Abstract
TGH (triacylglycerol hydrolase) catalyses the lipolysis of intracellular stored triacylglycerol. To explore the mechanisms that regulate TGH expression in adipose tissue, we studied the expression of TGH during the differentiation of 3T3-L1 adipocytes. TGH mRNA and protein levels increased dramatically in 3T3-L1 adipocytes compared with pre-adipocytes. Electrophoretic mobility shift assays demonstrated enhanced binding of nuclear proteins of adipocytes to the distal murine TGH promoter region (−542/−371 bp), yielding one adipocyte-specific migrating complex. Competitive and supershift assays demonstrated that the distal TGH promoter fragment bound C/EBPα (CCAAT/enhancer-binding protein α). Transient transfections of different mutant TGH promoter–luciferase constructs into 3T3-L1 adipocytes and competitive electromobility shift assays showed that the C/EBP-binding elements at positions −470/−459 bp and −404/−390 bp are important for transcriptional activation. Co-transfection with C/EBPα cDNA and TGH promoter constructs in 3T3-L1 pre-adipocytes demonstrated that C/EBPα increased TGH promoter activity. Ectopic expression of C/EBPα in NIH 3T3 cells activated TGH mRNA expression without causing differentiation into adipocytes. These experiments directly link increased TGH expression in adipocytes to transcriptional regulation by C/EBPα. This is the first evidence that C/EBPα participates directly in the regulation of an enzyme associated with lipolysis.
Keywords: 3T3-L1 adipocyte, CCAAT/enhancer-binding protein α (C/EBPα), lipolysis, NIH 3T3 cell, transient transfection, triacylglycerol hydrolase
Abbreviations: ADD, adipocyte determination- and differentiation-dependent factor; aP2, adipocyte lipid-binding protein; C/EBP, CCAAT/enhancer-binding protein; CREB, cAMP response element-binding protein; DMEM, Dulbecco's modified Eagle's medium; EMSA, electrophoretic mobility shift assay; HSL, hormone-sensitive lipase; LXR, liver X receptor; PPAR, peroxisome-proliferator-activated receptor; RT-PCR, reverse transcription–PCR; RXR, retinoid X receptor; RXRE, RXR response element; SREBP, sterol response element-binding protein; Stat 5, signal transducers and activators of transcription 5; TG, triacylglycerol; TGH, triacylglycerol hydrolase
INTRODUCTION
Fatty acids stored within the TGs (triacylglycerols) of adipose cells constitute the vast majority of energy reserves in animals. While the TG pool in adipocytes provides a reservoir of energy for times of restricted caloric intake, there is also a normal flux of fatty acids into and out of adipose tissue in response to meals and other factors, such as insulin [1]. Products of fatty acid metabolism can function as ligands for nuclear receptors, thereby controlling the transcription of genes involved in adipogenesis and lipid metabolism [2]. Thus the regulatory mechanisms underlying release of non-esterified (free) fatty acids from TGs stored in adipocytes are important for understanding the pathophysiology of various metabolic disorders, such as diabetes, obesity, hyperlipidaemia, hypertension and atherosclerosis.
HSL (hormone-sensitive lipase) is a neutral lipid lipase that shares only minimal overall sequence similarity with other known neutral lipases [3]. Overexpression of HSL prevents TG accumulation in adipocytes, and HSL is commonly considered to be the rate-limiting enzyme of lipolysis in adipocytes [4]. The lipolytic activities of HSL are under acute neuronal and hormonal control. Catecholamines and other lipolytic hormones stimulate these activities through the reversible phosphorylation of a serine residue by cAMP-dependent protein kinase. Conversely, insulin suppresses HSL activity by preventing phosphorylation. Therefore the activation of HSL in adipose tissues is thought to account for the elevation of plasma levels of non-esterified fatty acids in various conditions, such as starvation or diabetes mellitus [4,5]. However, the exclusivity of HSL being the only lipase responsible for TG hydrolysis in adipose tissue has been challenged by the generation of HSL null mice [5–7]. These mice did not develop obesity, and their white adipose tissue retained approx. 40% of TG lipolytic activity [6]. Thus HSL is not the only lipase hydrolysing TGs in adipose tissue.
Besides HSL, another intracellular neutral TG lipase identified within the murine adipocyte is the microsomal enzyme TGH (TG hydrolase), which has been demonstrated to hydrolyse stored hepatic TGs [8–11]. By functional proteomics analysis, it was shown that a major fraction of in vitro lipolytic activity in white adipose tissue is attributable to TGH [11]. In addition, inhibition of TGH reduced basal fatty acid release by 3T3-L1 adipocytes and circulating non-esterified fatty acid levels in hamsters by approx. 40% (W. Gao and R. Lehner, unpublished work). All of the above data suggest that TGH may play a significant role in the mobilization of adipocyte TG droplets.
Previous experiments showed that the expression of TGH was induced during 3T3-L1 adipocyte differentiation [12]. However, the mechanisms regulating its expression in this process are still unclear. In the present studies, we explored the transcriptional regulation of TGH expression during differentiation of 3T3-L1 adipocytes. CEBPα (CCAAT/enhancer-binding protein α), an adipogenic transcription factor, was shown to modulate the expression of TGH through enhanced transcription.
MATERIALS AND METHODS
Materials
Dexamethasone, 3-isobutyl-1-methylxanthine, bovine insulin, 4-methylumbelliferyl heptanoate and sodium taurodeoxycholate were purchased from Sigma (St. Louis, MO, U.S.A.). The luciferase reporter vector pGL3-Basic, that contains the cDNA encoding Photinus pyralis luciferase, the control pRL-CMV vector, that contains the cDNA encoding Renilla reniformis luciferase, the pSV-β-galactosidase vector, the dual luciferase reporter assay system and the β-galactosidase assay system were obtained from Promega (Madison, WI, U.S.A.). Lipofectamine™ Plus reagent, DMEM (Dulbecco's modified Eagle's medium), fetal bovine serum and pCDNA3.1(+) vector were from Invitrogen. Anti-C/EBPα goat, anti-C/EBPβ rabbit and anti-C/EBPδ rabbit antibodies were purchased from Santa Cruz Biotechnology. Anti-(protein disulphide isomerase) rabbit polyclonal antibody was from Stressgen Biotechnologies (Victoria, B.C., Canada). Horseradish peroxidase-labelled goat anti-(rabbit IgG) and horse-radish peroxidase-labelled rabbit anti-(goat IgG) were from Pierce. ECL® immunoblotting reagents were purchased from Amersham Biosciences. All other reagents were of analytical grade or higher.
Cell culture and differentiation of NIH 3T3-L1 fibroblasts
NIH 3T3 fibroblasts, Cos-7 African green monkey kidney cells and NIH 3T3-L1 fibroblasts (A.T.C.C.) were cultured in DMEM supplemented with penicillin G (100 units/ml), streptomycin (100 units/ml) and 10% (v/v) fetal bovine serum in a 5% CO2 humidified incubator at 37 °C. To induce cell differentiation, NIH 3T3-L1 fibroblasts were cultivated in growth media until confluent. At 2 days after reaching confluency, cells were induced to differentiate by the addition of 0.25 μM dexamethasone, 0.5 mM 3-isobutyl-1-methylxanthine and 3 μg/ml insulin for 3 days. Cells were then washed and incubated in normal growth media until differentiated. Media were changed every 2 days.
