Abstract
Escherichia coli MutS, MutL and MutH proteins act sequentially in the MMRS (mismatch repair system). MutH directs the repair system to the newly synthesized strand due to its transient lack of Dam (DNA-adenine methylase) methylation. Although Pseudomonas aeruginosa does not have the corresponding E. coli MutH and Dam homologues, and consequently the MMRS seems to work differently, we show that the mutL gene from P. aeruginosa is capable of complementing a MutL-deficient strain of E. coli. MutL from P. aeruginosa has conserved 21 out of the 22 amino acids known to affect functioning of E. coli MutL. We showed, using protein affinity chromatography, that the C-terminal regions of P. aeruginosa and E. coli MutL are capable of specifically interacting with E. coli MutH and retaining the E. coli MutH. Although, the amino acid sequences of the C-terminal regions of these two proteins are only 18% identical, they are 88% identical in the predicted secondary structure. Finally, by analysing (E. coli–P. aeruginosa) chimaeric MutL proteins, we show that the N-terminal regions of E. coli and P. aeruginosa MutL proteins function similarly, in vivo and in vitro. These new findings support the hypothesis that a large surface, rather than a single amino acid, constitutes the MutL surface for interaction with MutH, and that the N- and C-terminal regions of MutL are involved in such interactions.
Keywords: chimaeric MutL, complementation, Escherichia coli, mismatch repair system, MutH, Pseudomonas aeruginosa
Abbreviations: Dam, DNA-adenine methylase; LB, Luria–Bertani; MMRS, mismatch repair system; Pcp, phosphorylcholine phosphatase
INTRODUCTION
The MMRS (mismatch repair system) is an important process for the correction of replication errors that escape the polymerase proofreading activity and for preventing homeologous recombination events [1]. In Escherichia coli, this repair pathway is initiated by binding of MutS to a mismatch. After the recruitment of MutL, this complex activates the strand discriminating endonuclease MutH, which cleaves the newly synthesized, unmethylated daughter strand at the nearest hemimethylated d(GATC) site, and thereby marks it for a removal and a repair–synthesis process that involves a variety of other proteins. The delay in methylation of the newly replicated d(GATC) sequences by the E. coli Dam (DNA-adenine methylase) provides the strand discrimination signal.
Eubacteria and eukaryotes express conserved MutS and MutL homologues [1,2]. However, the E. coli MutH/d(GATC) methylation mechanism is found only in a small group of proteobacteria from the γ-subdivision [3] and not in eukaryotes. The mechanism of strand discrimination mentioned above seems to be different in most other prokaryotic and eukaryotic organisms and remains poorly understood.
MutL and its homologues are a family of proteins that have a conserved region of approx. 300 residues at the N-terminus and a divergent C-terminal region of 300–500 residues. A majority of reported mutations in E. coli MutL, which affect functionality, are within the conserved N-terminal region [4,5]. In humans, more than 50% of mutations found in the MutL homologous MLH1, in hereditary non-polyposis colon cancer, are within the equivalently conserved region [6,7].
The physical interaction between E. coli MutL and MutH has been demonstrated by two-hybrid assays [8,9], the ability of a MutH affinity column to retain MutL [8], MutL-mediated binding of MutH to a MutS column [10] and chemical and photo cross-linking [11,12]. Two different sites on MutL have been proposed as putative MutH-binding sites for MutL–MutH interaction. The hypothesis that MutL interacts with MutH through the N-terminal (LN40) domain is supported by the fact that LN40 activates endonuclease activity of MutH in the presence of heteroduplex DNA, ATP and MutS, and also by in vitro protein–protein chemical and photo cross-linking [11,12]. Based on crystallographic results, a concave surface of LN40 has been proposed to interact with and activate MutH [13]. However, mutations of solvent-exposed residues within this surface resulted in proteins that behaved like wild-type MutL in vitro and in vivo [5].
The second hypothesis is that the C-terminal region of E. coli MutL interacts with MutH. Studies using the yeast two-hybrid system showed that the C-terminal 218 amino acid region of MutL is sufficient for the interaction with MutH and that small deletions of the N- or C-terminal regions of this fragment completely eliminate two-hybrid interaction with MutH [8]. Various studies have indicated that the E. coli MutL C-terminal region interacts with the DNA helicase UvrD (DNA helicase II, also involved in the MMRS) [14,15] and mediates MutL dimerization [14,16].
Integrity of the C-terminal region of MutL appears to be crucial for the functioning of E. coli MutL because deletion of the last 66, or more, amino acids results in non-functional proteins and all except one of the mutations affecting this region of E. coli MutL were nonsense mutations [4].
These results, collectively, suggest that interaction between MutL and MutH involves a large surface of MutL, such that single amino acid substitutions are insufficient to disrupt it [5]. It remains unclear whether this interaction resides on the N- or C-terminal regions of MutL or on both.
In the present study, we used the E. coli MutL homologue from Pseudomonas aeruginosa to further characterize MutL functioning and MutL–MutH interaction. We analysed in vitro and in vivo functioning of E. coli and P. aeruginosa wild-type and chimaeric (P. aeruginosa/E. coli) MutL, and utilized protein affinity chromatography to analyse the interactions of the N- or C-terminal regions of P. aeruginosa or E. coli MutL with E. coli MutH.
EXPERIMENTAL
Bacterial strains, plasmids and chemicals
E. coli BL21(DE3) was from Novagen (Madison, WI, U.S.A.); E. coli ER 2566 was from New England Biolabs (Beverly, MA, U.S.A.); P. aeruginosa wild-type strain (Hex1T) was isolated and characterized in our laboratory [17]; and E. coli MutL-deficient strain GM4348 [F-mutL459 (KanR)] was generously provided by Dr M. G. Marinus (University of Massachusetts Medical School, Worcester, MA, U.S.A.). Plasmids pTX412, pTX417 and pTX418 (pET-15b derivatives containing the E. coli mutS, mutL and mutH genes respectively) were from Feng and Winkler [18]. Plasmids were purified using the Wizard Plus SV Miniprep DNA purification system (Promega, Madison, WI, U.S.A.). DNA extractions from agarose gels were performed using the QIAEX II Gel Extraction kit (Qiagen, Chatsworth, CA, U.S.A.). Restriction enzymes were from Promega and New England Biolabs. GoTaq DNA polymerase and T4 DNA ligase were from Promega.
Analysis of sequenced bacterial genomes by BLAST
Genomic BLAST (www.ncbi.nlm.nih.gov/sutils/genom_table.cgi) [19] with standard parameters was used to search genomes of bacteria completely sequenced against E. coli K12 MutS, MutL, MutH, Dam and UvrD proteins (E. coli K12 genome accession no. gi:48994873). In the case of Buchnera, Buchnera aphidicola str. Bp contains E. coli MutS, MutL, MutH and UvrD but not Dam homologues. B. aphidicola str. APS and B. aphidicola str. Sg have E. coli MutS, MutL and UvrD, but neither MutH nor Dam homologues [20].
