Abstract
TMPRSS2 is a type II transmembrane-bound serine protease that has gained interest owing to its highly localized expression in the prostate and its overexpression in neoplastic prostate epithelium. Once activated, the serine protease domain of TMPRSS2 is released from the cell surface into the extracellular space. PAR (protease-activated receptor)-2 belongs to a family of G-protein-coupled receptors (PAR-1–4) that are activated by specific serine proteases, which are expressed in many normal and malignant cell types. Previous in vitro studies on prostate cancer cells suggest a role for PAR-2 in prostate cancer metastasis. A polyclonal anti-human TMPRSS2 antibody was generated against the TMPRSS2 serine protease domain. The antibody showed specific reactivity with recombinant expressed TMPRSS2, and so was used to extract and purify the cleaved active TMPRSS2 protease from prostate cancer cells. Reverse transcriptase PCR and Western blot analysis were used to show the expression of both TMPRSS2 and PAR-2 in the androgen-dependent LNCaP prostate cancer cell line. Treatment of LNCaP cells with the cellular immunopurified TMPRSS2 protease induced a transient increase in intracellular calcium, which is indicative of G-protein-coupled-receptor activation. This calcium mobilization was inhibited by cellular pre-treatment with a specific PAR-2 antagonist, but not with a PAR-1 antagonist; inhibition of the protease activity also failed to mobilize calcium, suggesting that TMPRSS2 is capable of cleaving and thereby activating the PAR-2 receptor. The calcium mobilization was also inhibited by cellular pre-treatment with suramin or 2-APB (2-aminoethoxydiphenyl borate), indicating that a G-protein pathway is involved and that subsequent calcium release is mainly from intracellular stores. The present study describes how TMPRSS2 may contribute to prostate tumour metastasis via the activation of PAR-2.
Keywords: metastasis, protease-activated receptor-2 (PAR-2), prostate cancer, TMPRSS2, type II transmembrane serine protease
Abbreviations: AMC, 7-amino-4-methylcoumarin; 2-APB, 2-aminoethoxydiphenyl borate; BCA, bicinchoninic acid; Cbz, benzyloxycarbonyl; Cbz-Lys(OPh)2, benzyloxycarbonyl lysine diphenylphosphonate; FCS, foetal calf serum; Fmoc, fluoren-9-ylmethoxycarbonyl; HAT, human airway trypsin; MAP, multiple antigenic peptide; MMP, matrix metalloprotease; MT-SP1, membrane-type serine protease 1; PAR, protease-activated receptor; RT, reverse transcriptase; TBS, Tris-buffered saline; TTSP, type II transmembrane serine protease
INTRODUCTION
Since Paoloni-Giacobino et al. [1] originally cloned TMPRSS2 in 1997, this membrane-bound serine protease has gained interest owing to its high expression in the prostate and its potential role in carcinogenesis. Serine proteases have many physiological and pathological functions, which include inflammation, tumour growth and metastasis [2–4]. The TTSP (type II transmembrane serine protease) family, of which TMPRSS2 is a member, represents an emerging group of serine proteases with 12 members identified at present, including enteropeptidase, corin, HAT (human airway trypsin), MT-SP1 (membrane-type serine protease 1), hepsin and TMPRSS2–5 [5,6]. The TMPRSS2 gene maps to chromosome 21q22.3 and is expressed preferentially in normal prostate tissue, but is overexpressed in neoplastic prostate epithelium [7]. It is also expressed to a lesser extent in pancreas, liver, lung, kidney, colon and colon cancer tissue [8]. This high expression in prostate cancer has initiated speculation that TMPRSS2 could be a potential marker or therapeutic target for prostate cancer [9]. The TMPRSS2 protein is androgen-regulated [10]; it is expressed as a 70 kDa full-length protein, which contains a 32 kDa serine protease domain. Activation of the serine protease requires its cleavage, which is autocatalytic [10]. The active serine protease with trypsin-like specificity is then shed into the extracellular space, where it is predicted to interact with other proteins on the cell surface, as well as soluble proteins, matrix components and proteins on adjacent cells [7,10].
Previous studies on the related protein MT-SP1 have revealed that it is capable of activating urokinase-type plasminogen activator and PAR (protease-activated receptor)-2 on oocytes [11]. It has also been shown that HAT is expressed on human bronchial epithelial cells and is capable of activating PAR-2 on these cells [12]. PARs are a family of G-protein-coupled receptors (PAR-1–4) that are expressed in many normal and malignant cell types with varying physiological roles including inflammation, metastasis and invasion [13]. It has previously been shown that PARs are expressed on many tumour cells and tissues, including prostate tissue [14–17], where they are thought to be involved in metastasis [18]. The progression to metastatic diseases is particularly undesirable in prostate cancer owing to the lack of effective treatment, resulting in significant patient death [19].
Investigation into the biological and biochemical characteristics of TMPRSS2 could yield beneficial results in furthering our understanding of its role in normal and malignant tissue. In the present study, we have investigated the ability of TMPRSS2 to activate PAR-2 on human prostate cancer cells. These results provide a link between the increased expression of TMPRSS2 and the activation of PAR-2, which could involve this membrane-bound serine protease in signalling cascades that promote tumour progression.
MATERIALS AND METHODS
Cell culture
The androgen-dependent prostate cancer cell line LNCaP was maintained in RPMI 1640 (Gibco-Invitrogen, Paisley, U.K.) supplemented with 2 mM L-glutamine, 1 mM sodium pyruvate 10% FCS (foetal calf serum) and 10 mM Hepes (all obtained from Sigma, Poole, U.K.). The PC-3 prostate cancer cell line (metastasized to bone) was maintained in RPMI 1640 with 10% FCS, the DU 145 prostate cancer cell line (metastasized to brain) was maintained in Dulbecco's modified Eagle's medium (Gibco-Invitrogen) with 10% FCS and the RWPE-1 non-cancerous transformed prostate cell line was maintained in keratinocyte serum-free medium (Gibco-Invitrogen), supplemented with 5 ng/ml human recombinant epidermal growth factor and 0.05 mg/ml bovine pituitary extract. All cell lines were obtained from the American Type Culture Collection (A.T.C.C., Manassas, VA, U.S.A.).