Immunoblot analyses
Portions of cell homogenates (20 μg) from NIH 3T3-L1 cells during differentiation or NIH 3T3 cells after transfection were heated for 5 min at 95 °C in 62.5 mM Tris/HCl, pH 8.3, 10% (v/v) glycerol, 5% (v/v) 2-mercaptoethanol, 1% SDS and 0.004% Bromophenol Blue. The protein samples were electrophoresed on an SDS/10%-polyacrylamide gel in 25 mM Tris/HCl, pH 8.3, 192 mM glycine and 0.1% SDS. The proteins were transferred to nitrocellulose by electroblotting in transfer buffer [25 mM Tris/HCl, pH 8.3, 192 mM glycine, 20% (v/v) methanol]. Following transfer, the membrane was incubated overnight at 4 °C with 5% (w/v) skimmed milk in 20 mM Tris/HCl, pH 7.4, 150 mM NaCl and 0.1% Tween 20, and incubated for 1 h with antibody raised against the specified protein. Depending on the primary antibodies, the secondary antibodies used were goat anti-(rabbit IgG) or rabbit anti-(goat IgG), and were detected using the ECL™ system according to the manufacturer's instructions. Where indicated, primary and secondary antibodies were stripped from the membrane by incubation with 100 mM 2-mercaptoethanol/2% SDS/62.5 mM Tris/HCl (pH 6.7) for 30 min at 50 °C, and the blot was re-probed with another antibody. The primary antibodies used were affinity purified anti-TGH polyclonal antibody anti-(protein disulphide isomerase) polyclonal antibody, anti-C/EBPα polyclonal antibody and anti-C/EBPβ polyclonal antibody.
RNA isolation and RT-PCR (reverse transcription–PCR) analysis
Total RNA was isolated from NIH 3T3-L1 cells during differentiation, or NIH 3T3 cells after transfection, using Trizol® reagent (Life Technologies, Inc.) according to the manufacturer's instructions. First-strand cDNA synthesis from 2 μg of total RNA was performed using SuperScript™ II reverse transcriptase (Invitrogen) primed by oligo(dT)12–18 primers. PCR was performed using the TGH primers 5′-CACTGCTGCTCTGATTACAAC-3′ (Ex6F) and 5′-GCCTTCAGCGAGTGGATAGC-3′ (Ex10R). Primers for the aP2 (adipocyte lipid-binding protein) gene were 5′-GAACCTGGAAGCTTGTCTTCG-3′ and 5′-ACCAGCTTGTCACCATCTCG-3′. Primers for cyclophilin were 5′-TCCAAAGACAGCAGAAAACTTTCG-3′ and 5′-TCTTCTTGCTGGTCTTGCCATTCC-3′. The PCR programme included denaturation at 94 °C for 30 s, annealing at 59 °C for 30 s, and elongation at 72 °C for 40 s (25 cycles). The PCR product was separated by electrophoresis on 1% (w/v) agarose gels and visualized by its exposure to autoradiographic film.
Plasmid constructions
pCR (−542/+112) was obtained as described [13]. The promoter region was excised from pCR 2.1 TOPO by restriction digestion with EcoRV and XbaI. The insert was purified from the agarose gel using the Qiaex II gel extraction kit (Qiagen Inc., Mississauga, Ontario, Canada) according to the manufacturer's instructions, and directionally ligated into the luciferase reporter vector pGL3-Basic, that had been subjected to restriction digestion with BamHI and SmaI, and gel purified to generate −542 Luc. The fragment (−370/+112) was generated by standard PCR methods using −542 Luc as template and GLprimer 2 (Promega) as the reverse primer, with the forward primer CGCGGTACCTGAGCAGAGCTTTGTAGA. The PCR product was cloned into the T/A cloning site of pCR2.1 TOPO (Invitrogen) according to the manufacturer's instructions to create pCR (−370/+112). The promoter region was excised from pCR 2.1 TOPO by restriction digestion with EcoRV and XbaI. The insert was purified from agarose gel and directionally ligated into the luciferase reporter vector pGL3-Basic, that had been subjected to restriction digestion with BamHI and SmaI, and gel purified to generate −370 Luc. To prepare mutated promoter constructs, GTAA (−464/−461) was mutated to ATCC and named −542 M1, and GAAA (−397/−394) was changed to AACC and named −542 M2. These mutations were created from −542 Luc by PCR using QuikChange™ site-directed mutagenesis kits (Stratagene, La Jolla, CA, U.S.A.). The mutant complementary primers were used for making each mutant construct, and the resulting plasmids were used for transformation of JM109 cells. All deleted and mutant constructs were sequenced, and correct ones were selected for further experiments. Vectors enabling expression of recombinant C/EBPα proteins [pCDNA3.1(+)-C/EBPα] and C/EBPβ (pCDNA3-C/EBPβ) were obtained from Dr M. D. Lane (Department of Biological Chemistry, Johns Hopkins University School of Medicine, Baltimore, MD, U.S.A.) [14].
Nuclear extract preparation and EMSAs (electrophoretic mobility shift assays)
Total nuclear extracts of NIH 3T3-L1 cells during differentiation were prepared as described in [15]. Distal (−542/−371), medial (−370/−68) and proximal (−67/+112) promoter fragments were released from pCR (−542/+112) by restriction digestion with BlpI/NotI, HindIII/BlpI and HindIII respectively. Promoter fragments were purified from 2% (w/v) agarose gels and end-labelled using Klenow fragment in the presence of [α-32P]dCTP. Promoter-derived oligonucleotides were synthesized by the University of Alberta DNA Core Facility. Complementary oligonucleotides (100 μM of each) were heated at 90 °C for 5 min, then slowly cooled to room temperature, and 5 pmol of double-stranded oligonucleotide was 5′-end labelled using T4 kinase (Roche Diagnostics) and [γ-32P]ATP (PerkinElmer). The sequences of oligonucleotides used in EMSAs were as follows (coding strand): 5′-TCTAAAGGTTGGTAAGTGTGAGTT-3′ (−470/−459), 5′-CAGGAAGGCTGTGAAATGTGTCCG-3′ (−404/−390), 5′-TCTAAT(GT)(AGCT)(AGCT)G(AGCT)AA(GT)GAGTTC-3′ (C/EBP consensus oligonucleotide), 5′-AGAGATTGCCTGACGTCAGAGAGCTAG-3′ [CREB (cAMP response element-binding protein) consensus oligonucleotide], 5′-AGATTTCTAGGAATTCAATCC-3′ [Stat 5 (signal transducers and activators of transcription 5) consensus oligonucleotide] and 5′-ATTCGATCGGGCGGGGCGAGC-3′ (Sp1 consensus oligonucleotide). For each binding reaction (40 μl), 4 μg of poly(dI-dC), 8 μl of a 5×binding buffer [100 mM Tris/HCl (pH 7.4), 10 mM MgCl2, 250 mM NaCl, 5 mM EDTA, 50% glycerol, 0.5% Nonidet P-40, 5 mM dithiothreitol], 5 μg of nuclear extract and labelled probe (20000 c.p.m.) were incubated at 4 °C for 30 min. Competitive EMSAs were performed under identical conditions, except that 200 pmol of a non-labelled double-stranded oligonucleotide or 5 μg of commercially available antibody was added to the binding reactions 30 min prior to the addition of labelled probe. Binding reactions were terminated by the addition of 8 μl of gel loading buffer [30% (v/v) glycerol, 0.1% (w/v) Bromophenol Blue, 0.1% (w/v) xylene cyanol]. Protein–DNA complexes were resolved on 4% (for labelled promoter fragments) or 6% (for labelled double-stranded oligonucleotides) non-denaturing PAGE with a Tris/borate/EDTA buffer system (45 mM Tris, 44.5 mM borate, 1 mM EDTA, pH 8.0) at 4 °C, and were detected by autoradiography of the dried gel.