Amino acid sequence alignment and secondary structure prediction
P. aeruginosa and E. coli MutL amino acid sequence alignment was performed using CLUSTAL W (www.ebi.ac.uk/clustalw/). Protein sequences were obtained from NCBI (www.ncbi.nlm.nih.gov/). Secondary structure predictions were obtained using JPRED (www.compbio.dundee.ac.uk/~www-jpred/). Percentage homology was calculated by dividing the number of amino acids that are identical (amino acid comparison), or which have the same predicted secondary structure (secondary structure comparison), by the total number of amino acids.
Cloning, expression and disruption of the mutL gene in P. aeruginosa
The mutL gene from P. aeruginosa was amplified by PCR using primers designed based on the published complete genome sequence of P. aeruginosa PAO1 (www.pseudomonas.com/) [21]. The Lpf primer (5′-AGCATATGAGTGAAGCACCGCGTATCC-3′) contains an extra five nucleotides (underlined) to create an NdeI restriction site at the ATG initiation codon of the mutL gene. The Lpr primer (5′-CGAGCAACGCCTTGTAATAGAGCA-3′) extends the TGA stop codon 391 bp downstream. The 2298 bp amplified PCR fragment was cloned in p-Geam-T-easy cloning vector (Promega) to generate plasmid pG-Lp.
For expression of P. aeruginosa MutL protein in E. coli, the NdeI–EcoRI fragment from the pG-Lp plasmid, carrying the coding region of this gene, was cloned in the corresponding restriction sites of the T7 polymerase-driven expression vector pET15b (Novagen) to generate plasmid pET-Lp. This plasmid produced N-terminal His-tagged P. aeruginosa MutL protein.
A P. aeruginosa MutL-deficient strain (ΔMutL) was generated by replacing the endogenous mutL gene (Lp) by homologous recombination, with an interrupted Lp::Km allele. The suicide pKN-Lp::Km plasmid was constructed as follows. The SalI fragment carrying the kanamycin-resistant gene from pUT-miniTn5-Km [22] was cloned in the XhoI site of plasmid pG-Lp, located 673 bp downstream from the transcription start site of P. aeruginosa mutL gene, to generate plasmid pG-Lp::Km. The ApaI–SpeI fragment from plasmid pG-Lp::Km, carrying the Km gene flanked by mutL sequences on both sides, was cloned in the corresponding restriction sites of plasmid pKNG101 [23] to generate plasmid pKN-Lp::Km. This plasmid was inserted into an E. coli S17-1 λpir strain, which was used as the donor strain for conjugation in biparental mating with a wild-type strain of P. aeruginosa as recipient strain, as described previously [23]. Disruption of the chromosomal copy of the P. aeruginosa mutL gene was confirmed by PCR and by phenotypic analyses.
Complementation assay
For mutL expression in complementation assays, BglII–EcoRI fragments from plasmid pET-Lp (the present study) and pTX418 (a pET15b derivative plasmid containing the E. coli mutL gene) [18] were cloned in the corresponding restriction sites of the broad range replication plasmid PBBRIMCS-5 (p5) [24], to generate plasmids p5-Lp and p5-Lc, containing the mutL gene from P. aeruginosa and from E. coli respectively. We observed previously that fragments BglII–EcoRI or XbaI–EcoRI from plasmid pET-mutS, carrying the P. aeruginosa mutS gene, cloned in any direction within the plasmid p5, were capable of complementing a P. aeruginosa MutS-deficient strain, probably by basal expression of this plasmid [17].
Complementation assays were performed by transforming E. coli GM4348 [F-mutL459 (KmR)] and P. aeruginosa (present study) MutL-deficient strains with plasmid p5 (no insert), plasmid p5-Lp or plasmid p5-Lc. Overnight cultures of transformed strains were grown from single colonies in LB (Luria–Bertani) media containing 10 or 30 μg/ml gentamicin for p5-, p5-Lp- or p5-Lc-transformed E. coli or P. aeruginosa strains respectively. Non-transformed controls were grown in LB without antibiotic. To calculate mutation frequencies, appropriate dilutions of overnight cultures were plated on LB (for non-transformed strains) or LB containing 10 or 30 μg/ml gentamicin (for p5-, p5-Lp- or p5-Lc-transformed E. coli or P. aeruginosa strains respectively) to determine the number of viable cells, or in LB containing 40 μg/ml nalidixic acid (for non-transformed E. coli strains), LB containing 100 μg/ml rifampicin (for non-transformed P. aeruginosa strains), LB containing 10 μg/ml gentamicin with 40 μg/ml nalidixic acid (for p5-, p5-Lp- or p5-Lc-transformed E. coli strains) or LB containing 30 μg/ml gentamicin with 100 μg/ml rifampicin (for p5-, p5-Lp- or p5-Lc-transformed P. aeruginosa strains) to score for rifampicin or nalidixic acid resistant cells respectively.
For complementation assays using chimaeric (E. coli/P. aeruginosa) genes, a similar procedure was followed. Similar results were obtained for three independent complementation assays for all plasmid constructs.
Complementation of the P. aeruginosa MutL-deficient strain with the mutL gene from P. aeruginosa confirmed that the PCR-amplified P. aeruginosa mutL gene was functional and that the P. aeruginosa MutL-deficient strain had no mutation other than interruption of the mutL gene, which could affect the MMRS.
Expression and purification of E. coli MutH, E. coli and P. aeruginosa MutL, and E. coli and P. aeruginosa N- and C-terminal regions of MutL
His-tagged E. coli MutH and E. coli and P. aeruginosa MutL proteins were isolated after expression of plasmids pTX417, pTX418 [18] and pET-Lp respectively in a BL21(DE3) strain, as recommended by the pET system manual (Novagen).
For expression of Intein-tagged proteins, the following constructions were performed using the pTYB12 plasmid of the Impact-CN system (New England Biolabs). For an Intein–MutH protein, the NdeI–EcoRI restriction fragment from plasmid pTX417 was cloned in the corresponding restriction sites of plasmid pTYB12, to generate the plasmid pTYB12-H. For Intein–Pcp (phosphorylcholine phosphatase, P. aeruginosa PA5292), the NdeI–EcoRI restriction fragment from plasmid pG-Pcp (a pGEM-T Easy vector containing a PCR-amplified fragment of the complete P. aeruginosa PA5292 gene, in which an NdeI restriction site was added at the ATG start codon site) was cloned in the corresponding restriction sites of plasmid pTYB12 to generate plasmid pTYB12-Pcp. For Intein–NLc and Intein–NLp fusion proteins, containing the N-terminal region of MutL protein from E. coli and P. aeruginosa respectively, the NdeI–HincII restriction fragments from plasmids pTX418 (903 bp) and pET-Lp (912 bp) were cloned in the NdeI–SmaI linearized plasmid pTYB12 to generate plasmids pTYB12-NLc and pTYB12-NLp respectively. These constructions produce N-terminal regions with an extra glycine residue at the C-termini of these fragments, corresponding to the GGG codon of the SmaI site that precedes the TGA stop codon. For Intein–CLc and Intein–CLp fusion proteins, containing the C-terminal region of MutL protein from E. coli and P. aeruginosa respectively, the HincII–EcoRI and XhoI–EcoRV restriction fragments from plasmids pTX418 (1742 bp) and pET-Lp (1422 bp) respectively were cloned in the NruI–EcoRI and XhoI–SmaI linearized pTYB12 plasmid respectively to generate the plasmids pTYB12-CLc and pTYB12-CLp.