For protein extraction, LNCaP cells were grown until 80% confluent. The medium was then removed, and the cells were washed before being removed in 3 ml of PBS using a plate scraper. The cell suspension was spun down at 800 g for 3 min, and the supernatant was discarded. The pellet was resuspended in 250 μl of lysis buffer (10 mM Tris/HCl, 10 mM EDTA and 0.2% Triton X-100, pH 7.5) and placed on ice for 20 min, with vortex-mixing after 10 min. The resulting lysate containing all the cell proteins was then passaged through a 21-gauge needle to reduce viscosity, centrifuged at 1000 g for 5 min to remove debris, and stored at −20 °C until use.
RNA isolation and RT (reverse transcriptase)-PCR
Total RNA was isolated from LNCaP, PC-3, DU 145 and RWPE-1 cells using the StrataPrep® Total RNA Miniprep kit (Stratagene, Amsterdam, The Netherlands) according to the manufacturer's instructions. cDNA was synthesized from 1 μg of total RNA (from 106 cells) using ProSTAR HF Single-Tube RT-PCR system (Stratagene). Primers were designed based on the mRNA sequence of human TMPRSS2 (NM_005656) using Oligo Primer Analysis Software, version 6.41 (Molecular Biology Insights, Cascade, CO, U.S.A.), and were synthesized and purified by Gibco-BRL/Life Technologies Custom Primers. Synthetic oligonucleotides 5′-AATCGGTGTGTTCGCCTCTAC-3′ and 5′-GCGGCTGTCACGATCC-3′ were used to amplify the TMPRSS2 sequence by RT-PCR. The 5 μl RT-PCR product, containing 0.5 μg of cDNA, was analysed by Tris/borate/EDTA agarose (1%) gel electrophoresis, and DNA bands were visualized with ethidium bromide.
RT-PCR products were sequenced using an ABI 3100 capillary electrophoresis fluorescent sequencer, using BigDye Terminator Cycle Sequencing chemistry, version 3.0 (Applied Biosystems).
Generation of anti-human TMPRSS2 polyclonal antibody
The SwissProt amino acid sequence for human TMPRSS2 was used to select an antigenic sequence, using Kyte and Doolittle [20] hydropathy plots (Molecular Programming, Seq Q-C version 2.5.1 demo) and protein cross-reactivity searches (http://www.ncbi.nlm.nih.gov/blast). The sequence Val246–Ala262 of the secreted protease domain was found to be both hydrophilic and had low cross-reactivity with other human proteins. The corresponding synthetic MAP (multiple antigenic peptide) was synthesized using standard Fmoc (fluoren-9-ylmethoxycarbonyl) solid-phase chemistry as described by Walker [21]. MAPs are based on a central component, in this case lysine, which gives a branched architecture where multiple antigenic peptides can be attached [22]. As a result of this multimerization, the MAP is highly effective at generating a humoral response. After a pre-immune bleed had been obtained, an adult New Zealand white rabbit was immunized with 1 mg of MAP dissolved in 500 μl of sterile PBS and 500 μl of Imject® Alum (Pierce, Rockford, IL, U.S.A.) as an adjuvant. The rabbit's immune system was boosted every 2 weeks, and blood samples were taken to monitor antibody production. IgG was isolated from serum using a Protein A column (Pierce) according to the manufacturer's instructions, and the fraction containing IgG was determined using a BCA (bicinchoninic assay) protein assay (Pierce), according to the manufacturer's instructions.
Production and purification of the soluble domain of TMPRSS2
The complete TMPRSS2 coding sequence (designated SS2-topo), cloned into pCR-II TOPO (Invitrogen), was generated by RT-PCR using total RNA from the prostate cancer cell line DLD-1 and primers 5′-GCTGTTGATAACAGCAAGATGGC-3′ and 5′-CAAGGACGAAGACCATGTGGAT-3′. A bacterial expression construct (designated pET-solSS2-T7-His) encoding the soluble domain of TMPRSS2 (encompassing nucleotides 452–1603; GenBank® accession code AF329454) was generated by cloning the PCR product generated using the SS2-topo construct as template and primers 5′-GAGAATTCATGGGCAGCAAGTGCTCC-3′ and 5′-CCGCTCGAGGCCGTCTGCCCTCATTTG-3′ into the EcoRI/SalI restriction enzyme sites of the pET-21a(+) vector (Novagen, Madison, WI, U.S.A.). This construct was transformed into BL21(DE3) bacterial cells, and recombinant protein expression was induced by the addition of IPTG (isopropyl β-D-thiogalactoside) to 1 mM. The soluble domain of TMPRSS2 tagged at the N-terminal with a T7 tag and at the C-terminal with a His6 tag was purified through a nickel resin column (Novagen). Fractions showing >95% purity, as assessed by PAGE analysis, were pooled. Samples of recombinant truncated TMPRSS2 (1 μg) were analysed under reducing conditions by SDS/PAGE, and stained for protein using Colloidal Blue Staining kit (Novex, San Diego, CA, U.S.A.).
