Abstract
In the present paper, we describe a novel approach to map monoclonal antibody epitopes, using three new monoclonal antibodies that recognize h-TG2 (human transglutaminase 2) as an example. The target gene was fragmented and cloned upstream of an antibiotic-resistance gene, in the vector pPAO2, to select for in-frame polypeptides. After removal of the antibiotic-resistance gene by Cre/Lox recombination, an antigen fragment phage display library was created and selected against specific monoclonal antibodies. Using the h-TG2 fragment library, we were able to identify epitopes. This technique can also be broadly applied to the study of protein–protein interactions.
Keywords: epitope mapping, β-lactamase, phage display, transglutaminase
Abbreviations: gp-, guinea-pig; h-, human; IPTG, isopropyl β-D-thiogalactoside; m-, mouse; mAb, monoclonal antibody; MPBS, 2% non-fat milk in PBS; PBST, PBS with 0.1% Tween 20; scFv, single-chain variable fragment; TG2, transglutaminase 2; TGase, transglutaminase
INTRODUCTION
The identification of mAb (monoclonal antibody) epitopes, in particular, and specific domains involved in protein interaction, in general, is an important goal in the study of protein–protein interactions and, to this end, phage display and the yeast two-hybrid system have been the most commonly used technologies. In phage display, two broad approaches have been taken to the identification of protein epitopes: the use of random peptide libraries [1–6], in which consensus sequences selected by binding proteins are compared with the sequences of the recognized proteins, and the use of gene fragment libraries [2,7–11], in which gene fragments are displayed on phage, and the sequences of recognized fragments directly identify the domains responsible. In direct comparisons between the two methods, epitopes were more easily and accurately identified using the gene fragment approach for a number of different antibodies [12,13]. However, gene fragment display suffers from the problem that the vast majority of clones are non-functional, since only one clone in 18, if starting with DNA encoding an open reading frame, will be correctly in-frame (one clone in three will start correctly, one clone in three will end correctly, and one clone in two will have the correct orientation), although experiments with synthetic peptide libraries have indicated that stop codons do not always prevent display [14]. The odds are improved to one in six if C-terminal display vectors [15–20] are used, since clones do not need to end in frame. However, while such C-terminal intracellular vectors increase the likelihood that open reading frames will be displayed, they do not themselves provide any selective pressure for open reading frames. We recently described a vector in which DNA fragments are cloned upstream of β-lactamase, flanked by Lox sites [21]. The in-frame fusion with β-lactamase selects open reading frames directly, as expected, and, after removal of the β-lactamase by Cre recombination, the selected open reading frames are displayed directly. Surprisingly, preliminary experiments showed that there was a strong selective pressure for open reading frames derived from real genes, as opposed to ones from random open reading frames. This was thought to be due to the possibility that polypeptides derived from random open reading frames were less likely to be able to fold, and so would cause misfolding of the attached β-lactamase, so precluding antibiotic resistance. In the present paper, we explore the use of this vector to identify the epitopes that are recognized by three new mAbs that recognize h-TG2 (human transglutaminase 2).
TGases (transglutaminases) are a widely distributed group of enzymes that catalyse the post-translational modification of proteins by the formation of isopeptide bonds or by deamidation of proteins (for a review, see [22]). The formation of isopeptide bonds occurs either through protein cross-linking via ε-(γ-glutamyl)-lysine bonds or through the incorporation of primary amines at selected peptide bond glutamine residues, whereas the deamidation occurs by conversion of certain glutamine residues into glutamic acid. The cross-linked products, often of high molecular mass, are highly resistant to mechanical challenge and proteolytic degradation, and are commonly found in a number of tissues and processes where such properties are important, e.g. skin, hair, blood clotting and wound healing. Exogenous proteins, such as ingested cereal gluten, can also be converted into biologically more active forms by the action of TGases either via deamidation or by their cross-linking to other cellular proteins [23]. TGases are suspected to be involved in many pathologies [24], including neurodegenerative diseases, tissue scarring and autoimmune diseases, such as coeliac disease [25]. Despite the interest in the biological properties of TGases, few well-characterized antibodies are available for experimental studies. Three new mAbs that recognize h-TG2 are described in the present paper, and the epitopes they recognize were identified using filtered fragment phage display [21].
EXPERIMENTAL
Bacterial strains and enzymes
The bacterial strains used in this study were Escherichia coli DH5αF' (Gibco BRL), F'/endA1 hsd17 (rK− mK+) supE44 thi-1 recA1 gyrA (Nalr) relA1Δ (lacZYA-argF)U169 deoR (F80dlacD-(lacZ)M15), and BS1365, BS591 F' Kan [BS591: recA1 endA1 gyrA96 thi-1 D lacU169 supE44 hsdR17 (lambda1mm434 nin5 X1-cre)]. Molecular biology enzymes were purchased from New England Biolabs, Promega or Life Technologies.