Intein-tagged proteins were overexpressed in bacterial strain ER2566 as recommended by the Impact-CN system instruction manual (New England Biolabs). Purified proteins were >95% pure as judged by Coomassie Brilliant Blue-stained SDS/PAGE (results not shown). When cited in molar terms, protein concentrations are expressed as monomer equivalents.
Endonuclease activity test
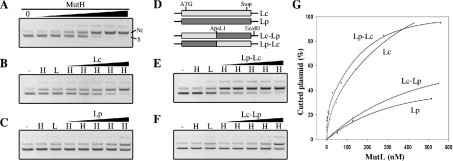
The endonuclease activity of E. coli MutH protein was assayed with a supercoiled d(GATC) unmethylated pBKS plasmid. The unmethylated plasmid was purified from an E. coli Dam-deficient strain. A supercoiled plasmid was isolated from the nicked species by agarose gel purification using agarACE (Promega; M1741). MutH (15, 30, 73, 145, 363, 725 and 1450 nM) was incubated with 0.3 μg of unmethylated pBKS plasmid/μl in 10 mM Tris/HCl, 10 mM MgCl2, 0.75 mM ATP and 0.05 mg/ml BSA (pH 7.5) for 10 min at 37 °C in the absence or presence of Lc (1, 16, 48, 143 and 430 nM), Lp (2, 21, 52, 129 and 517 nM), Lc–Lp (2, 21, 52, 129 and 550 nM) or Lp–Lc (2, 28, 70, 140 and 560 nM) protein, in a final volume of 35 μl. Cleavage reactions were analysed by 0.6% agarose gel electrophoresis and DNA was visualized by ethidium bromide staining. Three independent experiments were performed and all gave similar results. One representative experiment is shown in Figure 4.
Figure 4. In vitro activation of E. coli MutH endonuclease activity.
(A) Unmethylated d(GATC) plasmid was digested with increasing amounts of E. coli MutH protein (see the Experimental section), separated on 0.6% agarose gel and visualized with ethidium bromide. S, supercoiled plasmid; Nc, nicked circular plasmid. (B, C, E and F) Unmethylated d(GATC) plasmid was incubated with buffer (–), with 70 nM E. coli MutH (H), with 500 nM of the indicated MutL protein (L) or with 70 nM E. coli MutH and increasing amounts of the indicated MutL protein (H/MutL) (see the Experimental section), before separation on 0.6% agarose gel. (D) Schematic representation of E. coli (Lc), P. aeruginosa (Lp) and chimaeric MutL proteins containing the E. coli N-terminal region and P. aeruginosa C-terminal region (Lc–Lp) or vice versa (Lp–Lc). ApaLI and EcoRI indicate positions of the corresponding restriction sites used for the construction of chimaeric mutL genes. (G) Ethidium bromide-stained gels (B, C, E and F) were scanned and the amount of Nc plasmid was plotted as a percentage. The amount of Nc plasmid obtained with MutH alone (70 nM) was defined as 0%. The difference between the total amount of plasmid used and the amount of Nc plasmid obtained with MutH alone is taken as 100%. Negatives of ethidium bromide photographs are shown.
Cloning, expression and purification of (E. coli/P. aeruginosa) MutL chimaeric proteins
E. coli/P. aeruginosa mutL chimaeric genes were constructed as follows. The ApaLI–EcoRI restriction fragment from plasmid pTX418, containing the C-terminal region of E. coli MutL, was replaced by the same restriction fragment from plasmid pET-Lp, containing the C-terminal region of P. aeruginosa MutL, and vice versa, to generate plasmids pET-Lc-Lp and pET-Lp-Lc. Expression and purification of the chimaeric proteins were performed as recommended by the pET system manual.
Protein affinity columns
Cultures (500 ml) were used to overexpress the Intein-tagged proteins (I–H, I–Pcp, I–CLc, I–CLp, I–NLc and I–NLp) as recommended by the Impact-CN instruction manual. A clarified cell extract (supernatant), obtained after centrifugation at 12000 g for 30 min at 4 °C, of each crude cell extract was used to saturate 800 μl of chitin beads. Unbound material was eliminated by successive washes with 1 M NaCl, 20 mM Tris/HCl (pH 7.5) and 1 mM EDTA with 0.1% Triton X-100 and then without detergent. Finally, the chitin column containing the adsorbed Intein-tagged protein was equilibrated in buffer A [25 mM Tris/HCl, pH 7.5, 10% (v/v) glycerol, 2.5 mM 2-mercaptoethanol, 3 mM MgCl2 and 50 mM NaCl]. Similar amounts of different Intein-tagged proteins were overexpressed and then adsorbed on to chitin beads, as confirmed by SDS/PAGE analysis of samples from clarified cell extracts and from saturated columns (results not shown).
His-tagged proteins (300 μg in 2 ml of buffer A) were applied to a chitin/Intein-protein column thrice. Columns were washed with 1.6 ml of buffer A and 0.4 ml fractions were collected. Finally, columns were eluted with 0.8, 1.6 and 0.8 ml of buffer A containing NaCl (final concentration of 500 mM, 1 and 2 M respectively) and then 0.2 ml fractions were collected. Fractions (50 μl for each sample) were analysed by electrophoresis on SDS/PAGE. Stained gels (Coomassie Blue) were scanned and values obtained were plotted using the same arbitrary units for all samples (SigmaPlot program, Smooth 2D option).
Analysis of the LN40(dimer)–MutH docking models
The ten docking models previously selected for the LN40(dimer)–MutH complex, generously provided by Dr P. Friedhoff [12], were analysed using the RasMol program (www.rasmol.org). For this analysis, LN40 molecules of the ten models were analysed to identify those amino acids in which the C-α atom is not more than 4 or 6 Å (1=0.1 nm) from any C-α atom of MutH. A similar analysis was performed in MutL to identify those amino acids in which any atom is not more than 3 Å from any atom of MutH. The three analyses gave similar results.
