to only Mig1p-binding site (curve iii), repression due to a decrease in Gal4p concentration only (curve ii) and due to dual effects of both Mig1p binding and decrease in Gal4p concentration (curve i). It is clear from the analysis that if MEL1 were to get repressed only through Mig1p binding, even at a higher glucose concentration, it would not be completely repressed (up to 20% expression) at the given Mig1p concentration. On the other hand, the repression through reduction in Gal4p concentration alone represses to an extent of approx. 12–15%, whereas the repression is almost complete when both the above mechanisms operate. It is clear from the above analysis that the two-step repression (binding of Mig1p to structural as well as transcriptional activator gene) brought about by Mig1p amplifies the glucose signal. It is worth recalling that the maximum expression (in the absence of glucose) is approx. 70% for MEL1 in a wild-type strain. A similar analysis indicates that, in addition to amplification, sensitivity is also increased for a gene with two binding sites for Gal4p (see Figure 3d). The ultrasensitive response observed in this case is due to dimerization and co-operative binding of Gal4p [36]. Unlike MEL1, the maximum expression (in the absence of glucose) is 100%, since GAL1 has two binding sites for Gal4p [24].
Repression of GAL genes by glucose in a wild-type strain
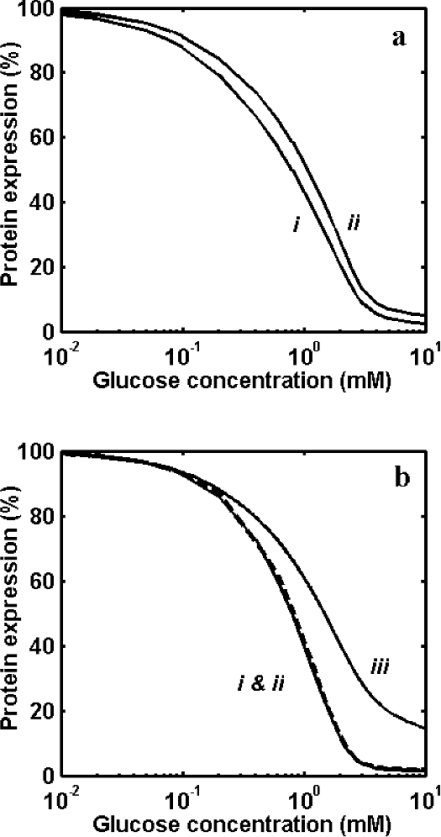
The steady-state analysis of the GAL genetic switch yielded percentage expression of wild-type strain for gene with one and two binding sites. Figure 4(a) shows the repression of GAL3 (genes with one binding site for Mig1p and Gal4p) and GAL80 (gene with only one binding site for Gal4p but no Mig1p-binding site). Repression of GAL3 is sensitive, with a K0.5 of 1.0 mM for glucose, while repression for Gal80p is less sensitive (Hill coefficient of 1.8), with a K0.5 of 1.5 mM for glucose. Figure 4(b) shows repression of GAL1 (gene with both Mig1p- and Gal4p-binding sites) and SUC2 (gene with two Mig1p-binding sites). However, a similar steady-state analysis indicates that repression of GAL1 and GAL7 coincides without showing the effect of Mig1p binding to GAL1. But a substantial amplification and increase in sensitivity were observed as compared with the expression from SUC2. Furthermore, the wild-type strain shows a sensitive response (see Figure 4b), with higher amplification as compared with expression in a mutant strain lacking Gal80p (see Figure 3b).
Figure 4. Repression of GAL genes in a wild-type strain.
(a) Repression by glucose of protein expression by GAL3/MEL1 and GAL80 having one binding site for Gal4p. Curve (i) is for GAL3 with one binding site for Mig1p, and curve (ii) is for GAL80 with no Mig1p-binding sites. (b) Repression by glucose of protein expression by GAL1, GAL7 and SUC2. Curve (i) is for expression by GAL1 with one binding site for Mig1p and two binding sites for Gal4p, curve (ii) is for GAL7 with no Mig1p-binding site, but with two binding sites for Gal4p, and curve (iii) is for SUC2 with two binding sites for Mig1p and no binding site for Gal4p.