TGases
h-TG2 was produced as described previously [26]. m-TG2 (mouse TG2) gene was obtained by amplifying cDNA from an intestinal specimen with specific primers (the restriction sites are underlined): Mttg-sense 5′-AGCTCGCTGCAGGTATGGCAGAGGAGCTGCTCCTG-3′ and Mttg-antisense 5′-GAAGCGAATTCTTAGGCCGGGCCGATGATAAC-3′, and cloned as a PstI/EcoRI fragment into pTrcHisB. Protein purification was performed as described in [27]. gp-TG2 (guinea-pig TG2) was purchased from Sigma.
mAb production and purification
Four 8-week-old BALB/c mice were intraperitoneally immunized with 50 μg of h-TG2, kindly provided by Professor T. Mothes (University Hospital, Leipzig, Germany), dissolved in PBS and mixed with complete Freund's adjuvant. Mice were later inoculated every 3 weeks for 3 months with 50 μg of h-TG2 mixed with incomplete Freund's adjuvant. Serum samples were collected, and reactivity was analysed by indirect immunofluorescence on monkey oesophagus sections. Staining revealed a similar immunofluorescence pattern to the characteristic coeliac EMA (endomysial antibody)-positive one. At 7 days after the last boost, serum samples were collected and analysed by indirect ELISA using Ca2+-activated h-TG2-coated plates (10 μg/ml, 50 μl/well) as described in [28]. The mouse having the highest anti-TG2 antibodies titre was chosen for the fusion protocol. mAbs were derived by somatic cell hybridization of NSO myeloma cells to spleen cells from the selected mouse as described by Kohler and Milstein [29] with minor modifications. Screening of cell culture supernatants was performed by indirect ELISA using plates coated with the Ca2+-activated TG2 (10 μg/ml, 50 μl/well). Positive cell cultures were expanded and cloned by limit dilution. According to their reactivity, three anti-TG2 mAbs were selected and called 4E1, 2G3 and 5G7. Cell culture supernatant was collected, and the pH of the crude antibody preparation was adjusted by adding 1/10 volume of 1.0 M Tris/HCl, pH 8.0. The antibody solution was passed through a pre-washed Protein G bead column, and the beads were washed with 10 column vol. of 100 mM Tris/HCl, pH 8.0, and 10 column vol. of 10 mM Tris/HCl, pH 8.0. The column was then eluted with 50 mM glycine, pH 3.0. The eluate was collected in tubes containing 1/10 volume of 1 M Tris/HCl, pH 8.0, and each tube was immediately mixed gently to bring the pH back to neutral. Protein concentration was measured using the Bradford reagent (Sigma).
Phage display library
Phage display of h-TG2 fragments was obtained using the method described in [21], with some differences in the cloning procedures. h-TG2 PCR-amplified DNA (10 μg) were sheared mechanically into fragments ranging in size 150–600 bp by sonication (1 min pulses at 100% power output). Fragments of 100–300 bp were separated by electrophoresis in a 2% agarose gel, recovered from the gel using the Qiaquick Gel Extraction kit (Qiagen), and repaired and blunt-ended by the addition of 1 unit/μg of T4 DNA polymerase in the presence of 100 μM dNTPs. The fragments were mixed with pPAO2 vector cut with StuI at a molar ratio of 8:1 and ligated with T4 ligase (New England Biolabs). The ligation mixture was extracted with an equal volume of 50:50 (v/v) phenol/chloroform, followed by ethanol precipitation, and the resulting DNA pellet was resuspended in water. A ligation reaction was then used to transform 50 μl of electrocompetent DH5αF' cells. To induce the removal of the β-lactamase gene following selection on ampicillin plates, phagemids were prepared from pooled selected clones, as described previously [30], and infected into BS1365 (bacteria constitutively expressing Cre) grown in 2xTY, 50 μg/ml kanamycin, 1% glucose at 37 °C to an A600 of 0.5. After adding chloramphenicol (25 μg/ml), the recombination was allowed to proceed by shaking overnight at 30 °C. The following day, bacteria were diluted 1/20 in the same medium, grown to an A600 of 0.5 at 37 °C, and infected with M13K07 helper phage at a multiplicity of infection of 20:1, before overnight growth at 30 °C. Colonies were derived from these phagemids by infection into DH5αF'. These represent pre-selected open reading frames, displayed on phagemid, in which β-lactamase has been removed. To characterize the transformants, randomly picked recombinants were analysed for inserts by PCR with primers VLPT2 5′-TACCTATTGCCTACGGCAGCCGCTGGATTGTTATTACTC-3′ and VHPT2 5′-TGGTGATGGTGAGTACTATCCAGGCCCAGCAGTGGGTTTG-3′ annealing external to the cloning site. PCR products were analysed by agarose gel electrophoresis.