Transient transfection assay
NIH 3T3-L1 fibroblasts at 50% confluency and differentiated 3T3-L1 cells (day 5) in 60-mm dishes were transfected with 4 μg of luciferase reporter constructs and 4 μg of pSV-β-galactosidase vector, with or without pCDNA3.1(+)-C/EBPα or control plasmid pCDNA3.1(+), using the calcium phosphate precipitation method. Cos-7 cells at 70% confluency in 35-mm dishes were transfected with 1 μg of luciferase reporter constructs and 1 μg of pSV-β-galactosidase vector with Lipofectamine Plus™ reagent. After 48 h, the cells were washed twice with PBS and lysed in 250 μl of reporter lysis buffer. Cell extracts were prepared by a 5-min centrifugation at 16000 g. Luciferase activity was assayed using commercial kits on a Microbeta liquid scintillation counter (PerkinElmer). β-Galactosidase activity was assayed spectrophotometrically according to the manufacturer's instructions. Luciferase activity was normalized to β-galactosidase activity to adjust for transfection efficiency. NIH 3T3 fibroblasts at 70% confluency in 60-mm dishes were transfected with or without pCDNA3.1(+)-C/EBPα or control plasmid pCDNA3.1(+) using Lipofectamine Plus™ reagent. After 48 h, cell homogenates or RNA were obtained for analysis of protein and mRNA levels.
RESULTS
Differentiation-dependent expression of TGH in 3T3-L1 adipocytes
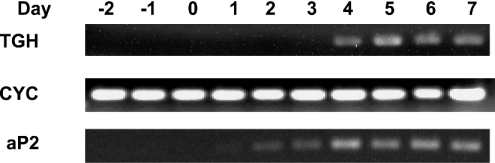
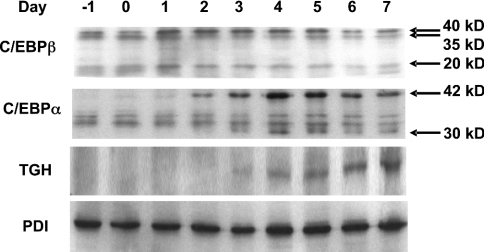
Murine NIH 3T3-L1 cells are widely used as an in vitro model for the study of gene regulation during adipogenesis [16,17]. Figure 1 presents the pattern and time course of expression of TGH and of aP2, an early marker of differentiation [18]. TGH expression had increased markedly by day 4 following the induction of differentiation, which is a late stage of differentiation of 3T3-L1 adipocytes.
Figure 1. TGH expression and activity are induced during adipocyte differentiation.
NIH 3T3-L1 fibroblasts were induced to differentiate into adipocytes. Day 0 represents 2 days post-confluence and the starting point of the differentiation programme. Cells were harvested at the indicated times, and RT-PCR analysis of TGH mRNA was performed. Total RNA (2 μg) was reverse-transcribed to cDNA, and TGH, aP2 and cyclophilin (CYC) mRNAs were detected by PCR. Results are representative of three independent experiments.
A distal murine TGH promoter fragment (−542/−371 bp) is important for expression in adipocytes
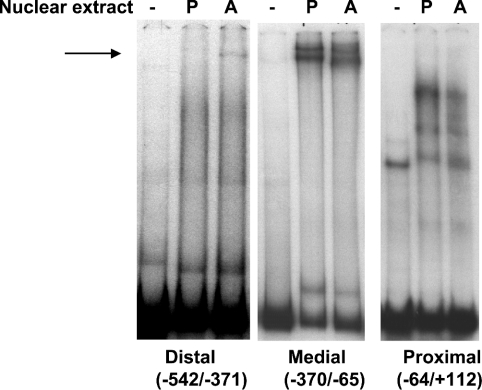
To address whether the increased expression of TGH mRNA in NIH 3T3-L1 adipocytes is due to transcriptional regulation, EMSA and promoter-activity assays were employed. A murine TGH promoter fragment (−542/+112 bp) was isolated from a bacterial artificial chromosome clone and digested with restriction enzymes, and three smaller fragments were obtained. They were designated as the distal (−542 to −371), medial (−370 to −65) and proximal (−64 to +112) promoter regions, and employed to evaluate their binding with nuclear extracts from NIH 3T3-L1 pre-adipocytes (day 0) and adipocytes (day 5). Not surprisingly, EMSAs (Figure 2) showed different results from those observed during liver development [13]. The medial promoter region bound nuclear extracts and yielded two major bands, which showed minimal differences between pre-adipocytes and adipocytes. After incubation of proximal promoter fragments with nuclear extracts, two specific migrating complexes were formed in pre-adipocytes, with lesser amounts in adipocytes. One possible explanation would be that these protein–DNA complexes inhibit TGH expression, and that fewer of these complexes in mature adipocytes would allow for more TGH expression. Unlike with hepatic nuclear extracts, the distal promoter region exhibited enhanced protein binding with nuclear extracts prepared from adipocytes compared with pre-adipocytes, yielding one specific migrating band, which disappeared in the presence of 20× unlabelled probe (results not shown). This result suggests that the distal promoter region is important for TGH expression in 3T3-L1 adipocytes.
Figure 2. Binding of nuclear extracts to the TGH promoter (−542/+112).
Cell nuclear extracts were prepared from 3T3-L1 pre-adipocytes (P; day 0) and adipocytes (A; day 5) and used in binding assays with each of three 32P-labelled promoter fragments. Protein–DNA complexes were resolved by non-denaturing 4%-PAGE with Tris/borate/EDTA buffer and were detected by autoradiography. The arrow shows the specific DNA–protein complex. These results are representative of three independent experiments, each using freshly isolated nuclear extracts.