TMPRSS2 purification from LNCaP cells
LNCaP cell lysate was mixed 2:1 with anti-human TMPRSS2 polyclonal antibody for 1 h at 37 °C, then an equal volume of binding buffer was added before the solution was passed through a Protein A column, pre-equilibrated with Immunopure® IgG binding buffer. After extensive washing to remove any unbound proteins, the bound TMPRSS2 was eluted with Immunopure® IgG elution buffer. The lower pH of the elution buffer causes both the antibody to dissociate from the Protein A beads and the enzyme to dissociate from the antibody. The fraction containing the dissociated antibody and TMPRSS2 enzyme was then be passed through the Protein A column again after it was equilibrated with binding buffer. The antibody, as before, interacted with the Protein A column, leaving the antibody-dissociated TMPRSS2 enzyme to pass through the column where it was collected and used for downstream procedures without the presence of the antibody. The fraction containing the purified TMPRSS2 protease was determined by BCA protein assay and then analysed under reducing conditions by SDS/PAGE and stained using the Colloidal Blue Staining kit. The purified enzyme was stored at −70 °C until use. The concentration of enzyme was measured using a BCA protein assay, and the activity was detected using a fluorescent trypsin substrate [Cbz-Gly-Gly-Arg-AMC (Sigma), where Cbz is benzyloxycarbonyl and AMC is 7-amino-4-methylcoumarin]. Purified TMPRSS2 (30 μl) was added to an equal volume of TBS (Tris-buffered saline, containing 50 mM Tris/HCl and 150 mM NaCl, pH 7.6) and 1 μl of 10 mM Cbz-Gly-Gly-Arg-AMC. Fluorescence was recorded at 455 nm (with excitation at 383 nm) on a Cytofluor multiwell plate reader (PE Biosystems, Framingham, MA, U.S.A.). Cleavage of the substrate by TMPRSS2 was also measured in the presence of the trypsin inhibitor Cbz-Lys(OPh)2 (benzyloxycarbonyl lysine diphenylphosphonate) [23] at a final concentration of 10 μM. The activity of 500 ng of trypsin was also analysed against the same concentration of fluorescent trypsin substrate. TMPRSS2 activity was also assessed in the presence of the synthetic PAR-2 antagonist, Phe-Ser-Leu-Leu-Arg-Tyr-NH2 at 200 μM [27].
Western blotting
Recombinant expressed enzyme (1 μg) was analysed under reducing conditions by SDS/PAGE and transferred on to a nitrocellulose membrane for immunoblotting. Samples were run in triplicate, with one sample being developed using T7 tag monoclonal antibody (Novagen), another using our generated anti-TMPRSS2 polyclonal antibody, and the third sample was developed using the anti-TMPRSS2 polyclonal antibody pre-absorbed with the expressed enzyme. After blotting, the nitrocellulose was blocked with 3% BSA in TBS for 1 h. T7 tag monoclonal antibody (1/10000) in TBS was incubated with the membrane for 30 min at room temperature (21 °C). After three 5 min incubations with TBS, goat anti-mouse secondary antibody conjugated to alkaline phosphatase (Sigma) was added at a 1/5000 dilution in TBS for 30 min at room temperature before alkaline phosphatase substrates (Sigma) were added to develop the blot. Anti-TMPRSS2 polyclonal antibody (1/500) was incubated with the membrane in TBS for 2.5 h at room temperature. After washing, goat anti-rabbit secondary antibody conjugated to alkaline phosphatase (Sigma) was added (1/15000) for 2 h at room temperature. The blot was again washed before substrates were added as before.
Active-site titration
The concentration of active TMPRSS2 was determined by titration of the enzyme with Cbz-Lys(OPh)2, an α-aminoalkyl diphenylphosphonate ester which is a known irreversible serine protease inhibitor [23]. The procedure was carried out as described by Barrett and Kirschke [24].
Kinetic inactivation studies
Studies were carried out with TMPRSS2 using the fluorogenic trypsin-like substrate Cbz-Gly-Gly-Arg-AMC. The inhibitor Cbz-Lys(OPh)2 was used at a final concentration of 0.5–1.5 μM. The kinetic constants were determined as described previously by Hawthorne et al. [23]. The steady-state inhibition constant (Ki), the first-order rate constant (ki) and the inhibitor rate constant (ki/Ki) were determined using the Grafit Data Analysis and Graphics program.
TMPRSS2 activation of PAR-2
Cells were grown at a concentration of 1×105 cells/ml, 100 μl/well, on a 96-well plate until confluent. Calcium-mobilization assays were performed as previously described by Hawthorne et al. [25]. PAR-2 was activated with 100 ng of sequencing-grade trypsin (Sigma), 3 μg of TMPRSS2 protein (40% active) or 50 μM PAR-2 activating peptide, SLIGKV. For inhibition studies, 10 μM PAR-1 antagonist, N-trans-cinnamoyl-p-fluoro-Phe-p-guanidinoPhe-Leu-Arg-NH2 [26], 200 μM PAR-2 antagonist, Phe-Ser-Leu-Leu-Arg-Tyr-NH2 [27], 100 μM Cbz-Lys(OPh)2 [23], 5 mM suramin (Calbiochem, Nottingham, U.K.) or 100 μM 2-APB (2-aminoethoxydiphenyl borate; Calbiochem) were used. Inhibitors were added before activation of PAR-2. PAR-1 and PAR-2 antagonists and PAR-2 agonist were synthesized using Fmoc chemistry as described by Walker [21].
Fluorescence was measured on a Cytofluor multiwell plate reader (PE Biosystems) at 516 nm (with excitation at 494 nm).