pPAO2/TG2 library selection
The pPAO2/TG2 library was selected using three mAbs against TG2 and CD 2.8, an scFv (single-chain variable fragment) antibody originally isolated from a phage display library [31]. The human scFv against TG2 was used as a minibody by adding mouse CH2 and CH3 Fc antibody domains to the scFv, cloning into the vector pCDNA3 (Invitrogen) and expressing in a HEK-293 (human embryonic kidney) cell culture (D. Sblattero, F. Ziller, R. Di Niro, F. Florian, S. Crovella, M. Stebel, M. Bestagno, O. Burrone, A. R. M. Bradbury and R. Marzari, unpublished work). Rescue of phagemid particles was as described by Marks et al. [32]. Panning was performed by adding phages diluted in MPBS (2% non-fat milk in PBS) to immunotubes (Nunc) coated with purified antibodies (10 μg/ml), washing 20 times with PBST (PBS with 0.1% Tween 20) and 20 times with PBS, followed by elution with 1 ml of DH5αF' E. coli cells at a A600 of 0.5 for 30 min, and plating and overnight growth on chloramphenicol plates at 30 °C. The panning procedure was repeated one or two times. After selection, 96 individual clones from each selection were screened for reactivity to their respective by microtitre plate ELISA.
Periplasmic extraction
Clones, each carrying a different TG2 fragment, were grown in 96-well plates until reaching a A600 of 0.5, and the protein expression was induced with 0.3 mM IPTG (isopropyl β-D-thiogalactoside) overnight at 30 °C. The plate was centrifuged at 500 g for 15 min, the supernatant was discarded, and the bacterial pellet was resuspended in 1/10 vol. of PPB buffer (200 mg/ml sucrose, 1 mM EDTA and 30 mM Tris/HCl) for 45 min at 4 °C to provide an osmotic shock. After centrifugation, periplasmic extracts containing TG2 fragments were recovered in the supernatant.
ELISA
ELISA was performed by coating ELISA plates with purified recombinant h-TG2, recombinant m-TG2 or gp-TG2 at 10 μg/ml, or with periplasmic extracts (from selected clones) diluted 1:10 in PBS for 15 h at 4 °C. Wells were blocked with MPBS and washed three times with PBST and three times with PBS. Purified mAbs were used as primary antibodies diluted 1:5000 with MPBS. CUB7402, a commercial anti-TG2 antibody (Neomarker, Fremont, CA, U.S.A.), was used as control. Secondary antibodies were goat anti-mouse conjugated with alkaline peroxidase (Dako) diluted 1:2000 in MPBS. The immunocomplexes were revealed with TMB (3,3′,5,5′-tetramethylbenzidine) (Pierce) and read at A450.
Western blotting
SDS/PAGE was performed according to standard techniques. h-TG2, m-TG2 and gp-TG2 were separated by SDS/PAGE and transferred on to nitrocellulose (Amersham Biosciences) by semi-dry blotting using the Multiphor II (Amersham Biosciences). The membrane was blocked using MPBS for 1 h at room temperature (20 °C). Purified mAb was used as a primary antibody. After 1 h of incubation at room temperature and extensive washing with PBST, the nitrocellulose was subsequently incubated with anti-mouse goat antibodies conjugated with alkaline phosphatase (Dako) and revealed by the chromogenic substrate BCIP (5-bromo-4-chloroindol-3-yl phosphate) and NBT (Nitro Blue Tetrazolium).