Model predicts the results available in the literature
The steady-state model was used to compare the transcriptional expression in various mutant strains with genotype MIG1::GAL80, mig1::GAL80, MIG1::gal80 and mig1::gal80 of S. cerevisiae grown on glucose and galactose (results are shown in Table 1). The model prediction matches well with experimental data reported by Nehlin et al. [12]. The maximum expressions of GAL4 and GAL1 observed were approx. 155% and 160% of wild-type respectively in the strain with genotype mig1::gal80 grown with galactose. A mutant strain with genotype mig1::gal80 (lacking both repressor genes) yields transcriptional expression of approx. 50% of wild-type for GAL genes having two binding sites, even in the presence of glucose. The reduction in transcriptional response of GAL4 and GAL1 in MIG1-deleted strains is due to Mig1p-independent glucose repression.
Table 1. Comparison of model-predicted transcriptional expression with experimental data for various genotypes of S. cerevisiae grown on galactose and glucose.
Numbers within parentheses are experimental data [12] given as percentages of wild-type expression.
| Genotype | Galactose | Glucose |
|---|---|---|
| MIG1::GAL80 | ||
| GAL4 | 100 (100) | 18 (18) |
| GAL1 | 100 (100) | 0 (0) |
| GAL2 | 100 (100) | 0 (0) |
| mig1::GAL80 | ||
| GAL4 | 155 (151) | 120 (127) |
| GAL1 | 113 (100) | 0.4 (0) |
| GAL2 | 113 (100) | 0.4 (0) |
| MIG1::gal80 | ||
| GAL4 | 100 (118) | 15 (19) |
| GAL1 | 153 (90) | 1.8 (6.3) |
| GAL2 | 156 (167) | 2 (4.7) |
| mig1::gal80 | ||
| GAL4 | 155 (164) | 118 (123) |
| GAL1 | 160 (98) | 50 (46) |
| GAL2 | 160 (147) | 50 (40) |
The steady-state model developed here is thus able to match all experimental observations reported in the literature for the set of parameter values reported in Table A1 (in the supplementary information available at http://www.BiochemJ.org/bj/388/bj3880843add.htm). Other than the specific parameters related to phosphorylation of Mig1p, binding of Mig1p to its cognate binding site and nucleocytoplasmic shuttling of Mig1p across the nuclear membrane were obtained from the literature. The sensitivity of the steady-state response due to changes in these parameters was obtained using the model (see the supplementary material at http://www.BiochemJ.org/bj/388/bj3880843add.htm). It was observed that extent of expression and half-saturation concentration of glucose to repress expression is sensitive to these parameter values. However, the hierarchical response of different genes under Mig1p control due to the unique structure is not affected either by the system parameters or the component concentrations.
DISCUSSION
S. cerevisiae has optimized its metabolic machinery towards utilizing glucose as the preferred carbon source. In order to achieve this, a large number of genes belonging to different catabolic pathways, which are not needed for glucose metabolism, are strongly repressed. In fact, S. cerevisiae has even evolved a mechanism of getting rid of the mitochondria (petite-positive phenotype) to optimize its energy resource available from glucose [37]. Such a stringent glucose repression mechanism must necessarily have features that allow the system to respond to a continuously changing environmental condition. This implies that with a changing glucose concentration in the medium, the repression of other sugar-utilizing genes should be gradual and not a switch-like response. Therefore the optimization of the catabolic machinery warrants a hierarchical transcriptional repression system by glucose in the presence of other sugars.
Consistent with the above, it has been observed that genes belonging to different families (such as SUC2, GAL genes, MAL genes and HAP) are de-repressed by inactivation of Mig1p to varying extents. This is due to responses having different sensitivity (steepness) and threshold glucose required to shut off expression for different subsystems. How is this differential response achieved through a common regulatory protein such as Mig1p? Our analysis of GAL and SUC2 systems indicates that the mechanism of repression either by binding of Mig1p directly to the structural gene (as in SUC2) or through the repression of transcriptional activator (as in GAL7) is sufficient to bring about a hierarchy in the expression of these genes. Furthermore, another level of regulation in the hierarchy of repression is achieved by having both of the above mechanisms operating in tandem, as in GAL1 [12,38,39] and GAL3 [5]. These transcriptional mechanistic features allow the cell to express SUC2 at a higher level than GAL1 (even at a saturating concentration of galactose) as a function of decreasing glucose concentration.