RESULTS AND DISCUSSION
TMPRSS2 mRNA expression
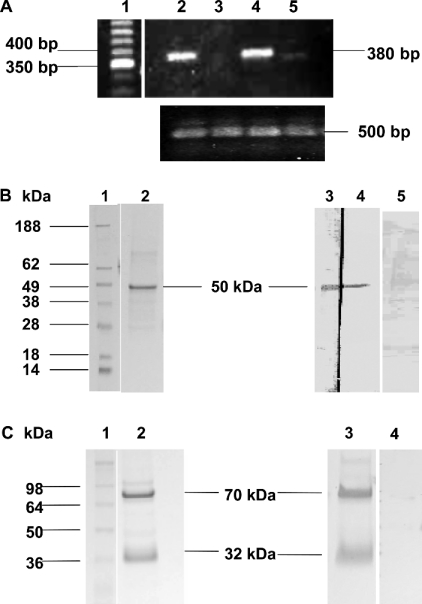
To detect the expression of TMPRSS2 mRNA in various prostate cancer cell lines (LNCaP, PC-3 and DU 145) and the non-cancerous transformed prostate cell line (RWPE-1), RT-PCR was performed on mRNA derived from these cell lines. Specific primers for human TMPRSS2 were used to generate cDNA products, which were visualized using an ethidium-bromide-containing gel and confirmed by sequencing. Results showed that high expression levels of TMPRSS2 3.8 kb transcript were detected in both the androgen-dependent LNCaP and androgen-independent PC-3 cell lines (Figure 1A), with very low levels found in RWPE-1, the non-malignant cell line. The prostate cancer cell line DU 145 showed no expression of TMPRSS2 mRNA. The presence of PAR1–4 mRNA in LNCaP cells has been shown previously by Wilson et al. [18]. The higher expression of TMPRSS2 mRNA in two out of the three prostate cancer cell lines compared with the normal prostate cell line is in accordance with its reported up-regulation in prostate cancer [7,9].
Figure 1. Detection of TMPRSS2 by RT-PCR, SDS/PAGE and Western blotting.
(A) RT-PCR analysis of TMPRSS2 mRNA distribution in PC-3, DU 145 and LNCaP human prostate cell lines and RWPE-1 human transformed prostate cell line. Predicted size of reaction product for TMPRSS2 is 3.8 kb. Upper panel: Lane 1, 50 bp cDNA ladder; lane 2, PC-3 RT-PCR product (380 bp); lane 3, DU145 RT-PCR product (none detected); lane 4, LNCaP RT-PCR product (380 bp); lane 5, RWPE-1 RT-PCR product (380 bp). Lower panel: lane 2, actin control PC-3 RT-PCR product (500 bp); lane 3, actin control DU145 RT-PCR product (500 bp); lane 4, actin control LNCaP RT-PCR product (500 bp); lane 5, actin control RWPE-1 RT-PCR product (500 bp). (B) Immunoblot analysis of recombinant TMPRSS2 extracellular domain containing a T7 tag. Lane 1, See Blue Plus 2 Pre-stained Standard molecular mass markers. Samples (1 μg) of recombinant expressed TMPRSS2 were analysed by SDS/PAGE and protein stain (lane 2) or Western blot (lanes 3–5). Lane 3 was developed with polyclonal anti-TMPRSS2 antibody (1/500), lane 4 was developed with anti-T7 tag monoclonal antibody (1/10000). A major band was detected at approx. 50 kDa in both lanes. Lane 5 was developed with polyclonal anti-TMPRSS2 antibody (1/500) pre-absorbed with antigen before use. (C) SDS/PAGE analysis of cellular immunopurified TMPRSS2. Lane 1. molecular mass standards. TMPRSS2 (3.75 μg of total protein) was analysed by SDS/PAGE and protein stain (lane 2), and Western blot (lanes 3 and 4). Bands were developed using polyclonal anti-TMPRSS2 antibody (1/500). Lane 4 shows polyclonal anti-TMPRSS2 antibody pre-absorbed with TMPRSS2. Full-length TMPRSS2 is shown as a major band at 70 kDa, and the soluble extracellular protease domain at 32 kDa.
Anti-TMPRSS2 antibody generation and specificity
An anti-human TMPRSS2 polyclonal antibody was generated using an antigenic peptide derived from the serine protease domain of TMPRSS2 from Val246 to Ala262. This antibody was then used for TMPRSS2 immunopurification. The specificity of the antibody was demonstrated using a purified recombinantly expressed extracellular domain of TMPRSS2 containing a T7 tag at the N-terminus.
The soluble extracellular domain of TMPRSS2 (47 kDa) lacking the transmembrane domain was expressed in bacteria purified and analysed by SDS/PAGE. Protein staining revealed a major band for the truncated TMPRSS2 at 50 kDa (Figure 1B). Western blotting and probing with both anti-TMPRSS2 antibody and anti-T7 tag antibody revealed that the 50 kDa was immunoreactive with the anti-TMPRSS2 polyclonal antibody and the anti-T7 antibody (Figure 1B) confirming the specificity of the anti-TMPRSS2 polyclonal antibody.
Immunopurification of TMPRSS2 from LNCaP cells
The TMPRSS2 serine protease was immunopurified from LNCaP cell lysates using the anti-TMPRSS2 polyclonal antibody and analysed by SDS/PAGE. Immunoreactive bands were detected at apparent molecular masses of 70 kDa and 32 kDa (Figure 1C). The molecular mass of the full-length TMPRSS2 protein was predicted by sequence analysis and in vitro translation to be 54 kDa; however, previous protein analysis has revealed it to be expressed in mammalian cells as a 70 kDa protein, with glycosylation being partly responsible for the difference [10]: TMPRSS2 has three potential N-glycosylation sites. The 32 kDa species is found with both in vitro translated TMPRSS2 and in cells and tissues expressing TMPRSS2. The 32 kDa species is the cleaved serine protease domain of TMPRSS2, identified previously by Afar et al. [10]. As has been shown with the other TTSPs, human MT-SP1 [28] and matriptase-2 [29], activation of the TMPRSS2 zymogen is autocatalytic, although the conditions under which the activation process occurs have yet to be elucidated.