RESULTS
mAbs and binding properties
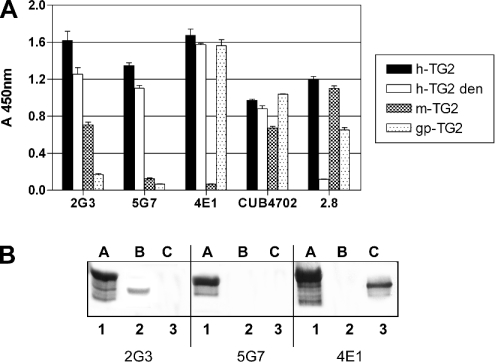
Spleen cells of mice immunized with human TG2 generated 207 hybridoma cells according to the visual inspection of macroscopic colonies in five 96-well plates. A total of 17 of the clone supernatants contained anti-TG2 antibodies, as assessed by ELISA on immobilized human TG2. Clones 2G3, 5G7 and 4E1, which produced the most reactive supernatants, were subcloned and thus selected for expansion. A preliminary characterization was attempted by immunorecognition using ELISA of recombinant h-TG2 in native or denatured form, recombinant m-TG2, as well as commercial gp-TG2. Commercial mAb CUB 4702 and one of the cloned antibodies isolated, in a previous study, from a phage display library (CD 2.8) obtained from a coeliac disease patient [31] were included as controls. The results are reported in Figure 1(A): all three mAbs were able to recognize denatured h-TG2; only 2G3 cross-reacted with m-TG2, whereas 4E1 recognized gp-TG2 but not m-TG2. These cross-reactivities are likely to be due to the high degree of similarity (>80%) between the rodent and human TG2s. mAb CUB 4702 and CD 2.8 showed the expected reactivity. The reactivity to h-TG2, m-TG2 and gp-TG2 separated by SDS/PAGE was also assessed (Figure 1B). The results confirm the recognition of the denatured h-TG2 protein and the specificity of recognition with homologous proteins from other species, with only 2G3 recognizing m-TG2 and 4E1 recognizing gp-TG2.
Figure 1. ELISA of three mAbs against TG2.
(A) Antigens: recombinant h-TG2 (native or denatured, den), recombinant m-TG2 and gp-TG2. Primary antibodies: mouse mAbs 2G3, 5G7 and 4E1 (each used at 1:5000 dilution). Controls: cloned human minibody 2.8 to TG2 (1:10 dilution) and mAb CUB 4702 (1:2000 dilution). Secondary antibodies: goat anti-mouse Ig conjugated with alkaline peroxidase used at 1:2000 dilution. The results are representative of three independent experiments. (B) Western blots of different species TG2. Antigens: A, h-TG2; B, m-TG2; C, gp-TG2. Primary antibodies: lanes 1–3, mAb 2G3; lanes 4–6, mAb 5G7; lanes 7–9, mAb 4E1. Secondary antibody: goat anti-mouse IgG conjugated with alkaline phosphatase.
Construction of TG2 phage display library
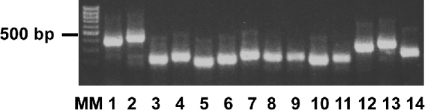
In order to analyse the amino acid sequences recognized by the anti-TG2 mAbs, we constructed a phage display library of random fragments of the TG2 gene using the newly developed phagemid vector pPAO2. This vector was originally designed to filter out functional open reading frames from DNA and display them on filamentous phages in such a way that they are amenable to subsequent selection or screening. A simpler blunt-end cloning procedure was used, rather than that based on ligation-independent cloning, in which blunt-ended TG2 fragments were cloned into the linearized pPAO2 cut at the blunt site. This method was preferred because of the low complexity of the analysed DNA. In the pPAO2 vector, the cloning site is located upstream of a β-lactamase gene flanked by two Lox recombination signals. The polylinker is out of frame with respect to the β-lactamase gene and can only be returned into frame if DNA containing an open reading frame is correctly cloned. Following insertion of the TG2 fragments and DH5αF' E. coli transformation, the bacterial cells were selected on plates containing 12 μg/ml ampicillin to ensure the selection of the in-frame sequences. A small aliquot of transformed cells was plated on chloramphenicol, and the ratio of the transformants on ampicillin/chloramphenicol was calculated. The result was 1:20, close to the theoretical 1:18 described above. The β-lactamase gene was removed by infecting phagemid made from these colonies into BS1365 (which expresses Cre recombinase), and re-infecting phagemid produced by these bacteria into DH5αF' cells. In order to examine the characteristics of the resultant library, 14 randomly picked bacterial colonies were analysed for the presence of inserts by PCR. The results are reported in Figure 2 and show that all the inserts were of different sizes, indicating that no single clone dominates, and range in length from 200 to 450 bp. A total of 94 random colonies were induced by IPTG and examined by dot blot, using a mAb recognizing the SV5 tag located downstream of the cloned sequence [21]. A total of 88 clones out of 94 (93.6%) were positive, indicating that almost all of the selected clones had the inserts in-frame (results not shown).
Figure 2. Electrophoresis of PCR-amplified insert DNA of randomly selected bacterial clones.
After transformation of TG2 fragment library, 14 clones were randomly selected, and the colony was picked and used directly as template for PCR amplification. Each clone expressing a different cloned TG2 fragment was amplified with VLPT2 and VHPT2 primers, and PCR products were run on a 2% agarose gel. MM, molecular mass (size in bp).