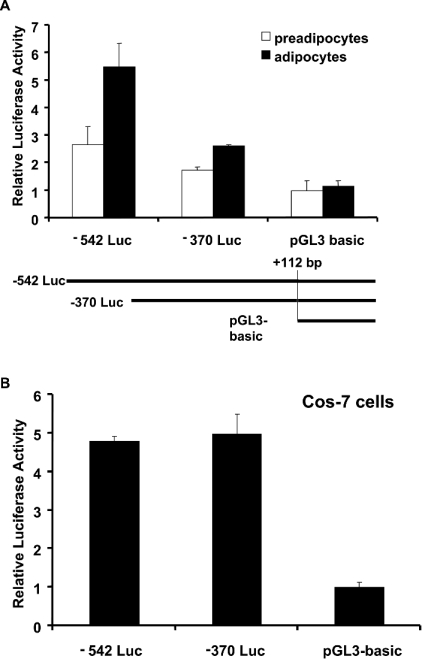
To determine if the distal TGH promoter region has any specific functional significance in 3T3-L1 adipocytes, the truncated TGH promoter reporter construct −370 Luc, which lacks the distal promoter fragment, was created, and transient transfection assays in 3T3-L1 pre-adipocytes and adipocytes were performed. Relative to the promoter-less parent vector (pGL3-Basic), transient transfection of the −542 Luc reporter into preadipocytes resulted in 2-fold enhanced luciferase expression (Figure 3A). However, the same construct induced luciferase expression 5-fold in adipocytes, indicating that this promoter region is more active in 3T3-L1 adipocytes than in pre-adipocytes. In contrast, the −370 promoter-luciferase vector yielded a small increase over pGL3-Basic, and there was no difference in expression between pre-adipocytes and adipocytes (Figure 3A). To evaluate further whether the distal promoter region is tissue specific, we transiently transfected the same constructs into Cos-7 cells. In comparison with empty vector (pGL3-Basic), the −542 luciferase construct could induce luciferase expression approx. 5-fold. However, deleting the distal fragment did not result in a decrease in luciferase expression (Figure 3B). In addition, previous results showed that the distal TGH promoter region is not required for luciferase expression in primary hepatocytes [13]. These data suggest that the TGH distal promoter region is important in adipocyte expression, but has not been identified as being important in other cells tested.
Figure 3. Tissue-specific activity of the TGH promoter.
(A) Schematic representation of the different truncated reporter constructs (−542 Luc, −370 Luc and pGL3-Basic) used in transfection assays. Transfections were performed as described in the Materials and methods section. Luciferase activity in 3T3-L1 pre-adipocytes and adipocytes is shown. Values are means±S.E.M. of three independent transfections, normalized to cotransfected β-galactosidase. (B) Luciferase activity in monkey Cos-7 cells. Each value is the mean±S.E.M. of three experiments.
C/EBPα binds the distal promoter region of TGH
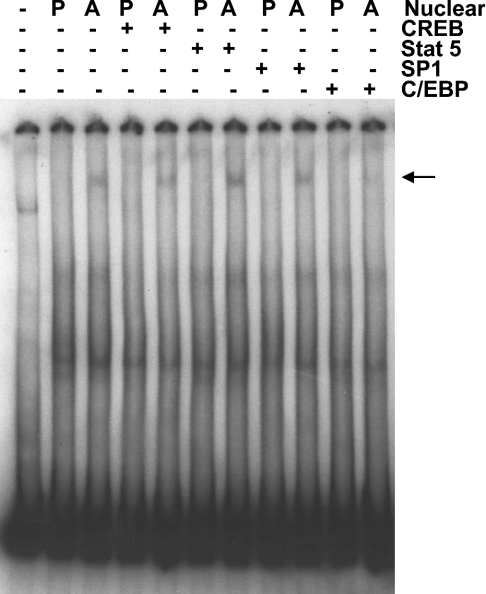
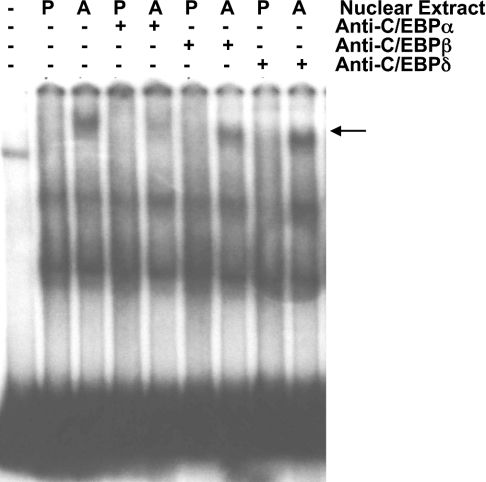
Adipogenesis is a complicated process, involving multiple transcription factors [14,18–28]. On analysing potential binding sites in the murine TGH distal promoter fragment for transcription factors by searching the TRANSFAC database, we detected two candidate binding sites for C/EBP, one for Sp1, one for CREB and three for Stat 5. Because each of these transcription factors has been shown to promote adipogenesis or to increase the expression of some adipocyte-specific genes [14,18–28], we explored the ability of these transcription factors to bind the distal TGH promoter region using competitive EMSA. As shown in Figure 4, the oligonucleotide corresponding to the C/EBP consensus blocked the formation of the adipocyte-specific protein–DNA complex, while the oligonucleotides corresponding to CREB, Stat 5 and Sp1 did not interfere with this interaction. Therefore C/EBP might be responsible for the regulation of TGH expression during 3T3-L1 adipocyte differentiation. To explore which isoform(s) of C/EBP binds to the distal TGH promoter region, we performed EMSAs with antibodies against C/EBPα, C/EBPβ and C/EBPδ. Antibodies to C/EBPβ and C/EBPδ did not block the binding of a protein to the TGH promoter region. Only the antibody against C/EBPα prevented the interaction of the distal TGH promoter region and nuclear extracts from adipocytes (Figure 5), suggesting that C/EBPα might participate in the regulation of TGH expression.
Figure 4. Analysis of protein–DNA interactions using competitive EMSA.
Band-shift assays were performed with nuclear extracts from 3T3-L1 pre-adipocytes (P; day 0) and adipocytes (A; day 5) using the distal (−542/−371 bp) promoter fragment as a probe. Unlabelled oligonucleotides (400 pmol) against CREB, Stat 5, Sp1 or C/EBP consensus sites were added to the binding assays as competitors, as indicated. The arrow shows the specific DNA–protein complex. The results are representative of three independent experiments.
Figure 5. EMSA-supershift analysis of proteins binding to the TGH promoter.
Cell nuclear extracts were prepared from 3T3-L1 preadipocytes (P; day 0) and adipocytes (A; day 5) and used in binding assays with the 32P-labelled distal (−542/−371 bp) promoter fragment with (+) or without (−) antibodies against C/EBPα, C/EBPβ or C/EBPδ. The arrow shows the specific DNA–protein complex. The results are representative of three independent experiments.
Expression of murine TGH is regulated directly by C/EBPα
To further address the functional significance of C/EBPs for the expression of TGH during 3T3-L1 cell differentiation, the protein expression patterns of C/EBPα, C/EBPβ and TGH were evaluated by immunoblot analysis (Figure 6). Consistent with previous reports [23], C/EBPβ was induced at a very early stage of differentiation (day 1). C/EBPβ governs the expression of two forms of C/EBPα, leading to increased levels of this transcription factor approx. 24 h following the induction of C/EBPβ. The appearance of TGH follows at 24 h after C/EBPα expression. The temporal expression of C/EBPs and TGH is consistent with a regulatory role for transcriptional activation of the TGH promoter by C/EBPα, but not by C/EBPβ.