RESULTS
Presence of E. coli MutS, MutL, MutH, Dam and UvrD homologues in bacterial sequenced genomes
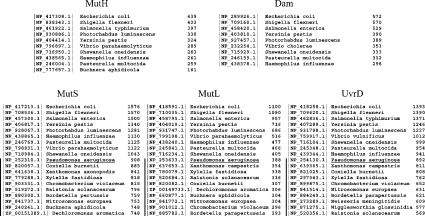
Completed bacterial genomes widely available were searched by BLAST [19] against the E. coli MutS, MutL, MutH, Dam and UvrD proteins. As expected, only a few bacterial species have homologues to the E. coli MutH protein (Figure 1). Each of these species also has the corresponding homologues of E. coli Dam, MutS, MutL and UvrD proteins, except for Buchnera, which does not have the corresponding Dam homologue (Figure 1; see the Experimental section). Within the species lacking the corresponding E. coli MutH and Dam homologues, P. aeruginosa has the highest sequence homology to E. coli MutS, MutL and UvrD proteins (59, 42 and 64% identical respectively) (Figure 1).
Figure 1. BLAST analysis of bacterial sequenced genomes.
BLAST search results using E. coli MutH, Dam, MutS, MutL and UvrD proteins as query for search of homologous proteins within completely sequenced bacterial genomes. Only the highest homologues to E. coli proteins for each bacterial species are indicated. Black vertical lines indicate bacterial species that do not have the MutH/Dam system and the first species within this group is underlined. For each protein the third column indicates the score (bits).
Absence of d(GATC) adenine methylation in P. aeruginosa
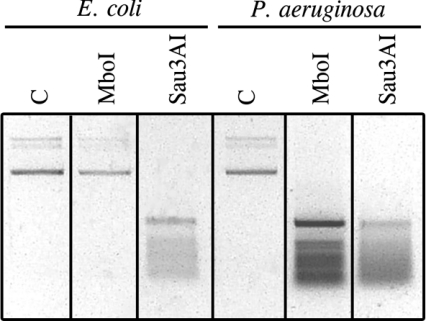
Homologues to the E. coli Dam methylation protein have not been found in the genome of P. aeruginosa [21]. We confirmed the absence of Dam methylation by analysing the methylation state of d(GATC) sequences in plasmids purified from P. aeruginosa, using restriction enzymes sensitive and non-sensitive to adenine methylation. MboI and Sau3AI restriction enzymes recognize the same sequence, d(GATC). MboI will not cut the sequence if the adenine residue is methylated, whereas Sau3AI is insensitive to adenine methylation. Plasmids purified from P. aeruginosa and from Dam+ E. coli were digested with Sau3AI and MboI restriction enzymes. While the plasmid purified from Dam+ E. coli DNA was digested only with Sau3AI, the plasmid purified from P. aeruginosa was digested equally with MboI and Sau3AI (Figure 2), confirming lack of Dam methylation in P. aeruginosa. Similar results were obtained when genomic DNA from wild-type P. aeruginosa and Dam+ E. coli were analysed with the same restriction enzymes (results not shown).
Figure 2. Analysis of adenine methylation within d(GATC) sequences.
Plasmids purified from E. coli or P. aeruginosa wild-type strains were digested with MboI or Sau3AI restriction enzymes, which are sensitive and insensitive to adenine methylation respectively, and then separated on 0.8% agarose gel. DNA was visualized by ethidium bromide staining. The negative of the photograph is shown. C, control plasmid from E. coli or P. aeruginosa not digested.
Comparative analysis of MutL proteins from E. coli and P. aeruginosa
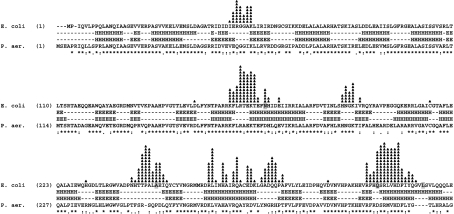
Analysis of sequence homology between E. coli and P. aeruginosa MutL proteins revealed, as expected, that the N-terminal regions (amino acids 1–335) are highly homologous (61% identical and 80% considering similar amino acids), whereas the C-terminal regions are only 18% identical (35% considering similar amino acids; Figure 3). However, the predicted secondary structure (see the Experimental section) gave very similar homology for the N- and C-terminal regions (91 and 88% identical respectively; Figure 3).
Figure 3. E. coli and P. aeruginosa MutL amino acid sequence comparison.
Amino acid sequences were aligned using CLUSTAL W. Amino acids for which mutation in E. coli MutL results in non-functional protein are indicated with grey boxes. Amino acids that are identical (*), strongly similar (:), weakly similar (.) or different () are indicated. Predicted secondary structures are shown between the amino acid sequences. H, helix; E, β-sheet. Black line indicates an unstructured linker region between the N- and C-terminal regions.
Amino acids important for the functioning of E. coli MutL protein have been identified after isolation of hydroxylamine-induced mutations, which impart a dominant-negative phenotype to the wild-type strain for increased spontaneous mutagenesis [4], or by amino acid substitution, guided by E. coli N-terminal MutL three-dimensional structure [5]. Among these mutations, 21 change amino acids that are located in the N-terminal region and one in the C-terminal region. The amino acids located in the N-terminal region are all conserved in the P. aeruginosa MutL protein (19 are identical and two have conservative changes), whereas the amino acid located in the C-terminal region is only weakly similar in P. aeruginosa (Figure 3).
Alanine scanning mutagenesis of Mlh1p revealed a region insensitive to mutagenesis (approx. 100 residues) [25]. It was postulated that this region, predicted to form a random coil or unstructured domain, could serve as a flexible linker between the C- and N-terminal domains. Analysis of E. coli MutL mutants in which different portions of this unstructured region are deleted revealed that part of this linker region can be eliminated without affecting the in vivo functioning of MutL [14]. The predicted secondary structure of P. aeruginosa MutL protein revealed the presence of a similar unstructured linker region (Figure 3).
In vivo complementation of MutL-deficient strains
Studies on complementation of MMRS-deficient E. coli strains with corresponding genes from other bacteria showed successful complementation in the case of mutS gene from Salmonella typhimurium [26] and mutH genes from Haemophilus influenzae and Vibrio cholerae [27], and partial complementation with an incomplete mutS gene from Pseudomonas putida [28] and mutL and mutS genes from H. influenzae [29]. No complementation was observed with the mutS or uvrD genes from P. aeruginosa [30], mutS or mutL genes from Streptococcus pneumoniae [31], or mutS gene from Thermus aquaticus [32].
It is not clear a priori if the mutL gene from P. aeruginosa will function in the context of the E. coli MMRS, since P. aeruginosa does not have the corresponding E. coli MutH and Dam homologues.