Selecting and screening using the pPAO/TG2 library
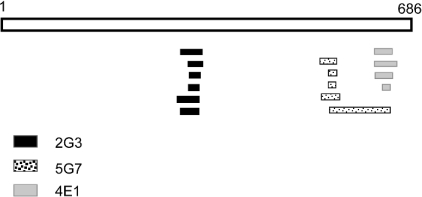
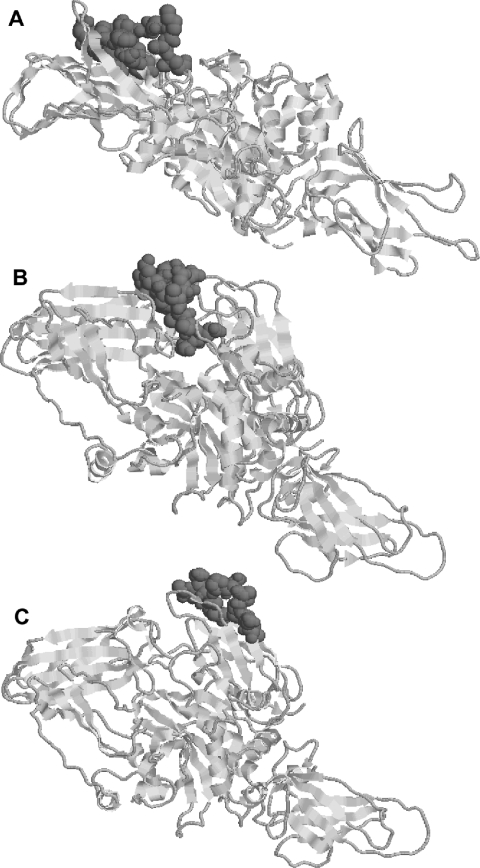
In order to determine the epitopes recognized by the three mAbs against TG2, the pPAO2/TG2 phage library was individually selected on the three antibody proteins. In addition, the minibody CD 2.8, which was previously isolated from a coeliac disease patient [31], was included as control. Purified antibody proteins were adsorbed on to the surface of immunotubes, the phage particles were added, and a single round of selection was performed. Periplasmic extracts from 48 individual clones of the eluted phages for each antibody were used as coating protein and analysed by ELISA with the respective mAb. Several different positive clones were identified, six for 2G3, five for 5G7 and four for 4E1. Many clones were represented with more than one copy in the selected population. Interestingly, no positive phages were obtained using the human CD 2.8, which was previously reported to recognize a conformational epitope [27]. All the selected clones underwent DNA sequencing and, after individual alignment, were shown to overlap each other, identifying a minimal region which can be considered to be the putative recognized epitope. The results are depicted in Figure 3 and summarized in Table 1. According to these results, 2G3 recognizes the sequence E314YFRNEFGEIQGDKSE329, 5G7 recognizes the sequence Y548EKYRDCLTES558, and 4E1 recognizes the sequence E637IPDPVEAGEEV648, although we cannot exclude that the real epitopes are restricted to even shorter sequences. In Figure 4, the position of the three epitopes on the TG2 molecular model is shown. As expected, the identified epitopes were all located on the surface of the TG2 protein. A comparison of the identified epitopes in the different species with their reactivity for the individual mAbs (Table 2) confirms the data obtained by ELISA and Western blot analysis. In fact, several differences with respect to the human sequence could be identified in each epitope for the different species. It is possible to identify specific conserved regions of each epitope likely to be important in recognition, as well as distributed mutations which probably inhibit binding.
Figure 3. Schematic representation of the TGase protein and the three mAbs against TG2-positive selected clones from the phage library.
TGase is a 686-amino-acid-long protein and is represented as open bar. Six selected positive clones for mAb 2G3 are represented as black bars, and the minimal epitope is located at amino acids 314–329; five clones for mAb 5G7 are represented as dotted bars, and the epitope is restricted to amino acids 548–558; four clones for mAb 4E1 are represented as grey bars, and the epitope is restricted to amino acids 637–648.
Table 1. Summary of minimal epitopes recognized by mAbs against TG2.
After individual aligment of positive clones for each identified sequence the beginning, the end and total length is indicated for both DNA and amino acid sequence.
| mAb | First base | Last base | Length (bases) | First amino acid | Last amino acid | Length (amino acids) |
|---|---|---|---|---|---|---|
| 2G3 | 939 | 988 | 50 | 314 | 329 | 16 |
| 5G7 | 1641 | 1675 | 35 | 548 | 558 | 11 |
| 4E1 | 1908 | 1945 | 38 | 637 | 648 | 12 |
Figure 4. Molecular model of TG2 in which the amino acids of the epitopes recognized by the three mAbs are highlighted.
(A) mAb 2G3. (B) mAb 5G7. (C) mAb 4E1. The representation of the structure was made using RasTop 2.1 software.
Table 2. Comparison between the amino acid sequences of recognized epitopes.