Figure 6. Analysis of C/EBPβ, C/EBPα and TGH proteins using immunoblotting.
NIH 3T3-L1 fibroblasts were induced to differentiate into adipocytes. Day 0 represents 2 days post-confluence and the starting point of the differentiation programme. Cells were harvested at the indicated times. Proteins of cell homogenates (50 μg) were resolved on SDS/10%-polyacrylamide gels, electroblotted on to nitrocellulose membranes, and probed with antibodies against C/EBPβ, C/EBPα, TGH or protein disulphide isomerase (PDI). The arrows show three isoforms of C/EBPβ (40, 35 and 20 kDa) and two isoforms of C/EBPα (42 and 30 kDa) separately [22,23]. The results are representative of two independent experiments.
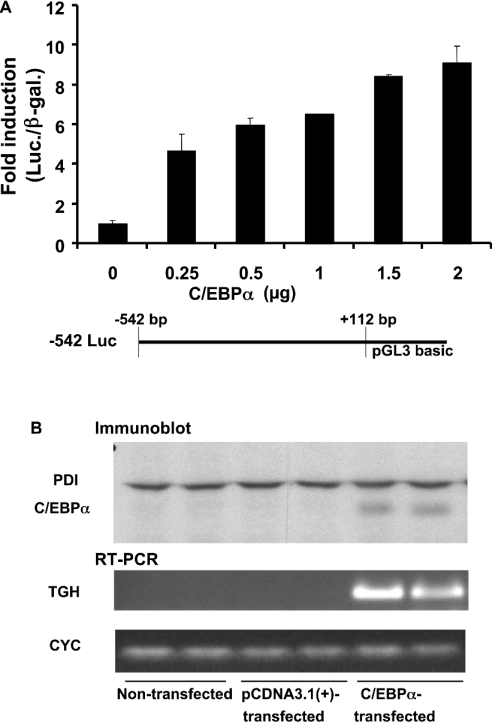
To test directly the ability of C/EBPα to trans-activate the murine TGH promoter, 3T3-L1 pre-adipocytes were transfected with TGH promoter constructs (−542 Luc) and expression constructs for C/EBPα. A concentration-dependent stimulation of TGH promoter activity by C/EBPα was demonstrated (Figure 7A).
Figure 7. Ectopic expression of C/EBPα stimulates TGH promoter activity and mRNA expression.
(A) The luciferase activity of the −542 Luc reporter vector is induced by C/EBPα in a dose-dependent fashion in 3T3-L1 pre-adipocytes. The TGH reporter plasmid −542 Luc (4 μg) was co-transfected into 3T3-L1 pre-adipocytes with pSV-β-galactosidase vector (4 μg) and the indicated concentrations of C/EBPα. Luciferase activity was measured relative to β-galactosidase activity. Values are means±S.E.M. of three experiments. (B) C/EBPα induces TGH mRNA expression in NIH 3T3 cells. C/EBPα or control vector pCDNA3.1(+) (2 μg) was transiently transfected into NIH 3T3 cells. After 48 h, cell homogenates were extracted and used to analyse C/EBPα and protein disulphide isomerase (PDI) expression by immunoblot analysis. TGH and cyclophilin (CYC) mRNAs were detected by RT-PCR. The results are representative of two independent experiments.
Overexpression of C/EBPα in 3T3-L1 pre-adipocytes is sufficient to induce differentiation independent of hormonal stimulation [14]. To ascertain if the increased TGH promoter activity is due to C/EBPα directly or the result of C/EBPα-induced differentiation, NIH 3T3 fibroblasts, which lack most adipogenic transcription factors [14,28] and have low potential to differentiate, were chosen. Fortunately, the high transfection efficiency in NIH 3T3 fibroblasts also made it possible to observe TGH mRNA abundance. As shown in Figure 7(B), ectopic expression of C/EBPα activated the expression of TGH, while parent plasmid pCDNA3.1(+) did not. Thus C/EBPα stimulates TGH promoter activity independent of its ability to induce cell differentiation.
Two C/EBP-binding sites within the distal TGH promoter region are required for high expression in adipocytes
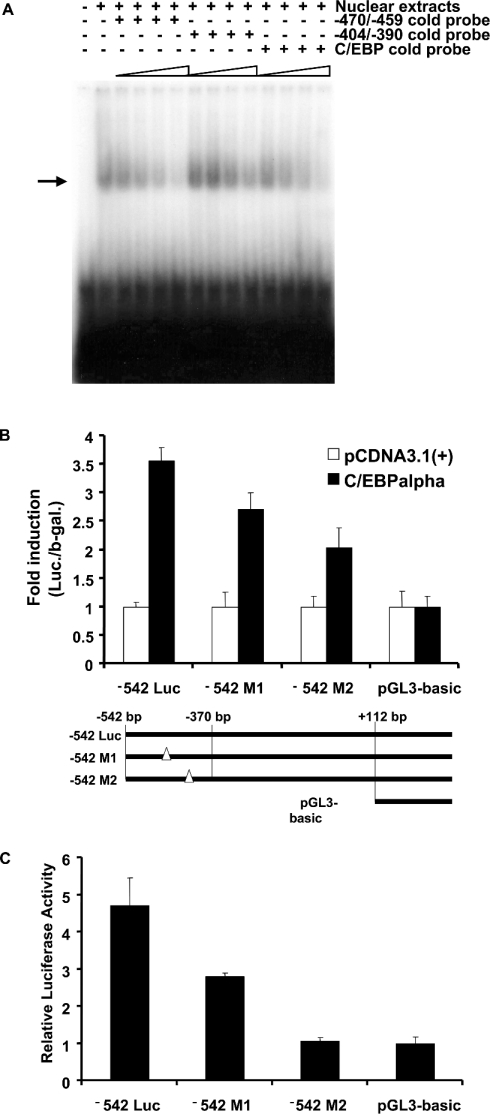
Two potential binding sites (−470/−459 bp and −404/−390 bp) in the distal murine TGH promoter show very high similarity to the C/EBP consensus sequence. To determine which site(s) is responsible for the regulation of TGH expression, a series of experiments were carried out. First, the ability of these two sites to bind nuclear extracts from 3T3-L1 adipocytes was examined. As shown in Figure 8(A), incubation of an oligonucleotide corresponding to the C/EBP consensus and nuclear extracts from 3T3-L1 adipocytes produced one specific complex. The addition of excess unlabelled oligonucleotides from either of the putative C/EBP sites blocked in a concentration-dependent fashion the formation of this specific band (Figure 8A), indicating both C/EBP-binding sites have the ability to bind C/EBPα protein.
Figure 8. C/EBP-binding sites (−470 to −459 bp and −404 to −390 bp) in the distal fragment are important for TGH promoter activity.