Functioning of MutL protein from P. aeruginosa was assessed by an in vivo complementation assay using a MutL-deficient E. coli strain. The complementation assay is based on the mutator phenotype that occurs when the bacterium lacks the MutL protein. As shown in Table 1, the mutL gene from P. aeruginosa complemented the E. coli MutL-deficient strain to a similar extent as the mutL gene from E. coli (92 and 99% respectively). The 10-fold increase of antibiotic-resistant cells in P. aeruginosa ΔMutL compared with E. coli ΔMutL (Table 1) is not due to the use of different antibiotics, since E. coli strains analysed with rifampicin gave results similar to those obtained with nalidixic acid (results not shown).
Table 1. In vivo complementation of E. coli and P. aeruginosa MutL-deficient strains with wild-type and chimaeric mutL genes.
| Strain | Plasmid* | Complemented strains (%) | No. of antibiotic-resistant cells/108 cells† | Complementation (%)‡ |
|---|---|---|---|---|
| E. coli WT | – | – | 1±0.8 | 99 |
| E. coli Δ MutL | p5 | 0 | 234±72 | 0 |
| E. coli Δ MutL | p5-Lc | 100 | 1±0.9 | 99 |
| E. coli Δ MutL | p5-Lp | 100 | 19±8 | 92 |
| E. coli Δ MutL | p5-Lc-Lp | 100 | 8±6 | 96 |
| E. coli Δ MutL | p5-Lp-Lc | 100 | 10±8 | 93 |
| P. aeruginosa WT | – | – | 5±3 | 99 |
| P. aeruginosa Δ MutL | p5 | 0 | 3.000±200 | 0 |
| P. aeruginosa Δ MutL | p5-Lp | 90 | 5±2 | 99§ |
| P. aeruginosa Δ MutL | p5-Lc | 74 | 25±6 | 99§ |
* p5, empty plasmid; p5-Lc, p5 plasmid with E. coli mutL gene; p5-Lp, p5 plasmid with P. aeruginosa mutL gene; p5-Lc-Lp, p5 plasmid with chimaeric (E. coli/P. aeruginosa) mutL gene; p5-Lp-Lc, p5 plasmid with chimaeric (P. aeruginosa/E. coli) mutL gene.
† Results are means±S.D. for three independent cultures. Antibiotic-resistant cells were scored using nalidixic acid for E. coli and rifampicin for P. aeruginosa.
‡ The percentage of complementation was calculated considering the number of antibiotic-resistant cells obtained in the absence of MutL as 0% of complementation, for each bacterial species.
§ The percentage of complementation was calculated considering only complemented strains.
The mutL gene from E. coli was likewise capable of complementing the P. aeruginosa MutL-deficient strain. In this case, complemented strains repaired more than 99% of the mutations. As negative and positive controls, E. coli and P. aeruginosa MutL-deficient strains were transformed with empty vector or complemented with mutL gene from E. coli and P. aeruginosa respectively (Table 1). These results show that MutL from P. aeruginosa restores functioning of the MMRS of an E. coli MutL-deficient strain.
P. aeruginosa MutL protein activates E. coli MutH endonuclease activity in vitro
In vivo complementation of the E. coli MutL-deficient strain indicates that the MutL protein from P. aeruginosa interacts with and activates the E. coli mutH protein. We analysed the in vitro capacity of P. aeruginosa MutL protein to stimulate E. coli MutH endonuclease activity. MutH endonuclease activity was monitored by studying the conversion of the supercoiled into the nicked circular form of d(GATC) unmethylated plasmid. At high enzyme concentrations, the E. coli MutH protein was able to cleave unmethylated plasmid DNA, whereas at low protein concentration, MutH alone showed almost no DNA cleavage activity (Figure 4A). As described previously [8,11], addition of E. coli MutL protein and ATP to the incubation system stimulates E. coli MutH endonuclease activity (Figures 4B and 4G). Addition of P. aeruginosa MutL protein and ATP also stimulated E. coli MutH endonuclease activity, although to a lesser extent (Figures 4C and 4G). These results show that MutL from P. aeruginosa activates E. coli MutH endonuclease activity in the in vitro mismatch-MutS independent system, although less efficiently compared with the E. coli MutL protein.
MutH is specifically retained by the C-terminal regions of E. coli and P. aeruginosa MutL proteins on affinity chromatography
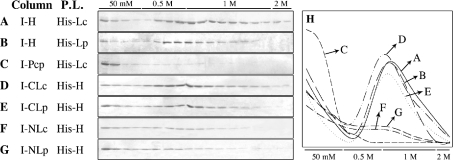
As described above, both the N- and C-terminal regions of E. coli MutL have been proposed to interact with MutH. We analysed in vitro interaction between E. coli MutH protein and the N- and C-terminal regions of P. aeruginosa and E. coli MutL proteins using protein affinity columns. MutH and the N- and C-terminal regions of P. aeruginosa and E. coli MutL proteins were expressed as fusion proteins. Intein-tagged proteins were overexpressed, adsorbed on to chitin beads through the Intein-tag and used as an affinity ligand. Purified His-tagged proteins (E. coli His–MutH, and P. aeruginosa and E. coli His–MutL) were loaded on to the affinity columns, then eluted with a series of salt washes as described previously [8] (see the Experimental section).
A fraction of the loaded E. coli MutL protein was retained on the MutH affinity column after the 50 mM NaCl wash step and was then eluted with 0.5–1 M NaCl (Figures 5A and 5H). A similar result was obtained when P. aeruginosa MutL protein was loaded on to a similar MutH affinity column (Figures 5B and 5H). As control, a similar experiment was performed but using the Intein–Pcp (Pcp from P. aeruginosa, PA5292) fusion protein adsorbed on to chitin beads as an affinity column. In this case, the loaded E. coli MutL protein was found principally in the flowthrough and 50 mM NaCl wash (Figures 5C and 5H).
Figure 5. Analysis of MutL–MutH interaction on protein affinity columns.
(A–G) Intein-tagged proteins were adsorbed on to chitin beads and used as affinity ligands. Different His-tagged proteins were loaded on to each column and then eluted with increasing amounts of NaCl, as indicated (see the Experimental section). Gels shown in (A–G) were scanned and the values plotted in (H) (see the Experimental section). Column, protein adsorbed on to chitin beads. P.L., protein loaded on to the column. I, Intein-tagged proteins; His, His-tagged proteins; H, E. coli MutH protein; CLc and CLp, MutL C-terminal regions of E. coli and P. aeruginosa; NLc and NLp, N-terminal regions of E. coli and P. aeruginosa.
To identify the MutL region responsible for the in vitro affinity interaction with MutH, the N- and C-terminal regions of MutL from P. aeruginosa (I–NLp and I–CLp) and E. coli (I–NLc and I–CLc) were overexpressed, fused to the Intein protein, adsorbed on to chitin beads through the Intein-tag and used as affinity ligands. A fraction of the loaded E. coli MutH protein was retained on affinity columns containing the C-terminal region of MutL from E. coli (I–CLc) or P. aeruginosa (I–CLp) after the 50 mM NaCl wash step and was then eluted with 0.5–1 M NaCl (Figures 5D, 5E and 5H). The amount of MutH retained by the C-terminal regions was similar to that retained by the full-length proteins. In contrast, when the E. coli MutH protein was loaded on to affinity columns containing the N-terminal region of MutL from E. coli (I–NLc) or P. aeruginosa (I–NLp), only a small fraction of the applied E. coli MutH protein was retained and subsequently eluted with 0.5–1 M NaCl (Figures 5F, 5G and 5H). These results show that the C-terminal regions of P. aeruginosa and E. coli MutL proteins specifically interact with and retain MutH in these protein affinity columns.