Our quantitative analysis clearly distinguishes the repression between genes having one and no binding site for Mig1p in a wild-type strain. For example, in the case of expression from GAL3 (one Gal4p-binding site and one Mig1p-binding site) and GAL80 (one Gal4p-binding site, but with no Mig1p-binding site), a distinct response to glucose was observed through our analysis (see Figure 4a). Since Gal3p and Gal80p are regulatory proteins, their expression has evolved to be leaky, and this is due to a single Gal4p-binding site [24,40]. Unlike the difference we observe with respect to expression of Gal80p and Gal3p, expression from GAL1 (with two Gal4p-binding sites and one Mig1p-binding site) and GAL7 (with two Gal4p-binding sites and no Mig1p-binding site) show no difference in response. This is due to (i) the presence of two Gal4p-binding sites, which masks the effect of Mig1p, and (ii) the repression caused by weak expression of Gal80p even in the presence of glucose. Accordingly, our analysis does not show a distinction in the response to glucose for GAL1 and GAL7 in a wild-type strain, but does show a distinction in a mutant strain lacking Gal80p (see Figures 3d and 4b). The hierarchical expression profile is not critically dependent on the absolute component concentrations, but is mainly due to the structure; however, the component concentrations will affect the extent of expression (results not shown).
Another important family of genes repressed by glucose through Mig1p is the genes that are involved in oxidative metabolism [5]. In this system, repression by glucose is brought about only through the transcriptional activator, the HAP system [5,41–46], without a binding site for Mig1p. Based on our analysis, we suggest that the expression of genes under HAP control is less sensitive to glucose repression (similar to the case of GAL80). Such a design allows the expression of these genes, even in the presence of glucose. For example, the genes which are under the control of the RTG family of transcriptional activators are repressed by glucose through the HAP system alone, when glutamate is provided as a nitrogen source. Instead, if ammonium sulphate is provided as the sole nitrogen source, glucose repression is overcome through the RTG family of transcriptional activators [47,48].
The on/off state of the gene expression to a stimulus can be regulated through an enzyme cascade mechanism such as in the MAPK (mitogen-activated protein kinase) system [49]. In this pathway, a phosphorylation cycle activates another downstream phosphorylation cycle to constitute a cascade system resulting in an amplification of the signal causing transcriptional activation [22,23] and leading to a switch-like response. The repression by glucose of the transcriptional activator (Gal4p) through Mig1p is akin to an enzyme cascade system, whereas a similar cascade structure is constituted in the case of repression through Mig1p, where an output of a phosphorylation cycle deactivates the synthesis of transcriptional activators to control expression. Our analysis suggests that, even in this case, properties such as amplification and sensitivity can be obtained similar to that of an enzyme cascade. If so, why did the cell evolve different mechanisms for obtaining amplification and sensitivity? The MAPK system [49] allows the cell to respond in a yes or no fashion to a given stimulus in certain situations such as cell-cycle regulation. However, such a response is not warranted, and may prove to be detrimental if the organism has to compete with other organisms for the limited supply of nutrients. Instead, the expression status is calibrated depending upon the various levels of the glucose stimulus. This is achieved by a hierarchical expression profile, as demonstrated in the Mig1p-mediated glucose repression system in S. cerevisiae. This implies that a transcriptional hierarchal expression profile is used in case of depleting nutritional status. Analogous to the glucose repression system, nitrogen catabolite repression also has to respond to depleting nitrogen concentrations. It is worthwhile to analyse the network structure of the nitrogen catabolite repression system to establish the generality of transcriptional hierarchal expression as a fundamental mechanism to regulate smooth transition from one transcriptional state to another.