Catalytic activity of immunopurified TMPRSS2
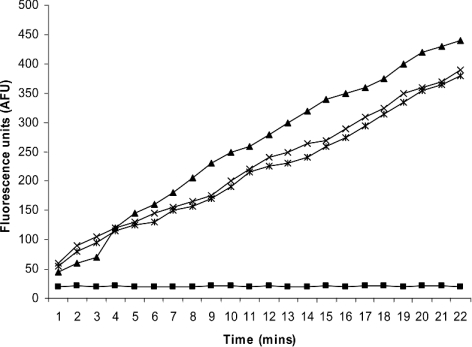
The activity of the purified TMPRSS2 was determined using the broad trypsin substrate Cbz-Gly-Gly-Arg-AMC. A 30 μl aliquot of TMPRSS2 containing 1.25 μg of active enzyme produced a change in fluorescence of over 400 AFU (arbitrary fluorescence units) over 22 min (Figure 2), 0.5 ng of trypsin produced a similar rate of substrate hydrolysis. The substrate hydrolysis produced by TMPRSS2 protease was completely inhibited by the addition of the trypsin inhibitor, Cbz-Lys(OPh)2 (10 μM). Addition of the PAR-2 synthetic antagonist (200 μM) did not affect the activity of TMPRSS2. The isolation, purification and preparation of TMPRSS2 probably compromised the enzyme structurally and catalytically, which is why it takes more than 1 μg to produce the same effect that is seen with 0.5 ng of trypsin. Under physiological situations, freshly secreted and activated TMPRSS2 is likely be active at nanogram levels, just like trypsin.
Figure 2. Cleavage of the substrate Cbz-Gly-Gly-Arg-AMC.
Cleavage of the substrate Cbz-Gly-Gly-Arg-AMC (100 μM) by 1.25 μg of active cellular immunopurified TMPRSS2 (×), 0.5 ng of trypsin (▲), 1.25 μg of active TMPRSS2 in the presence of 10 μM Cbz-Lys(OPh)2 (■) or 1.25 μg of active TMPRSS2 in the presence of 200 μM PAR-2 antagonist (Phe-Ser-Leu-Leu-Arg-Tyr-NH2) (*).
Active-site titration revealed that 1 μg of purified cellular TMPRSS2 contained 416 ng of active enzyme. The kinetic constants for TMPRSS2 inhibition by Cbz-Lys(OPh)2 were also calculated. The ki was calculated as 0.42 min−1, the Ki was 0.59 μM and the ki/Ki was 0.71 M−1·min−1. This shows that Cbz-Lys(OPh)2 is an effective inhibitor of TMPRSS2. These values can be compared with the kinetic constants for trypsin inhibition by Cbz-Lys(OPh)2, reported previously by Hawthorne et al. [23] as ki=0.14 min−1, Ki=12.1 μM and ki/Ki=12.0×103 M−1· min−1. Comparing the inhibitor rate constants for TMPRSS2 and trypsin shows that Cbz-Lys(OPh)2 is a better inhibitor for TMPRSS2 than it is for trypsin.
TMPRSS2 activation of PAR-2
The active site of TMPRSS2 contains the residues His296, Asp345 and Ser441. At the base of its S1 pocket, it contains Asp435, and therefore is predicted to cleave after lysine or arginine residues [1], giving it a trypsin-like specificity. PAR-2 is known to be activated by tryptase in vitro [30], and is endogenously activated by trypsin in the intestinal lumen [31].
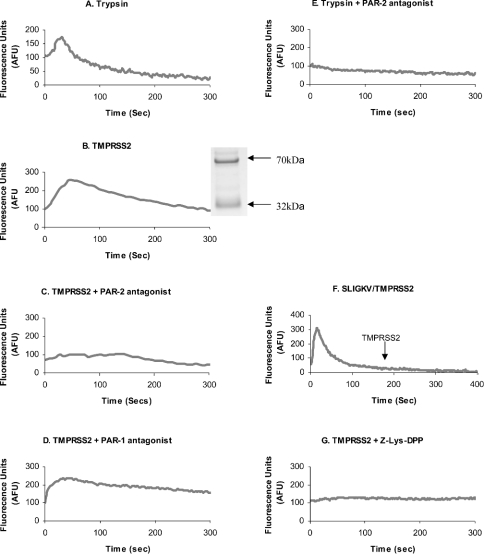
To determine whether TMPRSS2 could regulate PAR-2, active TMPRSS2 enzyme was isolated and used to treat PAR-2-containing prostate cancer cells; the activation of PAR-2 was monitored using a calcium-mobilization assay. PAR activation results in a rapid and transient increase in the intracellular calcium concentration which is detected using the calcium fluorophore, Fluo 4 [23]. As a positive control, 100 ng of trypsin, a potent PAR-2 activator was used (Figure 3A). Results showed that 1.25 μg of purified active TMPRSS2 produced a similar transient calcium peak after addition to cells (Figure 3B), suggesting that cellular PARs are being activated by TMPRSS2. To determine whether calcium mobilization occurred via PAR-1 or PAR-2 activation, specific antagonists were used to block each receptor. Cells were incubated with a PAR-1 antagonist (10 μM) or a PAR-2 antagonist (200 μM) for 5 min before enzyme treatments were initiated. TMPRSS2 produced calcium peaks in the presence of the PAR-1 antagonist (Figure 3D), but after the addition of the PAR-2 antagonist no calcium peak was manifested (Figure 3C). The addition of both PAR antagonists resulted in no calcium mobilization after TMPRSS2 addition (results not shown). In Figure 2, it could be seen that TMPRSS2 activity was unaffected by the presence of the PAR2 antagonist, therefore the PAR2 antagonist is not exerting its effect via inhibition of TMPRSS2 enzyme. The same concentration of PAR-2 antagonist was also effective in blocking calcium release by 100 ng of trypsin (Figure 3E). Since blocking PAR-1 has no suppressive effect on the TMPRSS2-dependent calcium mobilization, it is evidently not responsible for the calcium peaks described. This was confirmed further by activating PAR-2 with 50 μM SLIGKV, a specific PAR-2 activating peptide, which desensitizes the cell to PAR-2 activation. Subsequent addition of TMPRSS2 resulted in no calcium mobilization, therefore TMPRSS2 does not appear to activate any other PARs (Figure 3F). Complete inhibition of calcium mobilization occurred when PAR-2 was blocked by either antagonist binding or receptor desensitization, thus PAR-4 and PAR-3 are unlikely to be involved. This implies that PAR-2 is responsible for TMPRSS2-dependent calcium mobilization. To confirm that the catalytic activity of TMPRSS2 was responsible for PAR activation, the enzyme was pre-incubated with the irreversible active-site trypsin-like inhibitor [Cbz-Lys(OPh)2] shown previously to inactivate TMPRSS2 (Figure 2). Figure 3(G) shows that loss of TMPRSS2 catalytic activity results in no calcium mobilization.