Analysis of E. coli–P. aeruginosa MutL chimaeric proteins
Based on the above results, it is not clear why, in vitro, MutL from P. aeruginosa stimulates E. coli MutH endonuclease activity less efficiently compared with E. coli MutL protein (see Figures 4B, 4C and 4G). One possibility is that although the C-terminal region of P. aeruginosa interacts with E. coli MutH protein, the highly divergent amino acid sequence of P. aeruginosa MutL protein affects stimulation of MutH endonuclease activity, performed by the MutL N-terminal region. However, the P. aeruginosa N-terminal region could also be involved. It was proposed recently that the interaction site between the N-terminal region of E. coli MutL and MutH could be mapped on to a region comprising Asn169, Ala251, Gln314 and Leu327 residues of MutL [12]; however, these amino acids are only weakly conserved in P. aeruginosa MutL protein (Figure 6).
Figure 6. Identification of putative amino acids involved in MutL–MutH interaction.
Selected docking models for interaction of E. coli LN40(dimer)–MutH [12] were analysed using the RasMol program (see the Experimental section). MutL amino acids in which the C-α is not more than 6 Å from any C-α of MutH are indicated. The number of black triangles indicates how many times each amino acid was identified by this analysis. The four amino acids identified on E. coli MutL protein by cross-linking (Asn169, Ala251, Gln314 and Leu327) [12] are indicated with grey boxes. Amino acids that are identical (*), strongly similar (:), wealhy similar (.) or different () are indicated. Predicted secondary structures are shown between the amino acid sequences. H, helix; E, β-sheet.
To analyse whether the N- or C-terminal region of P. aeruginosa MutL protein was responsible for the decreased stimulatory effect of E. coli MutH endonuclease activity in vitro, we generated chimaeric proteins containing the E. coli N-terminal region and the P. aeruginosa C-terminal region (Lc–Lp), or vice versa (Lp–Lc) (Figure 4D). In vivo analysis showed that both these chimaeric proteins are capable of complementing the MMRS of an E. coli MutL-deficient strain. Transformed E. coli MutL-deficient strains expressing Lc–Lp or Lp–Lc were capable of repairing 96 and 93% of the mutations respectively (Table 1).
In vitro, the Lp–Lc chimaeric protein stimulated E. coli MutH endonuclease activity similar to E. coli MutL protein (Figures 4E and 4G). The Lc–Lp chimaeric protein also stimulated E. coli MutH endonuclease activity, but to a lesser extent, similar to P. aeruginosa MutL protein (Figures 4F and 4G). These results indicate that (i) the C-terminal region of P. aeruginosa MutL protein is responsible for the decreased in vitro stimulatory effect on E. coli MutH endonuclease activity, and (ii) the N-terminal region of P. aeruginosa MutL protein functions, in vivo and in vitro, similarly to the N-terminal region of E. coli MutL protein.
Analysis of the MutH interface of the N-terminal region of MutL
It was recently proposed, based on a combination of site-directed mutagenesis and site-specific cross-linking, that the MutH interaction site of LN40 maps on to a region comprising Asn169, Ala251, Gln314 and Leu327 residues [12]. From 10000 docking models generated for the complex LN40(dimer)–MutH, ten were selected on the basis of cross-linking results [12].
The chimaeric Lp–Lc protein behaved similar to wild-type E. coli MutL protein in vivo and in vitro, showing that the N-terminal regions of P. aeruginosa and E. coli MutL proteins function in the same manner. The N-terminal region of P. aeruginosa MutL protein has conserved the 21 amino acids essential for the functioning of E. coli MutL protein (see Figure 3). However, the four amino acids identified on the surface of the E. coli LN40 dimer by cross-linking experiments with MutH (positions 169, 251, 314 and 327) are only weakly conserved in P. aeruginosa MutL protein (Figure 6). Our analysis of the ten models previously selected for the LN40(dimer)–MutH complex [12] revealed interesting features. LN40 molecules of the ten docking models were analysed to identify amino acids in which the C-α atom is not more than 4 or 6 Å from any C-α atom of MutH, or amino acids in which any atom is no more than 3 Å from any atom of MutH. Considering these distances (3–6 Å), the amino acids identified could also be involved in the interaction between LN40 and MutH. The three analyses gave similar results. In Figure 6 (showing only amino acids in which the C-α atom is not more than 6 Å from any C-α atom of MutH), the identified amino acids are clustered in several groups. Some of these clusters are in regions that are highly conserved between E. coli and P. aeruginosa (Figure 6), suggesting that selected models involving these regions could also model the interaction between the N-terminal region of P. aeruginosa MutL protein and E. coli MutH protein. Comparative analysis between E. coli and P. aeruginosa MutL proteins will better identify regions and/or amino acids important for interaction between MutL and MutH proteins.
DISCUSSION
We have shown that MutL from P. aeruginosa is capable of complementing an E. coli MutL-deficient strain. This result was surprising considering that (i) P. aeruginosa does not have E. coli MutH and Dam homologues and therefore the mechanism of DNA strand discrimination for mismatch repair in P. aeruginosa is presumably different from that of E. coli; (ii) MutS and UvrD proteins from P. aeruginosa do not complement the corresponding E. coli mutant strains [30]. In fact, MutS produced a dominant-negative effect when introduced into a wild-type E. coli strain, increasing the mutation frequency 50 times [30]. Oliver et al. [30] reported that P. aeruginosa mutL gene failed to complement an E. coli MutL-deficient strain. However, in that study, the P. aeruginosa mutL gene was introduced with its own promoter region into the E. coli MutL-deficient strain, and it is possible that the P. aeruginosa mutL promoter was non-functional in E. coli or that the expression level obtained was insufficient to complement the E. coli MutL-deficient strain.
It has been proposed that the E. coli mutH methylation-based system evolved from a restriction modification system [3]. If this evolution was recent, perhaps the E. coli and P. aeruginosa MutL proteins did not have time to diverge and so they are both capable of complementing E. coli and P. aeruginosa MutL-deficient strains. Another possibility is that the regions that allow these proteins to work in both species are structurally important for MutL, or for interaction with other proteins (MutS, UvrD, Vsr endonuclease, etc.), and are therefore stable during evolution.