(A) EMSA was performed with nuclear extracts from 3T3-L1 adipocytes (day 5). A 32P-labelled C/EBP consensus oligonucleotide was used as a probe. Unlabelled oligonucleotides (50×, 100×, 200× or 400×) derived from the TGH promoter (−470/−459 bp, 5′-TCTAAAGGTTGGTAAGTGTGAGTT-3′; −404/−390 bp, 5′-CAGGAAGGCTGTGAAATGTGTCCG-3′) or the C/EBP consensus [5′-TCTAAT(GT)(AGCT)(AGCT)G(AGCT)AA(GT)GAGTTC-3′] were added to the binding assays as competitors. The arrow shows the specific DNA–protein complex. The results are representative of two independent experiments. (B) Schematic representation of the different mutated reporter constructs (−542 Luc, −542 M1, −542 M2 and pGL3-Basic) used in transfection assays. Also shown are the results of trans-activation assays with different mutated TGH promoter–reporter constructs co-transfected with C/EBPα in 3T3-L1 pre-adipocytes. Values are means±S.E.M. of three independent transfections, normalized to co-transfected β-galactosidase. (C) Trans-activation assays of different mutated TGH promoter–reporter constructs transfected into 3T3-L1 adipocytes. Values are means±S.E.M. of three experiments.
We next investigated the involvement of these two cis-binding domains in the induction of TGH promoter activity. Mutant reporters bearing C/EBP mutations were constructed (Figure 8B). Mutation of either of the two C/EBP sites in the TGH reporter constructs decreased the stimulation of luciferase activity when co-transfected with C/EBPα in 3T3-L1 pre-adipocytes compared with the pCDNA3.1(+) vector (Figure 8B). To further assess whether or not endogenous C/EBPα contributes to the basal activity of the TGH promoter during adipogenesis, the wild-type and mutated reporter constructs were transfected into 3T3-L1 adipocytes and their activities were compared (Figure 8C). These data demonstrate the abrogation of reporter activity in adipocytes following mutagenesis of either of the C/EBP sites in the −542 Luc construct.
DISCUSSION
A hepatic TGH was purified and is speculated to be involved in mobilizing stored TGs for very-low-density lipoprotein assembly via a lipolysis and re-esterification pathway [8–10,29]. TGH is also abundantly expressed in adipose tissue and is induced during 3T3-L1 adipocyte differentiation [8,12]. In a few animal models of obesity, the expression of TGH is reduced significantly (V. W. Dolinsky, R. Lehner and D. E. Vance, unpublished work). These data suggest that TGH is associated with energy metabolism in adipose tissue. Recently, it was found that TGH is responsible for a major part of non-HSL lipase activity in white adipose tissue in vitro, and may mediate some or all HSL-independent lipolysis [11]. Inhibition of TGH reduced dramatically basal fatty acid release by 3T3-L1 adipocytes and circulating non-esterified fatty acid levels in hamsters (W. Gao and R. Lehner, unpublished work). These results further verified that TGH is an important enzyme promoting lipolysis and participating in the regulation of lipid homoeostasis in adipose tissue. Microarray analysis of 3T3-L1 adipocytes has revealed that induction of the genes involved in the ability of the adipocyte to mobilize energy stores is a late event during the acquisition of the adipocyte phenotype [19]. Therefore the pattern of TGH expression during the differentiation of 3T3-L1 pre-adipocytes is consistent with the proposed role for TGH in the basal TG lipase activity in adipocytes.
Most genes are regulated, at least partially, at the transcriptional level, and it has been reported that the transcription factor Sp1 induces the expression of TGH during liver development [13]. The induction of mRNA and protein for TGH suggests that a transcriptional regulatory mechanism also participates in its expression during 3T3-L1 adipocyte differentiation. Incubating nuclear extracts from 3T3-L1 pre-adipocytes and adipocytes with three different TGH promoter regions demonstrated different binding abilities. Thus a transcriptional mechanism is apparently involved in the regulation of TGH during the differentiation of 3T3-L1 pre-adipocytes. The results are different from those observed during liver development [13], suggesting that some specific transcription factors for adipogenesis are required for the expression of TGH in adipocytes. Incubation of nuclear extracts from 3T3-L1 pre-adipocytes and adipocytes with the medial TGH promoter fragment resulted in the formation of two complexes; however, the amounts and sizes of these bands did not differ. These data suggest that the medial TGH promoter region is not sufficient for the expression of TGH in adipocytes. PPARγ (peroxisome-proliferator-activated receptor γ), LXR (liver X receptor) and ADD1 (adipocyte determination- and differentiation-dependent factor)/SREBP1c (sterol response element-binding protein 1c) are important nuclear factors that promote adipogenesis and regulate the expression of a range of genes involved in lipid metabolism [19,30–32]. Because one RXRE (retinoid X receptor response element) and one SRE exist in the medial TGH promoter region, the roles of PPARγ, LXR and ADD1/SREBP1c in the regulation of TGH were studied. An effect of PPARγ was excluded because fatty acids and synthetic PPARγ agonists did not influence the expression of TGH in 3T3-L1 adipocytes [12]. In other experiments, 3T3-L1 adipocytes were incubated with LXR and RXR agonists for 24 h, followed by EMSA analyses. No changes in the ability of these nuclear receptors to bind the TGH promoter were found (results not shown). Thus it appears that LXR/RXR at least is not involved in the transcriptional regulation of TGH expression. Addition of cholesterol and fatty acids to the standard lab diet resulted in increased murine hepatic TGH expression [12]. However, the enforced expression of a nuclear form of SREBP1a in transgenic mice failed to affect TGH protein expression (R. Lehner and D. E. Vance, unpublished work). Thus it is unclear whether or not TGH expression is regulated directly by SREBPs.
Members of the C/EBP family represent another important class of transcription factors that play essential roles in the adipocyte differentiation programme. Expression of C/EBPβ and C/EBPδ occurs early in clonal expansion, followed by that of C/EBPα as the cells exit the cell cycle and begin to express adipocyte genes [22,23]. The results in the present study strongly support the proposal that C/EBPα induces the expression of TGH during the conversion of 3T3-L1 pre-adipocytes into adipocytes. C/EBPα serves as transcriptional activator of many adipocyte genes, such as those encoding aP2, leptin, PEPCK (phosphoenoylpyruvate carboxy-kinase) and others [20–23]. Chemical assays and functional analyses both indicated that C/EBPα induced TGH transcription through binding to two C/EBP-binding sites (−470/−459 bp and −404/−390 bp) in the distal promoter region. The trans-activation of TGH mRNA by C/EBPα in the low-adipogenic-potential NIH 3T3 fibroblasts further verified that the increase in TGH is derived from the direct induction of C/EBPα, independent of cell differentiation. Different from other C/EBPα-regulated adipogenic gene products, such as aP2 and PEPCK, which increase immediately subsequent to C/EBPα inducation, the expression of TGH was delayed by 24 h. Other mechanisms may account for the delay in TGH expression. For example, dephosphorylation of C/EBPα by insulin in differentiated 3T3-L1 adipocytes was speculated to cause a decrease in transcriptional activity and to result in decreased transcription of target genes, such as the GLUT4 gene [33,34]. Whether post-translational regulation of C/EBPα is involved in the trans-activation of TGH is a question of interest. In addition, mutational analysis of the TGH promoter showed that the responses of two C/EBP-binding sites to endogenous C/EBPα were stronger than to exogenous C/EBPα, suggesting that some other factors in 3T3-L1 adipocytes also participate in the regulation of TGH expression.