Figure 3. Intracellular calcium mobilization via activation of PAR-2 on LNCaP cells.
Cells were loaded with Fluo 4 and protease-triggered calcium release was assessed. Cells were treated with 100 ng of trypsin (A), 1.25 μg of soluble TMPRSS2 (B) (the inset shows a sample of immunopurified TMPRSS2 used for activation of PAR-2 analysed by SDS/PAGE from Figure 1C, with molecular mass sizes indicated in kDa), TMPRSS2 in the presence of PAR-2 antagonist (200 μM) (C) and PAR-1 antagonist (10 μM) (D), 100 ng of trypsin in the presence of PAR-2 antagonist (200 μM) (E), TMPRSS2 addition to cells 180 s after a PAR-2-desensitizing treatment of 50 μM SLIGKV (F), and TMPRSS2 pre-incubated with the irreversible serine protease inhibitor Cbz-Lys(OPh)2 (10 μM) (G). Treatments were added at zero-time, except in (F) when they were added at zero-time and at 180 s. Fluorescence was measured at 516 nm (with excitation at 494 nm).
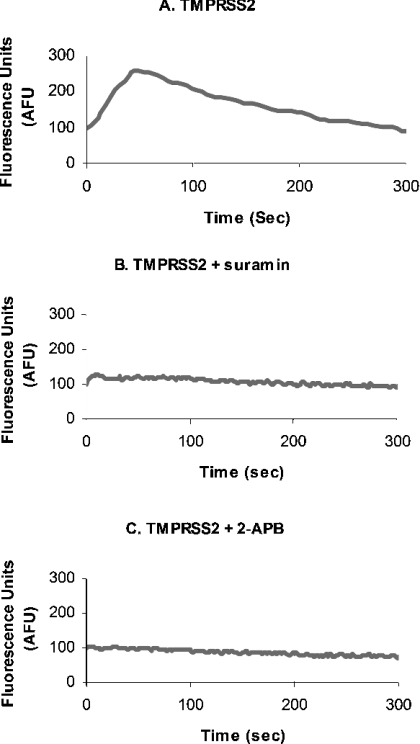
In order to determine whether G-proteins were involved in the activation of PAR-2 by TMPRSS2, cells were pre-treated with 5 mM suramin for 30 min before PAR activation. Suramin inhibits G-protein-coupled receptors by uncoupling the G-proteins from the receptor, preventing the downstream effects of G-protein activation, such as intracellular calcium mobilization. There was an inhibition of the TMPRSS2-dependent calcium peak following suramin treatment (Figures 4A and 4B). To confirm the intracellular increase was due to the release of calcium from intracellular stores and not from an influx of calcium from the medium, cells were pre-treated with the Ins(1,4,5)P3 receptor inhibitor, 2-APB (100 μM). This treatment also inhibited the calcium response and thus confirms that Ins(1,4,5)P3-regulated calcium stores within the cell are responsible for the release of calcium seen with G-protein-coupled-receptor activation (Figure 4C).
Figure 4. Intracellular calcium mobilization via activation of PAR-2 on LNCaP cells.
LNCaP cells were treated with TMPRSS2 (A) as before, and also pre-treated with 5 mM suramin (B) or 100 μM 2-APB before the addition of TMPRSS2 (C).
The TTSPs are a relatively newly discovered family of membrane-bound serine proteases, and their physiological functions and natural substrates have not yet been fully characterized. MT-SP1/matriptase, similar to TMPRSS2, possesses trypsin-like activity, as well as gelatinolytic activity [32], and has been implicated in tumour growth and metastasis. MT-SP1 also is capable of activating PAR-2 on oocytes [11]. More recently, HAT also has been shown to activate PAR-2 on human bronchial epithelial cells [12]. As TMPRSS2, in addition to being anchored on the cell membrane, is also released from the cell surface, it is likely to interact with molecules on the cell surface in an autocrine or paracrine manner. The PAR-2 receptor could provide a potential endogenous substrate for TMPRSS2. PAR-2 is activated by trypsin [33] and tryptase [34] in vitro, but these are unlikely to be the only endogenous activators, as they are frequently not expressed in tissues that express PAR-2 [35]. PARs have generated interest in recent years owing to their involvement in tumour progression and metastasis [2]. PAR-2 is expressed in prostate tissue, and its activation has been shown to increase levels of MMP (matrix metalloprotease)-2 and MMP-9 in prostate cancer cells [18]. These gelatinases are known to be key proteases enabling tumour cells to metastasize [36–38]. The above findings suggest that TMPRSS2 may have a role to play in the complex process surrounding prostate cancer progression.
Owing to the tissue-specific expression of TMPRSS2, and its location on the cell membrane, it provides a potential therapeutic target for prostate cancer. Treatment of invasive cancer depends increasingly more on therapeutic strategies for prevention and intervention before metastatic dissemination. TMPRSS2 is likely to have many physiological and pathological roles, giving it the potential to be more than a marker for prostate cancer. The present study provides an indication of just one potential function of TMPRSS2 in the process of prostate cancer.
Acknowledgments
S. W. was funded by DEL (Department of Employment and Learning), Northern Ireland. This work was also supported in part by the Wellcome Trust (Grant Reference 053002/HF/JPS).