The E. coli MutL protein and the chimaeric construct having P. aeruginosa N-terminal region were both functional in vivo and also capable of stimulating in vitro the E. coli MutH endonuclease activity, showing that the N-terminal regions of P. aeruginosa and E. coli MutL proteins function similarly. Recently, a model for LN40–MutH complex was proposed [12]. Although the four amino acids of E. coli LN40 proposed for MutL–MutH interaction are only weakly conserved in P. aeruginosa MutL, our analysis revealed that some of the proposed LN40(dimer)–MutH models could involve amino acid regions that are highly conserved in P. aeruginosa MutL.
E. coli MutL mutants with deletion of the C-terminal region (amino acids 67–285) have a dominant-negative effect when introduced into a wild-type strain [4]. MutH and UvrD interact with the last 218 amino acid segment of E. coli MutL [8,14,15], and removal of a small number of residues from either the N- or C-terminus of this segment terminates this interaction [8]. Our results are consistent with these and support the hypothesis that the C-terminal region of MutL is involved in the interaction with MutH. We showed, using protein affinity chromatography, that the C-terminal regions of E. coli and P. aeruginosa MutL are capable of specifically interacting with and retaining E. coli MutH. Although the amino acid sequences of the C-terminal regions of E. coli and P. aeruginosa MutL have low homology (18%), the predicted secondary structure showed high homology (88%). These results, and the fact that no MutL point mutation capable of preventing MutL–MutH interaction has been isolated so far, support the hypothesis that the MutH interface on MutL involves a large surface or a particular structure.
MutH is capable of interacting with the C-terminal region of MutL without any particular requirement (ATP, MutS, DNA, etc.) (the present study and [8]), suggesting that a part of cellular MutL forms a complex with MutH. Such a MutL–MutH cellular soluble complex would have functional implications.
According to the sliding clamp model [33], several sliding clamps are generated at a mismatch site after the binding of MutS and exchange of ADP with ATP. MutL, with no binding to ATP, then interacts with MutS(ATP) sliding clamps. The function of MutL is to physically connect the MutS sliding clamps with MutH endonuclease and UvrD helicase. MutS(ATP) and MutS-(ATP)–MutL sliding clamps are both capable of ATP hydrolysis-independent diffusion along duplex DNA. However, MutL binding enhances unloading of MutS sliding clamps 10-fold [33]. MutH does not affect MutS mispair binding activity, MutS or MutL ATPase activity, or formation/release of MutS–MutL complex [33]. Therefore interaction of MutS with preformed MutL–MutH complex, rather than two-stage interaction (first with MutL, then with MutH), will make MutH more likely to exert its enzymatic activity before dissociation of the clamp.
This hypothesis can be extended to UvrD, which, similar to MutH, interacts with the last 218 amino acid segment of E. coli MutL without any particular requirement [14,15]. The use of uvrD mutant strains to obtain purified MutL, not contaminated with UvrD further supports the putative MutL–UvrD soluble complexes in vivo [10].
The E. coli MMRS proceeds through recognition of a hemimethylated d(GATC) site by MutH or through the presence of a nick within the double-stranded DNA molecule [34,35]. Therefore UvrD function is independent of the formation and/or activation of a MutS–MutL–MutH complex. The proposed model is capable of explaining both cases. MutS–MutL–MutH sliding clamps will slide through the DNA until a hemimethylated d(GATC) site is found, generating a nick in the unmethylated DNA strand, or until it dissociates. If a MutS–MutL–UvrD sliding clamp is formed, this complex will slide through the DNA until a nick is found or until it dissociates. Both kinds of complex can be generated continuously and can operate independently or consecutively.
Results of the present and previous studies suggest that both the N- and C-terminal regions of MutL are involved in the interaction with MutH. However, further genetic and biochemical experiments are necessary to understand how MutL interacts with and activates MutH at the molecular level within the E. coli MMRS.
Acknowledgments
We thank Dr P. Friedhoff for generously providing us docking models for interaction of E. coli LN40 with MutH [12] and for a critical reading of this paper. We also thank Dr M. E. Alvarez for a critical reading of the paper, Dr C. R. Mas for assistance with artwork, Dr M.A. Villarreal for assistance with RasMol analysis, M.I. Burgos for help in the construction of the chimaeric mutL genes and Dr S. Anderson for editing. This work was supported by grants from the Fundación Antorchas, Agencia Córdoba Ciencia, SECyT-UNC, ANPCyT and CONICET (Argentina).
References
- 1.Modrich P., Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 2.Kolodner R. Biochemistry and genetics of eukaryotic mismatch repair. Genes Dev. 1996;10:1433–1442. doi: 10.1101/gad.10.12.1433. [DOI] [PubMed] [Google Scholar]
- 3.Eisen J. A., Hanawalt P. C. A phylogenomic study of DNA repair genes, proteins, and processes. Mutat. Res. 1999;435:171–213. doi: 10.1016/s0921-8777(99)00050-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aronshtam A., Marinus M. G. Dominant negative mutator mutations in the mutL gene of Escherichia coli. Nucleic Acids Res. 1996;24:2498–2504. doi: 10.1093/nar/24.13.2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Junop M. S., Yang W., Funchain P., Clendenin W., Miller J. H. In vitro and in vivo studies of MutS, MutL and MutH mutants: correlation of mismatch repair and DNA recombination. DNA Repair. 2003;2:387–405. doi: 10.1016/s1568-7864(02)00245-8. [DOI] [PubMed] [Google Scholar]
- 6.Peltomaki P., Vasen H. F. Mutations predisposing to hereditary nonpolyposis colorectal cancer: database and results of a collaborative study. The International Collaborative Group on Hereditary Nonpolyposis Colorectal Cancer. Gastroenterology. 1997;113:1146–1158. doi: 10.1053/gast.1997.v113.pm9322509. [DOI] [PubMed] [Google Scholar]
- 7.Umar A., Kunkel T. A. DNA-replication fidelity, mismatch repair and genome instability in cancer cells. Eur. J. Biochem. 1996;238:297–307. doi: 10.1111/j.1432-1033.1996.0297z.x. [DOI] [PubMed] [Google Scholar]
- 8.Hall M. C., Matson S. W. The Escherichia coli MutL protein physically interacts with MutH and stimulates the MutH-associated endonuclease activity. J. Biol. Chem. 1999;274:1306–1312. doi: 10.1074/jbc.274.3.1306. [DOI] [PubMed] [Google Scholar]