Besides adipose tissue, C/EBP factors are also expressed abundantly in the liver, and are regarded as pivotal regulators of liver functions such as nutrient metabolism and its control by hormones, the acute-phase response and hepatic regeneration [35]. Interestingly, the protein expression patterns of C/EBP and TGH during rat liver development are similar [35,36]. These data suggest that the role of C/EBP family members in the regulation of TGH expression may have broader significance in the liver.
In conclusion, the present study shows that the distal promoter region of TGH is essential for TGH expression in 3T3-L1 adipocytes, and that C/EBPα transcriptionally regulates TGH expression.
Acknowledgments
This research was supported by a contract from GlaxoSmithKline and a grant from the Canadian Institutes of Health Research. E. W. is a CIHR/HSFC Strategic Training Fellow in Stroke, Cardiovascular, Obesity, Lipid, Atherosclerosis Research (SCOLAR). D. E. V. holds the Canada Research Chair in Molecular and Cell Biology of Lipids, and is a Heritage Medical Scientist of the Alberta Heritage Foundation for Medical Research. R. L. is a Senior Scholar of the Alberta Heritage Foundation for Medical Research.
References
- 1.Londos C., Brasaemle D. L., Schultz C. J., Adler-Wailes D. C., Levin D. M., Kimmel A. R., Rondinone C. M. On the control of lipolysis in adipocytes. Ann. N.Y. Acad. Sci. 1999;892:155–168. doi: 10.1111/j.1749-6632.1999.tb07794.x. [DOI] [PubMed] [Google Scholar]
- 2.Krey G., Braissant O., L'Horset F., Kalkhoven E., Perroud M., Parker M. G., Wahli W. Fatty acids, eicosanoids, and hypolipidemic agents identified as ligands of peroxisome proliferator-activated receptors by coactivator-dependent receptor ligand assay. Mol. Endocrinol. 1997;11:779–791. doi: 10.1210/mend.11.6.0007. [DOI] [PubMed] [Google Scholar]
- 3.Holm C., Kirchgessner T. G., Svenson K. L., Fredrikson G., Nilsson S., Miller C. G., Shively J. E., Heinzmann C., Sparkes R. S., Mohandas T., et al. Hormone-sensitive lipase: sequence, expression, and chromosomal localization to 19 cent-q13.3. Science. 1988;241:1503–1506. doi: 10.1126/science.3420405. [DOI] [PubMed] [Google Scholar]
- 4.Carey G. B. Mechanisms regulating adipocyte lipolysis. Adv. Exp. Med. Biol. 1998;441:157–170. doi: 10.1007/978-1-4899-1928-1_15. [DOI] [PubMed] [Google Scholar]
- 5.Okazaki H., Osuga J., Tamura Y., Yahagi N., Tomita S., Shionoiri F., Iizuka Y., Ohashi K., Harada K., Kimura S., et al. Lipolysis in the absence of hormone-sensitive lipase: evidence for a common mechanism regulating distinct lipases. Diabetes. 2002;51:3368–3375. doi: 10.2337/diabetes.51.12.3368. [DOI] [PubMed] [Google Scholar]
- 6.Osuga J., Ishibashi S., Oka T., Yagyu H., Tozawa R., Fujimoto A., Shionoiri F., Yahagi N., Kraemer F. B., Tsutsumi O., Yamada N. Targeted disruption of hormone-sensitive lipase results in male sterility and adipocyte hypertrophy, but not in obesity. Proc. Natl. Acad. Sci. U.S.A. 2000;97:787–792. doi: 10.1073/pnas.97.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haemmerle G., Zimmermann R., Strauss J. G., Kratky D., Riederer M., Knipping G., Zechner R. Hormone-sensitive lipase deficiency in mice changes the plasma lipid profile by affecting the tissue-specific expression pattern of lipoprotein lipase in adipose tissue and muscle. J. Biol. Chem. 2002;277:12946–12952. doi: 10.1074/jbc.M108640200. [DOI] [PubMed] [Google Scholar]
- 8.Dolinsky V. W., Sipione S., Lehner R., Vance D. E. The cloning and expression of a murine triacylglycerol hydrolase cDNA and the structure of its corresponding gene. Biochim. Biophys. Acta. 2001;1532:162–172. doi: 10.1016/s1388-1981(01)00133-0. [DOI] [PubMed] [Google Scholar]
- 9.Lehner R., Vance D. E. Cloning and expression of a cDNA encoding a hepatic microsomal lipase that mobilizes stored triacylglycerol. Biochem. J. 1999;343:1–10. [PMC free article] [PubMed] [Google Scholar]
- 10.Gilham D., Ho S., Rasouli M., Martres P., Vance D. E., Lehner R. Inhibitors of hepatic microsomal triacylglycerol hydrolase decrease very low density lipoprotein secretion. FASEB J. 2003;17:1685–1687. doi: 10.1096/fj.02-0728fje. [DOI] [PubMed] [Google Scholar]
- 11.Soni K. G., Lehner R., Metalnikov P., O'Donnell P., Semache M., Gao W., Ashman K., Pshezhetsky A. V., Mitchell G. A. Carboxylesterase 3 (EC 3.1.1.1) is a major adipocyte lipase. J. Biol. Chem. 2004;279:40683–40689. doi: 10.1074/jbc.M400541200. [DOI] [PubMed] [Google Scholar]
- 12.Dolinsky V. W., Gilham D., Hatch G. M., Agellon L. B., Lehner R., Vance D. E. Regulation of triacylglycerol hydrolase expression by dietary fatty acids and peroxisomal proliferator-activated receptors. Biochim. Biophys. Acta. 2003;1635:20–28. doi: 10.1016/j.bbalip.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 13.Douglas D. N., Dolinsky V. W., Lehner R., Vance D. E. A role for Sp1 in the transcriptional regulation of hepatic triacylglycerol hydrolase in the mouse. J. Biol. Chem. 2001;276:25621–25630. doi: 10.1074/jbc.M103874200. [DOI] [PubMed] [Google Scholar]
- 14.Lin F. T., Lane M. D. CCAAT/enhancer binding protein alpha is sufficient to initiate the 3T3-L1 adipocyte differentiation program. Proc. Natl. Acad. Sci. U.S.A. 1994;91:8757–8761. doi: 10.1073/pnas.91.19.8757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deryckere F., Gannon F. A one-hour minipreparation technique for extraction of DNA-binding proteins from animal tissues. Biotechniques. 1994;16:405. [PubMed] [Google Scholar]
- 16.Ntambi J. M., Young-Cheul K. Adipocyte differentiation and gene expression. J. Nutr. 2000;130:3122S–3126S. doi: 10.1093/jn/130.12.3122S. [DOI] [PubMed] [Google Scholar]