References
- 1.Paoloni-Giacobino A., Chem H., Peitsch M. C., Rossier C., Antonarakis S. E. Cloning of the TMPRSS2 gene, which encodes a novel serine protease with transmembrane, LDLRA, and SRCR domains and maps to 21q22.3. Genomics. 1997;44:309–320. doi: 10.1006/geno.1997.4845. [DOI] [PubMed] [Google Scholar]
- 2.Del Rosso M., Fibbi G., Pucci M., D'Alessio S., Del Rosso A., Magnelli L., Chiarugi V. Multiple pathways of cell invasion are regulated by multiple families of serine proteases. Clin. Exp. Metastasis. 2002;19:193–207. doi: 10.1023/a:1015531321445. [DOI] [PubMed] [Google Scholar]
- 3.Cocks T. M., Moffatt J. D. Protease-activated receptors: sentries for inflammation? Trends Pharmacol. Sci. 2000;21:103–108. doi: 10.1016/s0165-6147(99)01440-6. [DOI] [PubMed] [Google Scholar]
- 4.Darmoul D., Gratio V., Devaud H., Lehy T., Laburthe M. Aberrant expression and activation of the thrombin receptor protease-activated receptor-1 induces cell proliferation and motility in human colon cancer cells. Am. J. Pathol. 2003;162:1503–1513. doi: 10.1016/S0002-9440(10)64283-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hooper J. D., Clements J. A., Quigley J. P., Antalis T. M. Type II transmembrane serine proteases: insights into an emerging class of cell surface proteolytic enzymes. J. Biol. Chem. 2001;276:857–860. doi: 10.1074/jbc.R000020200. [DOI] [PubMed] [Google Scholar]
- 6.Netzel-Arnett S., Hooper J. D., Szabo R., Madison E. L., Quigley J. P., Bugge T. H., Antalis T. M. Membrane anchored serine proteases: a rapidly expanding group of cell surface proteolytic enzymes with potential roles in cancer. Cancer Metastasis Rev. 2003;22:237–258. doi: 10.1023/a:1023003616848. [DOI] [PubMed] [Google Scholar]
- 7.Vaarala M. H., Porvari K., Kyllonen A., Lukkarinen O., Vihko P. The TMPRSS2 gene encoding transmembrane serine protease is overexpressed in a majority of prostate cancer patients: detection of mutated TMPRSS2 form in a case of aggressive disease. Int. J. Cancer. 2001;94:705–710. doi: 10.1002/ijc.1526. [DOI] [PubMed] [Google Scholar]
- 8.Lin B., Ferguson C., White J. T., Wang S., Vessella R., True L. D., Hood L., Nelson P. S. Prostate-localized and androgen-regulated expression of the membrane-bound serine protease TMPRSS2. Cancer Res. 1999;59:4180–4184. [PubMed] [Google Scholar]
- 9.Vaarala M. H., Porvari K., Kyllonen A., Vihko P. Differentially expressed genes in two LNCaP prostate cancer cell lines reflecting changes during prostate cancer progression. Lab. Invest. 2000;80:1259–1268. doi: 10.1038/labinvest.3780134. [DOI] [PubMed] [Google Scholar]
- 10.Afar D. E., Vivanco I., Hubert R. S., Kuo J., Chen E., Saffran D. C., Raitano A. B., Jakobovits A. Catalytic cleavage of the androgen-regulated TMPRSS2 protease results in its secretion by prostate and prostate cancer epithelia. Cancer Res. 2001;61:1686–1692. [PubMed] [Google Scholar]
- 11.Takeuchi T., Harris J. L., Huang W., Yan K. W., Coughlin S. R., Craik C. S. Cellular localization of membrane-type serine protease 1 and identification of protease-activated receptor-2 and single-chain urokinase-type plasminogen activator as substrates. J. Biol. Chem. 2000;275:26333–26342. doi: 10.1074/jbc.M002941200. [DOI] [PubMed] [Google Scholar]
- 12.Miki M., Nakamura Y., Takahashi A., Nakaya Y., Eguchi H., Masegi T., Yoneda K., Yasuoka S., Sone S. Effect of human airway trypsin-like protease on intracellular free Ca2+ concentration in human bronchial epithelial cells. J. Med. Invest. 2003;50:95–107. [PubMed] [Google Scholar]
- 13.MacFarlane S. R., Seatter M. J., Dande T., Hunter G. D., Plevin R. Proteinase-activated receptors. Pharmacol. Rev. 2001;53:245–282. [PubMed] [Google Scholar]
- 14.Li Y., Sarkar F. H. Down-regulation of invasion and angiogenesis-related genes identified by cDNA microarray analysis of PC3 prostate cancer cells treated with genistein. Cancer Lett. 2002;186:157–164. doi: 10.1016/s0304-3835(02)00349-x. [DOI] [PubMed] [Google Scholar]
- 15.Chay C. H., Cooper C. R., Gendernalik J. D., Dhanasekaran S. M., Chinnaiy A. M., Rubin M. A., Schmaier A. H., Pienta K. J. A functional thrombin receptor (PAR1) is expressed on bone-derived prostate cancer cell lines. Urology. 2002;60:760–765. doi: 10.1016/s0090-4295(02)01969-6. [DOI] [PubMed] [Google Scholar]
- 16.Greenberg D. L., Mize G. J., Takayama T. K. Protease-activated receptor mediated RhoA signalling and cytoskeletal reorganization in LNCaP cells. Biochemistry. 2003;42:702–709. doi: 10.1021/bi027100x. [DOI] [PubMed] [Google Scholar]
- 17.Cooper C. R., Chay C. H., Gendernalik J. D., Lee H.-L., Bhatia J., Taichman R. S., McCauley L. K., Keller E. T., Pienta K. J. Stromal factors involved in prostate carcinoma metastasis to bone. Cancer. 2003;97(suppl.):739–747. doi: 10.1002/cncr.11181. [DOI] [PubMed] [Google Scholar]