The sequences of human, mouse and guinea pig protein are shown. Amino acid differences from the human epitope are indicated in bold and underlined. For each species, ELISA and Western blot positive recognition by the monoclonal is indicated.
| 2G3 | Recognition | 5G7 | Recognition | 4E1 | Recognition | |
|---|---|---|---|---|---|---|
| Human | EYFRNEFGEIQGDKSE | + | YEKYRDCLTES | + | EIPDPVEAGEEV | + |
| Mouse | EYFRNEFGELESNKSE | + | YEKYSGCLTES | − | EVSDPVPAGDLV | − |
| Guinea-pig | EYFRNESGEIEGNKSE | − | YEKYGDYLTES | − | EVPDPVEAGEQA | + |
DISCUSSION
Phage display peptide libraries are a commonly used method to determine the epitopes recognized by mAbs with the mAb fixed to a solid phase and interacted with either peptide [6] or gene-specific libraries [7]. After a number of cycles of binding, the sequence of selected peptides is determined by sequencing the DNA of the bound phages. Usually, one or more different motifs are obtained, and such motifs may be similar to a sequence found within the antigen. If the identity of the antigen is unknown, screening the sequences of selected peptides against protein databases may allow the identification of the protein recognized, although the degeneracy of the motif usually obtained can make this difficult, and experimental confirmation is always required. When the motif has no homologue within the antigen, it is termed a ‘mimotope’, and is thought to mimic a conformational or discontinuous epitope.
The use of gene fragment libraries, rather than peptide libraries, has generally been more successful in the identification of antibody epitopes [12,13]. In order to increase the chance that open reading frames are displayed, gene fragments have been displayed at the C-terminus of a Jun peptide which interacts with Fos displayed at the N-terminus of p3 [16], at the C-terminus of the phage coat proteins p3 [17], p8 [18] and p6 [19]. However, none of these vectors actually select open reading frames, but just increase the chance that open reading frames are displayed.
In order to overcome this problem, we recently described a vector, pPAO2, in which resistance to ampicillin was initially used to select open reading frames [21], with subsequent removal of the β-lactamase gene by recombination, allowing full display of the selected fragments. According to the results reported here, epitope mapping based on pPAO2 allowed the identification of epitopes as small as 11 amino acids, as shown for 5G7. Interestingly, all the identified epitopes correspond to structural loops exposed on the surface of the protein. This is not surprising, since the hybridoma clones were screened against the whole antigen, so ignoring all those antibodies directed to internal portions of the molecule. Notwithstanding the success of this approach to identify these three epitopes, this method was unable to identify the conformational or discontinuous epitope recognized by the minibody CD 2.8, although it is possible that the fragments used were too small to re-create the conformational epitope.
As far as ELISA recognition of the cloned TG2s is concerned, 2G3 cross-reacted with m-TG2, while 4E1 reacted with gp-TG2 and 5G7 did not recognize either of the two rodent TG2s. These different recognition patterns account for the differences between the antibodies and, from a practical point of view, may be useful for experimental purposes in animal models. Furthermore, these recognition results confirm and augment the epitope identification found using this new phage display method. For 2G3 and 4E1, a contiguous portion of the epitope which appears to be required for recognition can be identified (2G3, the first ten amino acids; 4E1, the central eight amino acids), while for 5G7, mouse and guinea-pig transglutaminase contain central differences which inhibit binding. Similar results were obtained in Western blotting, with the additional finding that all three mAbs were able to interact with the appropriate denatured TG2, almost certainly as a result of the recognition of linear epitopes, as outlined by the epitope mapping experiments.
These experiments provide excellent validation of the use of this filtering display system in the identification of antibody epitopes, and it is likely that it can also be used in the study of other protein–protein interactions, both to identify interaction partners, as well as the identification of the specific epitopes recognized, as described in the present paper. By using antibiotic selection pressure to ensure the display of open reading frames, the sizes of the libraries required are smaller, and there is less likelihood that selections are overtaken by faster growing non-displaying clones, rendering the procedure more efficient.