- 9.Mansour C. A., Doiron K. M., Cupples C. G. Characterization of functional interactions among the Escherichia coli mismatch repair proteins using a bacterial two-hybrid assay. Mutat. Res. 2001;485:331–338. doi: 10.1016/s0921-8777(01)00071-4. [DOI] [PubMed] [Google Scholar]
- 10.Spampinato C., Modrich P. The MutL ATPase is required for mismatch repair. J. Biol. Chem. 2000;275:9863–9869. doi: 10.1074/jbc.275.13.9863. [DOI] [PubMed] [Google Scholar]
- 11.Ban C., Yang W. Crystal structure and ATPase activity of MutL: implications for DNA repair and mutagenesis. Cell (Cambridge, Mass.) 1998;95:541–552. doi: 10.1016/s0092-8674(00)81621-9. [DOI] [PubMed] [Google Scholar]
- 12.Giron-Monzon L., Manelyte L., Ahrends R., Kirsch D., Spengler B., Friedhoff P. Mapping protein-protein interactions between MutL and MutH by cross-linking. J. Biol. Chem. 2004;279:49338–49345. doi: 10.1074/jbc.M409307200. [DOI] [PubMed] [Google Scholar]
- 13.Yang W. Structure and function of mismatch repair proteins. Mutat. Res. 2000;460:245–256. doi: 10.1016/s0921-8777(00)00030-6. [DOI] [PubMed] [Google Scholar]
- 14.Guarne A., Ramon-Maiques S., Wolff E. M., Ghirlando R., Hu X., Miller J. H., Yang W. Structure of the MutL C-terminal domain: a model of intact MutL and its roles in mismatch repair. EMBO J. 2004;23:4134–4145. doi: 10.1038/sj.emboj.7600412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall M. C., Jordan J. R., Matson S. W. Evidence for a physical interaction between the Escherichia coli methyl-directed mismatch repair proteins MutL and UvrD. EMBO J. 1998;17:1535–1541. doi: 10.1093/emboj/17.5.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drotschmann K., Aronshtam A., Fritz H. J., Marinus M. G. The Escherichia coli MutL protein stimulates binding of Vsr and MutS to heteroduplex DNA. Nucleic Acids Res. 1998;26:948–953. doi: 10.1093/nar/26.4.948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pezza R. J., Smania A. M., Barra J. L., Argarana C. E. Nucleotides and heteroduplex DNA preserve the active conformation of Pseudomonas aeruginosa MutS by preventing protein oligomerization. Biochem. J. 2002;361:87–95. doi: 10.1042/0264-6021:3610087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng G., Winkler M. E. Single-step purifications of His6-MutH, His6-MutL and His6-MutS repair proteins of Escherichia coli K-12. Biotechniques. 1995;19:956–965. [PubMed] [Google Scholar]
- 19.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 20.van Ham R. C., Kamerbeek J., Palacios C., Rausell C., Abascal F., Bastolla U., Fernandez J. M., Jimenez L., Postigo M., Silva F. J., et al. Reductive genome evolution in Buchnera aphidicola. Proc. Natl. Acad. Sci. U.S.A. 2003;100:581–586. doi: 10.1073/pnas.0235981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stover C. K., Pham X. Q., Erwin A. L., Mizoguchi S. D., Warrener P., Hickey M. J., Brinkman F. S., Hufnagle W. O., Kowalik D. J., Lagrou M., et al. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature (London) 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 22.de Lorenzo V., Timmis K. N. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 1994;235:386–405. doi: 10.1016/0076-6879(94)35157-0. [DOI] [PubMed] [Google Scholar]
- 23.Kaniga K., Delor I., Cornelis G. R. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene. 1991;109:137–141. doi: 10.1016/0378-1119(91)90599-7. [DOI] [PubMed] [Google Scholar]
- 24.Kovach M. E., Phillips R. W., Elzer P. H., Roop R. M., II, Peterson K. M. pBBR1MCS: a broad-host-range cloning vector. Biotechniques. 1994;16:800–802. [PubMed] [Google Scholar]
- 25.Argueso J. L., Kijas A. W., Sarin S., Heck J., Waase M., Alani E. Systematic mutagenesis of the Saccharomyces cerevisiae MLH1 gene reveals distinct roles for Mlh1p in meiotic crossing over and in vegetative and meiotic mismatch repair. Mol. Cell. Biol. 2003;23:873–886. doi: 10.1128/MCB.23.3.873-886.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haber L. T., Pang P. P., Sobell D. I., Mankovich J. A., Walker G. C. Nucleotide sequence of the Salmonella typhimurium mutS gene required for mismatch repair: homology of MutS and HexA of Streptococcus pneumoniae. J. Bacteriol. 1988;170:197–202. doi: 10.1128/jb.170.1.197-202.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Friedhoff P., Sheybani B., Thomas E., Merz C., Pingoud A. Haemophilus influenzae and Vibrio cholerae genes for mutH are able to fully complement a mutH defect in Escherichia coli. FEMS Microbiol. Lett. 2002;208:123–128. doi: 10.1111/j.1574-6968.2002.tb11071.x. [DOI] [PubMed] [Google Scholar]
- 28.Kurusu Y., Narita T., Suzuki M., Watanabe T. Genetic analysis of an incomplete mutS gene from Pseudomonas putida. J. Bacteriol. 2000;182:5278–5279. doi: 10.1128/jb.182.18.5278-5279.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joseph N., Sawarkar R., Rao D. N. DNA mismatch correction in Haemophilus influenzae: characterization of MutL, MutH and their interaction. DNA Repair. 2004;3:1561–1577. doi: 10.1016/j.dnarep.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 30.Oliver A., Baquero F., Blazquez J. The mismatch repair system (mutS, mutL and uvrD genes) in Pseudomonas aeruginosa: molecular characterization of naturally occurring mutants. Mol. Microbiol. 2002;43:1641–1650. doi: 10.1046/j.1365-2958.2002.02855.x. [DOI] [PubMed] [Google Scholar]
- 31.Prudhomme M., Martin B., Mejean V., Claverys J. P. Nucleotide sequence of the Streptococcus pneumoniae hexB mismatch repair gene: homology of HexB to MutL of Salmonella typhimurium and to PMS1 of Saccharomyces cerevisiae. J. Bacteriol. 1989;171:5332–5338. doi: 10.1128/jb.171.10.5332-5338.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Biswas I., Hsieh P. Identification and characterization of a thermostable MutS homolog from Thermus aquaticus. J. Biol. Chem. 1996;271:5040–5048. doi: 10.1074/jbc.271.9.5040. [DOI] [PubMed] [Google Scholar]
- 33.Acharya S., Foster P. L., Brooks P., Fishel R. The coordinated functions of the E. coli MutS and MutL proteins in mismatch repair. Mol. Cell. 2003;12:233–246. doi: 10.1016/s1097-2765(03)00219-3. [DOI] [PubMed] [Google Scholar]
- 34.Langle-Rouault F., Maenhaut-Michel G., Radman M. GATC sequences, DNA nicks and the MutH function in Escherichia coli mismatch repair. EMBO J. 1987;6:1121–1127. doi: 10.1002/j.1460-2075.1987.tb04867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lahue R. S., Au K. G., Modrich P. DNA mismatch correction in a defined system. Science. 1989;245:160–164. doi: 10.1126/science.2665076. [DOI] [PubMed] [Google Scholar]