- 18.Wilson S. R., Gallagher S., Warpeha K., Hawthorne S. J. Amplification of MMP-2 and MMP-9 production by prostate cancer cell lines via activation of protease-activated receptors. Prostate. 2004;60:168–174. doi: 10.1002/pros.20047. [DOI] [PubMed] [Google Scholar]
- 19.Gopalkrishnan R. V., Kang D.-C., Fisher P. B. Molecular markers and determinants of prostate cancer metastasis. J. Cell. Physiol. 2001;189:245–256. doi: 10.1002/jcp.10023. [DOI] [PubMed] [Google Scholar]
- 20.Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 21.Walker B. Solid phase peptide synthesis. In: Wisdom G. B., editor. Peptide Antigens A Practical Approach. Oxford: IRL Press/Oxford University Press; 1994. pp. 27–81. [Google Scholar]
- 22.Tam J. P. Recent advances in multiple antigen peptides. J. Immunol. Methods. 1996;196:17–32. doi: 10.1016/0022-1759(96)00066-x. [DOI] [PubMed] [Google Scholar]
- 23.Hawthorne S., Hamilton R., Walker B. J., Walker B. Utilization of biotinylated diphenyl phosphonates for disclosure of serine proteases. Anal. Biochem. 2004;326:273–275. doi: 10.1016/j.ab.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 24.Barrett A. J., Kirschke H. Cathepsin B, cathepsin H, and cathepsin L. Methods Enzymol. 1981;80:C535–C561. doi: 10.1016/s0076-6879(81)80043-2. [DOI] [PubMed] [Google Scholar]
- 25.Hawthorne S. J., Macey M., Howells G., Walker B. A high-throughput microtiter plate-based calcium assay for the study of protease-activated receptor 2 activation. Anal. Biochem. 2001;290:378–379. doi: 10.1006/abio.2001.4964. [DOI] [PubMed] [Google Scholar]
- 26.Bernatowicz M. S., Klimas C. E., Hartl K. S., Peluso M., Allegretto N. J., Seiler S. M. Development of potent thrombin receptor antagonist peptides. J. Med. Chem. 1996;39:4879–4887. doi: 10.1021/jm960455s. [DOI] [PubMed] [Google Scholar]
- 27.Al-Ani B., Saifeddine M., Wijesuriya S. J., Hollenberg M. D. Modified proteinase-activated receptor-1 and -2 derived peptides inhibit proteinase-activated receptor-2 activation by trypsin. J. Pharmacol. Exp. Ther. 2002;300:702–708. doi: 10.1124/jpet.300.2.702. [DOI] [PubMed] [Google Scholar]
- 28.Takeuchi T., Shuman M. A., Craik C. S. Reverse biochemistry: use of macromolecular protease inhibitors to dissect complex biological processes and identify a membrane-type serine protease in epithelial cancer and normal tissue. Proc. Natl. Acad. Sci. U.S.A. 1999;96:11054–11061. doi: 10.1073/pnas.96.20.11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Velasco G., Cal S., Quesada V., Sanchez L. M., Lopez-Otin C. Matriptase-2, a membrane-bound mosaic serine proteinase predominantly expressed in human liver and showing degrading activity against extracellular matrix proteins. J. Biol. Chem. 2002;277:37637–37646. doi: 10.1074/jbc.M203007200. [DOI] [PubMed] [Google Scholar]
- 30.Molino M., Barnathan E. S., Numerof R., Clark J., Dreyer M., Cumashi A., Hoxie J. A., Schechter N., Woolkalis M., Brass L. F. Interactions of mast cell tryptase with thrombin receptors and PAR-2. J. Biol. Chem. 1997;272:4043–4049. doi: 10.1074/jbc.272.7.4043. [DOI] [PubMed] [Google Scholar]
- 31.Kong W., McConalogue K., Khitin L. M., Hollenberg M. D., Payan D. G., Bohm S. K., Bunnett N. W. Luminal trypsin may regulate enterocytes through proteinase-activated receptor 2. Proc. Natl. Acad. Sci. U.S.A. 1997;94:8884–8889. doi: 10.1073/pnas.94.16.8884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi Y. E., Torri J., Yieh L., Wellstein A., Lippman M. E., Dickson R. B. Identification and characterisation of a novel matrix-degrading protease from hormone-dependent human breast cancer cells. Cancer Res. 1993;53:1409–1415. [PubMed] [Google Scholar]
- 33.Nystedt S., Emilsson K., Wahlestedt C., Sundelin J. Molecular cloning of a potential protease activated receptor. Proc. Natl. Acad. Sci. U.S.A. 1994;91:9208–9212. doi: 10.1073/pnas.91.20.9208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schechter N. M., Brass L. F., Lavker R. M., Jensen P. J. Reaction of mast cell proteases tryptase and chymase with protease activated receptors (PARs) on keratinocytes and fibroblasts. J. Cell. Physiol. 1998;176:365–373. doi: 10.1002/(SICI)1097-4652(199808)176:2<365::AID-JCP15>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 35.Hollenberg M. D. Protease-mediated signalling: new paradigms for cell regulation and drug development. Trends Pharmacol. Sci. 1996;17:3–6. doi: 10.1016/0165-6147(96)81562-8. [DOI] [PubMed] [Google Scholar]
- 36.Stearns M., Stearns M. E. Evidence for increased activated metalloproteinase 2 (MMP-2a) expression associated with human prostate cancer progression. Oncol. Res. 1996;8:69–75. [PubMed] [Google Scholar]
- 37.Lokeshwar B. L. MMP inhibition in prostate cancer. Ann. N.Y. Acad. Sci. 1999;878:271–289. doi: 10.1111/j.1749-6632.1999.tb07690.x. [DOI] [PubMed] [Google Scholar]
- 38.Egeblad M., Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat. Rev. Cancer. 2002;2:161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]