References
- 1.Parmley S. F., Smith G. P. Antibody-selectable filamentous fd phage vectors: affinity purification of target genes. Gene. 1988;73:305–318. doi: 10.1016/0378-1119(88)90495-7. [DOI] [PubMed] [Google Scholar]
- 2.Parmley S. F., Smith G. P. Filamentous fusion phage cloning vectors for the study of epitopes and design of vaccines. Adv. Exp. Med. Biol. 1989;251:215–218. doi: 10.1007/978-1-4757-2046-4_21. [DOI] [PubMed] [Google Scholar]
- 3.Cwirla S. E., Peters E. A., Barrett R. W., Dower W. J. Peptides on phage: a vast library of peptides for identifying ligands. Proc. Natl. Acad. Sci. U.S.A. 1990;87:6378–6382. doi: 10.1073/pnas.87.16.6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yayon A., Aviezer D., Safran M., Gross J. L., Heldman Y., Cabilly S., Givol D., Katchalski-Katzir E. Isolation of peptides that inhibit binding of basic fibroblast growth factor to its receptor from a random phage-epitope library. Proc. Natl. Acad. Sci. U.S.A. 1993;90:10643–10647. doi: 10.1073/pnas.90.22.10643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balass M., Heldman Y., Cabilly S., Givol D., Katchalski-Katzir E., Fuchs S. Identification of a hexapeptide that mimics a conformation-dependent binding site of acetylcholine receptor by use of a phage-epitope library. Proc. Natl. Acad. Sci. U.S.A. 1993;90:10638–10642. doi: 10.1073/pnas.90.22.10638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith G. P., Scott J. K. Libraries of peptides and proteins displayed on filamentous phage. Methods Enzymol. 1993;217:228–257. doi: 10.1016/0076-6879(93)17065-d. [DOI] [PubMed] [Google Scholar]
- 7.Bluthner M., Bautz E. K., Bautz F. A. Mapping of epitopes recognized by PM/Scl autoantibodies with gene-fragment phage display libraries. J. Immunol. Methods. 1996;198:187–198. doi: 10.1016/s0022-1759(96)00160-3. [DOI] [PubMed] [Google Scholar]
- 8.Bluthner M., Schafer C., Schneider C., Bautz F. A. Identification of major linear epitopes on the sp100 nuclear PBC autoantigen by the gene-fragment phage-display technology. Autoimmunity. 1999;29:33–42. doi: 10.3109/08916939908995970. [DOI] [PubMed] [Google Scholar]
- 9.Du Plessis D. H., Romito M., Jordaan F. Identification of an antigenic peptide specific for bluetongue virus using phage display expression of NS1 sequences. Immunotechnology. 1995;1:221–230. doi: 10.1016/1380-2933(95)00026-7. [DOI] [PubMed] [Google Scholar]
- 10.Petersen G., Song D., Hugle-Dorr B., Oldenburg I., Bautz E. K. Mapping of linear epitopes recognized by monoclonal antibodies with gene-fragment phage display libraries. Mol. Gen. Genet. 1995;249:425–431. doi: 10.1007/BF00287104. [DOI] [PubMed] [Google Scholar]
- 11.Wang L. F., Du Plessis D. H., White J. R., Hyatt A. D., Eaton B. T. Use of a gene-targeted phage display random epitope library to map an antigenic determinant on the bluetongue virus outer capsid protein VP5. J. Immunol. Methods. 1995;178:1–12. doi: 10.1016/0022-1759(94)00235-o. [DOI] [PubMed] [Google Scholar]
- 12.Fack F., Hugle-Dorr B., Song D., Queitsch I., Petersen G., Bautz E. K. Epitope mapping by phage display: random versus gene-fragment libraries. J. Immunol. Methods. 1997;206:43–52. doi: 10.1016/s0022-1759(97)00083-5. [DOI] [PubMed] [Google Scholar]
- 13.Matthews L. J., Davis R., Smith G. P. Immunogenically fit subunit vaccine components via epitope discovery from natural peptide libraries. J. Immunol. 2002;169:837–846. doi: 10.4049/jimmunol.169.2.837. [DOI] [PubMed] [Google Scholar]
- 14.Carcamo J., Ravera M. W., Brissette R., Dedova O., Beasley J. R., Alam-Moghe A., Wan C., Blume A., Mandecki W. Unexpected frameshifts from gene to expressed protein in a phage-displayed peptide library. Proc. Natl. Acad. Sci. U.S.A. 1998;95:11146–11151. doi: 10.1073/pnas.95.19.11146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crameri R., Jaussi R., Menz G., Blaser K. Display of expression products of cDNA libraries on phage surfaces: a versatile screening system for selective isolation of genes by specific gene-product/ligand interaction. Eur. J. Biochem. 1994;226:53–58. doi: 10.1111/j.1432-1033.1994.tb20025.x. [DOI] [PubMed] [Google Scholar]