Multimedia adjunct
References
- 1.Hanes S. D., Bostian K. A. Control of cell growth and division in Saccharomyces cerevisiae. CRC Crit. Rev. Biochem. 1986;21:153–220. doi: 10.3109/10409238609113611. [DOI] [PubMed] [Google Scholar]
- 2.Johnston M. A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol. Rev. 1987;51:458–476. doi: 10.1128/mr.51.4.458-476.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnston M., Carlson M. Regulation of carbon and phosphate utilization. In: Johnes E. W., Pringle J. R., Broach J. R., editors. The Molecular and Cellular Biology of the Yeast Saccharomyces. Plainview: Cold Spring Harbor Laboratory Press; 1992. pp. 193–281. [Google Scholar]
- 4.Gancedo J. M. Yeast carbon catabolite repression. Eur. J. Biochem. 1992;206:297–313. doi: 10.1111/j.1432-1033.1992.tb16928.x. [DOI] [PubMed] [Google Scholar]
- 5.Lundin M., Nehlin J. O., Ronne H. Importance of a flanking AT-rich region in target site recognition by the GC box-binding zinc finger protein MIG1. Mol. Cell. Biol. 1994;14:1979–1985. doi: 10.1128/mcb.14.3.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Entian K. D. Glucose repression: a complex regulatory system in yeast. Microbiol. Sci. 1986;3:366–371. [PubMed] [Google Scholar]
- 7.Carlson M. Regulation of sugar utilization in Saccharomyces cerevisiae species. J. Bacteriol. 1987;169:4873–4877. doi: 10.1128/jb.169.11.4873-4877.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gancedo J. M., Gancedo C. Catabolite repression mutants of yeast. FEMS Microbiol. Rev. 1986;32:179–187. [Google Scholar]
- 9.Klein C. J. L., Olsson L., Nielsen J. Glucose control in Saccharomyces cerevisiae: the role of MIG1 in metabolic functions. Microbiology. 1998;144:13–24. doi: 10.1099/00221287-144-1-13. [DOI] [PubMed] [Google Scholar]
- 10.Lutfiyya L. L., Johnston M. Two zinc-finger-containing repressors are responsible for glucose repression of SUC2 expression. Mol. Cell. Biol. 1996;16:4790–4797. doi: 10.1128/mcb.16.9.4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turkel S., Turgut T., Savapcidlu Y. Analysis of the effects of transcription factors Gcr2p and Sgc1p on the control of the SUC2 gene expression in Saccharomyces cerevisiae. Turk. J. Biol. 2003;27:233–239. [Google Scholar]
- 12.Nehlin J. O., Carlberg M., Ronne H. Control of yeast GAL genes by MIG1 repressor: a transcriptional cascade in the glucose response. EMBO J. 1991;11:3373–3377. doi: 10.1002/j.1460-2075.1991.tb04901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yin Z., Smith R. J., Brown A. J. P. Multiple signaling pathways trigger the exquisite sensitivity of yeast gluconeogenic mRNAs to glucose. Mol. Microbiol. 1996;20:751–764. doi: 10.1111/j.1365-2958.1996.tb02514.x. [DOI] [PubMed] [Google Scholar]
- 14.Finley R. L., Jr, Chen S., Ma J., Byrne P., West R. W., Jr Opposing regulatory functions of positive and negative elements in UASG control transcription of the yeast GAL genes. Mol. Cell. Biol. 1990;10:5663–5670. doi: 10.1128/mcb.10.11.5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J., Sirenko O., Needleman R. Genomic footprinting of Mig1p in the MAL62 promoter. J. Biol. Chem. 1997;272:4613–4622. doi: 10.1074/jbc.272.7.4613. [DOI] [PubMed] [Google Scholar]
- 16.Treitel M. A., Carlson M. Repression by SSN6-TUP1 is directed by MIG1, a repressor/activator protein. Proc. Natl. Acad. Sci. U.S.A. 1995;92:3132–3136. doi: 10.1073/pnas.92.8.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeVit M. J., Waddle J. A., Johnston M. Regulated nuclear translocation of the Mig1 glucose repressor. Mol. Biol. Cell. 1997;8:1603–1618. doi: 10.1091/mbc.8.8.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Treitel M. A., Kuchin S., Carlson M. Snf1 protein kinase regulates phosphorylation of the Mig1 repressor in Saccharomyces cerevisiae. Mol. Cell. Biol. 1998;18:6273–6280. doi: 10.1128/mcb.18.11.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnston M. Feasting, fasting and fermenting glucose sensing in yeast and other cells. Trends Genet. 1998;15:29–33. doi: 10.1016/s0168-9525(98)01637-0. [DOI] [PubMed] [Google Scholar]