- 17.Camp H. S., Ren D., Leff T. Adipogenesis and fat-cell function in obesity and diabetes. Trends Mol. Med. 2002;8:442–447. doi: 10.1016/s1471-4914(02)02396-1. [DOI] [PubMed] [Google Scholar]
- 18.Christy R. J., Yang V. W., Ntambi J. M., Geiman D. E., Landschulz W. H., Friedman A. D., Nakabeppu Y., Kelly T. J., Lane M. D. Differentiation-induced gene expression in 3T3-L1 preadipocytes: CCAAT/enhancer binding protein interacts with and activates the promoters of two adipocyte-specific genes. Genes Dev. 1989;3:1323–1335. doi: 10.1101/gad.3.9.1323. [DOI] [PubMed] [Google Scholar]
- 19.Ross S. E., Erickson R. L., Gerin I., DeRose P. M., Bajnok L., Longo K. A., Misek D. E., Kuick R., Hanash S. M., Atkins K. B., et al. Microarray analyses during adipogenesis: understanding the effects of Wnt signaling on adipogenesis and the roles of liver X receptor alpha in adipocyte metabolism. Mol. Cell. Biol. 2002;22:5989–5999. doi: 10.1128/MCB.22.16.5989-5999.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller S. G., De Vos P., Guerre-Millo M., Wong K., Hermann T., Staels B., Briggs M. R., Auwerx J. The adipocyte specific transcription factor C/EBPalpha modulates human ob gene expression. Proc. Natl. Acad. Sci. U.S.A. 1996;93:5507–5511. doi: 10.1073/pnas.93.11.5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J. W., Klemm D. J., Vinson C., Lane M. D. Role of CREB in transcriptional regulation of CCAAT/enhancer-binding protein beta gene during adipogenesis. J. Biol. Chem. 2004;279:4471–4478. doi: 10.1074/jbc.M311327200. [DOI] [PubMed] [Google Scholar]
- 22.Tang Q. Q., Lane M. D. Activation and centromeric localization of CCAAT/enhancer-binding proteins during the mitotic clonal expansion of adipocyte differentiation. Genes Dev. 1999;13:2231–2241. doi: 10.1101/gad.13.17.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lane M. D., Tang Q. Q., Jiang M. S. Role of the CCAAT enhancer binding proteins (C/EBPs) in adipocyte differentiation. Biochem. Biophys. Res. Commun. 1999;266:677–683. doi: 10.1006/bbrc.1999.1885. [DOI] [PubMed] [Google Scholar]
- 24.Zhu Q., Zhu J., Liao K. An SP1-like cis-element is the major DNA motif for differential expression regulation of the adipocyte amino acid transporter. Biochem. Biophys. Res. Commun. 2000;271:91–99. doi: 10.1006/bbrc.2000.2585. [DOI] [PubMed] [Google Scholar]
- 25.Barth N., Langmann T., Scholmerich J., Schmitz G., Schaffler A. Identification of regulatory elements in the human adipose most abundant gene transcript-1 (apM-1) promoter: role of SP1/SP3 and TNF-alpha as regulatory pathways. Diabetologia. 2002;45:1425–1433. doi: 10.1007/s00125-002-0895-5. [DOI] [PubMed] [Google Scholar]
- 26.Harp J. B., Franklin D., Vanderpuije A. A., Gimble J. M. Differential expression of signal transducers and activators of transcription during human adipogenesis. Biochem. Biophys. Res. Commun. 2001;281:907–912. doi: 10.1006/bbrc.2001.4460. [DOI] [PubMed] [Google Scholar]
- 27.Nanbu-Wakao R., Morikawa Y., Matsumura I., Masuho Y., Muramatsu M. A., Senba E., Wakao H. Stimulation of 3T3-L1 adipogenesis by signal transducer and activator of transcription 5. Mol. Endocrinol. 2002;16:1565–1576. doi: 10.1210/mend.16.7.0862. [DOI] [PubMed] [Google Scholar]
- 28.Floyd Z. E., Stephens J. M. STAT5A promotes adipogenesis in nonprecursor cells and associates with the glucocorticoid receptor during adipocyte differentiation. Diabetes. 2003;52:308–314. doi: 10.2337/diabetes.52.2.308. [DOI] [PubMed] [Google Scholar]
- 29.Lehner R., Verger R. Purification and characterization of a porcine liver microsomal triacylglycerol hydrolase. Biochemistry. 1997;36:1861–1868. doi: 10.1021/bi962186d. [DOI] [PubMed] [Google Scholar]
- 30.Brun R. P., Spiegelman B. M. PPAR gamma and the molecular control of adipogenesis. J. Endocrinol. 1997;155:217–218. doi: 10.1677/joe.0.1550217. [DOI] [PubMed] [Google Scholar]
- 31.Suzawa M., Takada I., Yanagisawa J., Ohtake F., Ogawa S., Yamauchi T., Kadowaki T., Takeuchi Y., Shibuya H., Gotoh Y., et al. Cytokines suppress adipogenesis and PPAR-gamma function through the TAK1/TAB1/NIK cascade. Nat. Cell Biol. 2003;5:224–230. doi: 10.1038/ncb942. [DOI] [PubMed] [Google Scholar]
- 32.Seo J. B., Moon H. M., Noh M. J., Lee Y. S., Jeong H. W., Yoo E. J., Kim W. S., Park J., Youn B. S., Kim J. W., et al. Adipocyte determination- and differentiation-dependent factor 1/sterol regulatory element-binding protein 1c regulates mouse adiponectin expression. J. Biol. Chem. 2004;279:22108–22117. doi: 10.1074/jbc.M400238200. [DOI] [PubMed] [Google Scholar]
- 33.MacDougald O. A., Cornelius P., Liu R., Lane M. D. Insulin regulates transcription of the CCAAT/enhancer binding protein (C/EBP) alpha, beta, and delta genes in fully-differentiated 3T3-L1 adipocytes. J. Biol. Chem. 1995;270:647–654. doi: 10.1074/jbc.270.2.647. [DOI] [PubMed] [Google Scholar]
- 34.Hemati N., Ross S. E., Erickson R. L., Groblewski G. E., MacDougald O. A. Signaling pathways through which insulin regulates CCAAT/enhancer binding protein alpha (C/EBPalpha) phosphorylation and gene expression in 3T3-L1 adipocytes. Correlation with GLUT4 gene expression. J. Biol. Chem. 1997;272:25913–25919. doi: 10.1074/jbc.272.41.25913. [DOI] [PubMed] [Google Scholar]
- 35.Diehl A. M., Michaelson P., Yang S. Q. Selective induction of CCAAT/enhancer binding protein isoforms occurs during rat liver development. Gastroenterology. 1994;106:1625–1637. doi: 10.1016/0016-5085(94)90420-0. [DOI] [PubMed] [Google Scholar]
- 36.Lehner R., Cui Z., Vance D. E. Subcellullar localization, developmental expression and characterization of a liver triacylglycerol hydrolase. Biochem. J. 1999;338:761–768. [PMC free article] [PubMed] [Google Scholar]