- 16.Crameri R., Blaser K. Cloning Aspergillus fumigatus allergens by the pJuFo filamentous phage display system. Int. Arch. Allergy Immunol. 1996;110:41–45. doi: 10.1159/000237308. [DOI] [PubMed] [Google Scholar]
- 17.Fuh G., Sidhu S. S. Efficient phage display of polypeptides fused to the carboxy-terminus of the M13 gene-3 minor coat protein. FEBS Lett. 2000;480:231–234. doi: 10.1016/s0014-5793(00)01946-3. [DOI] [PubMed] [Google Scholar]
- 18.Fuh G., Pisabarro M. T., Li Y., Quan C., Lasky L. A., Sidhu S. S. Analysis of PDZ domain-ligand interactions using carboxyl-terminal phage display. J. Biol. Chem. 2000;275:21486–21491. doi: 10.1074/jbc.275.28.21486. [DOI] [PubMed] [Google Scholar]
- 19.Jespers L. S., Messens J. H., De Keyser A., Eeckhout D., Van den Brande I., Gansemans Y. G., Lauwereys M. J., Vlasuk G. P., Stanssens P. E. Surface expression and ligand-based selection of cDNAs fused to filamentous phage gene VI. Biotechnology (N.Y.) 1995;13:378–382. doi: 10.1038/nbt0495-378. [DOI] [PubMed] [Google Scholar]
- 20.Weiss G. A., Sidhu S. S. Design and evolution of artificial M13 coat proteins. J. Mol. Biol. 2000;300:213–219. doi: 10.1006/jmbi.2000.3845. [DOI] [PubMed] [Google Scholar]
- 21.Zacchi P., Sblattero D., Florian F., Marzari R., Bradbury A. R. Selecting open reading frames from DNA. Genome Res. 2003;13:980–990. doi: 10.1101/gr.861503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Griffin M., Casadio R., Bergamini C. M. Transglutaminases: Nature's biological glues. Biochem. J. 2002;368:377–396. doi: 10.1042/BJ20021234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fleckenstein B., Qiao S. W., Larsen M. R., Jung G., Roepstorff P., Sollid L. M. Molecular characterization of covalent complexes between tissue transglutaminase and gliadin peptides. J. Biol. Chem. 2004;279:17607–17616. doi: 10.1074/jbc.M310198200. [DOI] [PubMed] [Google Scholar]
- 24.Kim S. Y., Jeitner T. M., Steinert P. M. Transglutaminases in disease. Neurochem. Int. 2002;40:85–103. doi: 10.1016/s0197-0186(01)00064-x. [DOI] [PubMed] [Google Scholar]
- 25.Dieterich W., Ehnis T., Bauer M., Donner P., Volta U., Riecken E. O., Schuppan D. Identification of tissue transglutaminase as the autoantigen of celiac disease. Nat. Med. 1997;3:797–801. doi: 10.1038/nm0797-797. [DOI] [PubMed] [Google Scholar]
- 26.Sblattero D., Berti I., Trevisiol C., Marzari R., Tommasini A., Bradbury A., Fasano A., Ventura A., Not T. Human recombinant tissue transglutaminase ELISA: an innovative diagnostic assay for celiac disease. Am. J. Gastroenterol. 2000;95:1253–1257. doi: 10.1111/j.1572-0241.2000.02018.x. [DOI] [PubMed] [Google Scholar]
- 27.Sblattero D., Florian F., Azzoni E., Zyla T., Park M., Baldas V., Not T., Ventura A., Bradbury A., Marzari R. The analysis of the fine specificity of celiac disease antibodies using tissue transglutaminase fragments. Eur. J. Biochem. 2002;269:5175–5181. doi: 10.1046/j.1432-1033.2002.03215.x. [DOI] [PubMed] [Google Scholar]
- 28.Sulkanen S., Halttunen T., Laurila K., Kolho K. L., Korponay-Szabo I. R., Sarnesto A., Savilahti E., Collin P., Maki M. Tissue transglutaminase autoantibody enzyme-linked immunosorbent assay in detecting celiac disease. Gastroenterology. 1998;115:1322–1328. doi: 10.1016/s0016-5085(98)70008-3. [DOI] [PubMed] [Google Scholar]
- 29.Kohler G., Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature (London) 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- 30.Sblattero D., Bradbury A. Exploiting recombination in single bacteria to make large phage antibody libraries. Nat. Biotechnol. 2000;18:75–80. doi: 10.1038/71958. [DOI] [PubMed] [Google Scholar]
- 31.Marzari R., Sblattero D., Florian F., Tongiorgi E., Not T., Tommasini A., Ventura A., Bradbury A. Molecular dissection of the tissue transglutaminase autoantibody response in celiac disease. J. Immunol. 2001;166:4170–4176. doi: 10.4049/jimmunol.166.6.4170. [DOI] [PubMed] [Google Scholar]
- 32.Marks J. D., Hoogenboom H. R., Bonnert T. P., McCafferty J., Griffiths A. D., Winter G. By-passing immunization: human antibodies from V-gene libraries displayed on phage. J. Mol. Biol. 1991;222:581–597. doi: 10.1016/0022-2836(91)90498-u. [DOI] [PubMed] [Google Scholar]