- 20.Nehlin J. O., Ronne H. Yeast MIG1 repressor is related to the mammalian early growth response and Wilms' tumour finger proteins. EMBO J. 1990;9:2891–2898. doi: 10.1002/j.1460-2075.1990.tb07479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lohr D., Venkov P., Zlatanova J. Transcriptional regulation in the yeast GAL gene family: a complex genetic network. FASEB J. 1995;9:777–786. doi: 10.1096/fasebj.9.9.7601342. [DOI] [PubMed] [Google Scholar]
- 22.Goldbeter A., Koshland D. E., Jr Ultrasensitivity in biochemical systems controlled by covalent modification. J. Biol. Chem. 1984;259:14441–14447. [PubMed] [Google Scholar]
- 23.Goldbeter A., Koshland D. E., Jr An amplified sensitivity arising from covalent modification in biological systems. Proc. Natl. Acad. Sci. U.S.A. 1981;78:6840–6844. doi: 10.1073/pnas.78.11.6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verma M., Bhat P. J., Venkatesh K. V. Quantitative analysis of GAL genetic switch of Saccharomyces cerevisiae reveals that nucleocytoplasmic shuttling of Gal80p results in a highly sensitive response to galactose. J. Biol. Chem. 2003;278:48764–48769. doi: 10.1074/jbc.M303526200. [DOI] [PubMed] [Google Scholar]
- 25.Xu H. E., Kodadek T., Johnston S. A. A single dimer of GAL4 protein can maximally stimulate transcription under physiological conditions. Proc. Natl. Acad. Sci. U.S.A. 1995;92:7677–7680. doi: 10.1073/pnas.92.17.7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ren B., Robert F., Wyrick J. J., Aparicio O., Jennings E. G., Simon I., Zeitlinger J., Schreiber J., Hannet N., Kanin E., et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 27.DeVit M. J., Johnston M. The nuclear exportin Msn5 is required for nuclear export of the Mig1 glucose repressor of Saccharomyces cerevisiae. Curr. Biol. 1999;9:1231–1241. doi: 10.1016/s0960-9822(99)80503-x. [DOI] [PubMed] [Google Scholar]
- 28.Peng G., Hopper J. E. Gene activation by interaction of an inhibitor with a cytoplasmic signaling protein. Proc. Natl. Acad. Sci. U.S.A. 2002;99:8548–8553. doi: 10.1073/pnas.142100099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hofmeyr J.-H. S., Cornish-Bowden A., Rohwer J. M. Taking enzyme kinetics out of control; putting control into regulation. Eur. J. Biochem. 1993;212:833–837. doi: 10.1111/j.1432-1033.1993.tb17725.x. [DOI] [PubMed] [Google Scholar]
- 30.Fell D. A. Beyond genomics. Trends Genet. 2001;17:680–682. doi: 10.1016/s0168-9525(01)02521-5. [DOI] [PubMed] [Google Scholar]
- 31.Torchia T. E., Hamilton R. W., Cano C. L., Hopper J. Disruption of regulatory gene GAL80 in Saccharomyces cerevisiae: effects on carbon-controlled regulation of galactose/melibiose pathway genes. Mol. Cell. Biol. 1984;4:1521–1527. doi: 10.1128/mcb.4.8.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Griggs D. W., Johnston M. Regulated expression of GAL4 activator gene in yeast provides a sensitive genetic switch for glucose repression. Proc. Natl. Acad. Sci. U.S.A. 1991;88:8597–8601. doi: 10.1073/pnas.88.19.8597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Post-Beittenmiller M. A., Hamilton R. W., Hopper J. E. Regulation of basal and induced levels of the MEL1 transcript in Saccharomyces cerevisiae. Mol. Cell. Biol. 1984;4:1235–1245. doi: 10.1128/mcb.4.7.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rose M., Botstein D. Regulation of basal and induced levels of the MEL1 transcript in Saccharomyces cerevisiae. Methods Enzymol. 1983;101:167–180. [Google Scholar]
- 35. Reference deleted.
- 36.Verma M., Bhat P. J., Venkatesh K. V. Expression of GAL genes in a mutant strain of Saccharomyces cerevisiae lacking GAL80: quantitative model and experimental verification. Biotechnol. Appl. Biochem. 2004;39:89–97. doi: 10.1042/BA20030119. [DOI] [PubMed] [Google Scholar]
- 37.Moller K., Olsson L., Pikur J. Ability for anaerobic growth is not sufficient for development of the petite phenotype in Saccharomyces kluyveri. J. Bacteriol. 2001;183:2485–2489. doi: 10.1128/JB.183.8.2485-2489.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Flick J. S., Johnston M. Two system of glucose repression of the GAL1 promoter in Saccharomyces cerevisiae. Mol. Cell. Biol. 1990;10:4757–4769. doi: 10.1128/mcb.10.9.4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flick J. S., Johnston M. Analysis of URSG-mediated glucose repression of the GAL1 promoter of Saccharomyces cerevisiae. Genetics. 1992;130:295–304. doi: 10.1093/genetics/130.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Melcher K., Xu H. E. Gal80–Gal80 interaction on adjacent Gal4p binding sites is required for complete GAL gene repression. EMBO J. 2001;20:841–851. doi: 10.1093/emboj/20.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Forsburg S. L., Guarente L. Identification and characterization of HAP4: a third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes Dev. 1989;3:1166–1178. doi: 10.1101/gad.3.8.1166. [DOI] [PubMed] [Google Scholar]
- 42.de Winde J. H., Crauwels M., Hohmann S., Thevelein J. M., Winderickx J. Differential requirement of the yeast sugar kinases for sugar sensing in establishing the catabolite-repressed state. Eur. J. Biochem. 1996;241:633–643. doi: 10.1111/j.1432-1033.1996.00633.x. [DOI] [PubMed] [Google Scholar]
- 43.De Winde J. H., Grivell L. A. Global regulation of mitochondrial biogenesis in Saccharomyces cerevisiae. Prog. Nucleic Acids Res. 1993;46:51–91. doi: 10.1016/s0079-6603(08)61018-1. [DOI] [PubMed] [Google Scholar]
- 44.Rosenkrantz M., Kell C. S., Pennell E. A., Devenish L. J. The HAP2,3,4 transcriptional activator is required for derepression of the yeast citrate synthase gene, CIT1. Mol. Microbiol. 1994;13:119–131. doi: 10.1111/j.1365-2958.1994.tb00407.x. [DOI] [PubMed] [Google Scholar]
- 45.Schlaepfer I. R., Mattoon J. R., Bajszar G. The sequence and potential regulatory elements of the HEM2 promoter of Saccharomyces cerevisiae. Yeast. 1994;10:227–229. doi: 10.1002/yea.320100209. [DOI] [PubMed] [Google Scholar]
- 46.Crawford M. J., Sherman D. R., Goldberg D. E. Regulation of Saccharomyces cerevisiae flavohemoglobin gene expression. J. Biol. Chem. 1995;270:6991–6996. doi: 10.1074/jbc.270.12.6991. [DOI] [PubMed] [Google Scholar]
- 47.Dang V. D., Bohn C., Bolotin-Fukuhara M., Dangnan-Fornier B. The CCAAT box-binding factor stimulates ammonium assimilation in Saccharomyces cerevisiae, defining a new cross-pathway regulation between nitrogen and carbon metabolisms. J. Bacteriol. 1996;178:1842–1849. doi: 10.1128/jb.178.7.1842-1849.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Z., Butow A. B. A transcriptional switch in the expression of yeast tricarboxylic acid cycle genes in response to a reduction or loss of respiratory function. Mol. Cell. Biol. 1999;19:6720–6728. doi: 10.1128/mcb.19.10.6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ferrell J. E., Jr, Machleder E. M. The biochemical basis of an all-or-none cell fate switch in Xenopus oocytes. Science. 1998;280:895–898. doi: 10.1126/science.280.5365.895. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.