Abstract
RNases P and MRP are ribonucleoprotein complexes involved in tRNA and rRNA processing, respectively. The RNA subunits of these two enzymes are structurally related to each other and play an essential role in the enzymatic reaction. Both of the RNAs have a highly conserved helical region, P4, which is important in the catalytic reaction. We have used a bioinformatics approach based on conserved elements to computationally analyze available genomic sequences of eukaryotic organisms and have identified a large number of novel nuclear RNase P and MRP RNA genes. For MRP RNA for instance, this investigation increases the number of known sequences by a factor of three. We present secondary structure models of many of the predicted RNAs. Although all sequences are able to fold into the consensus secondary structure of P and MRP RNAs, a striking variation in size is observed, ranging from a Nosema locustae MRP RNA of 160 nt to much larger RNAs, e.g. a Plasmodium knowlesi P RNA of 696 nt. The P and MRP RNA genes appear in tandem in some protists, further emphasizing the close evolutionary relationship of these RNAs.
INTRODUCTION
RNases P and MRP are ribonucleoprotein complexes that are involved in RNA processing (1). RNase P is found in all kingdoms of life and endonucleolytically cleaves a pre-tRNA to generate the mature 5′ end of the tRNA. MRP is found only in eukaryotes. It has been shown to be important for rRNA processing at a specific site, A3, cleaving pre-rRNA to release 5.8S rRNA (2–5). MRP probably has additional functions. For example, it was recently shown that RNase MRP cleaves the CLB2 mRNA in yeast (6,7). MRP RNA is associated with the genetic disease cartilage hair hypoplasia (8).
Some of the protein subunits found in the RNase P from yeast and man are also found in the MRP counterpart (1). Furthermore, the P and MRP RNA components may be folded into very similar secondary structures (9). For these reasons, it is likely that RNases P and MRP are evolutionary related and that the RNA subunits have a common ancestor.
The RNA subunit plays an essential role in the enzymatic reaction. In bacterial RNase P, the RNA component is catalytically active without any protein component (10). In eukaryotes and Archaea, one or more protein subunits are required for catalysis (11). Secondary structure models have been presented for the bacterial P RNA and they are similar to models for eukaryotic P RNA and MRP RNA (Figure 1) (9). The bacterial P RNA is organized into two different domains, one catalytic domain and one specificity (S) domain known to bind pre-tRNA substrates. Also, the eukaryotic RNAs seem to have a similar organization (domains 1 and 2 in Figure 1, where domain 1 is the catalytic domain). The structure of a bacterial P RNA S domain was recently solved (12,13) and a low-resolution model of the bacterial catalytic domain has been presented (14).
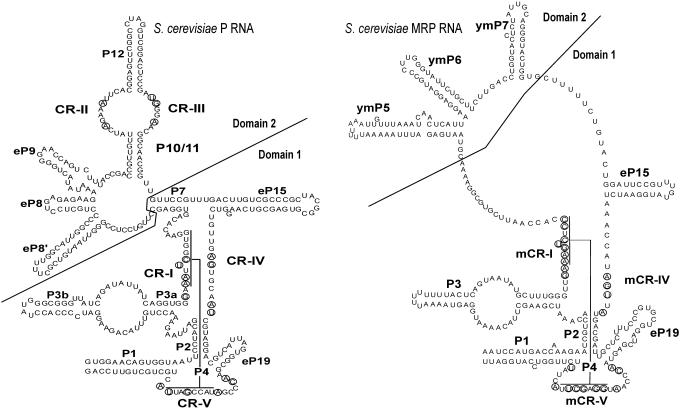
Figure 1.
Models of S.cerevisiae RNase P and MRP RNAs. Nucleotides that are conserved in all known P and MRP RNAs, respectively, are circled. P RNA is labeled as described previously by Frank et al. (44). CR-I through CR-V are conserved regions. CR-I and CR-V form the helix P4 as indicated. CR-II, CR-III and CR-IV have consensus sequences AGARA, UGNNA and AGNNNNAU, respectively. The nomenclature for the helices P1, P2, P3, P4, P7, P10/11 and P12 is based on homologous bacterial counterparts. In the case of the helices eP8, eP9, eP15 and eP19 the homology to the bacterial counterparts is tentative only. In addition to commonly accepted nomenclature, the helix 5′ of eP8 is here labeled as eP8′. MRP RNA is labeled in a manner similar to P RNA using the nomenclature in Li et al. (15). The structure includes helices ymP5, ymP6 and ymP7 shown to be present in yeast MRP RNA. Conserved regions labeled mCR-I, mCR-IV and mCR-V are counterparts to the CRs in P RNA. mCR-IV has the consensus sequence AGNNA. Figure is in part based on drawings in Walker and Avis (41), although the potential P7 interactions in MRP RNA are not shown here. For details as to CR-I, CR-V, mCR-I and mCR-V see also legend to Figure 2.
A number of secondary structure elements are shared between P and MRP RNAs (Figure 1). The P4 helix, formed by pairing of bases from the regions CR-I and CR-V (Figure 1), is essential for catalytic activity and is probably an important part of the enzyme active site. Very similar P4 helices may be formed in P and MRP RNAs (15) and the sequences of the regions CR-I and CR-V are conserved in evolution. Another conserved region in domain 1 is CR-IV (Figure 1) with the consensus sequence AGNNNNA for P RNA and AGNNA for MRP RNA.
There are also conserved elements in the domain 2 of P RNA, CR-II and CR-III (Figure 1). In many species, the CR-II sequence is AGARA. Mutational analysis of CR-II in yeast indicates that this region of the RNA is important in catalytic efficiency (16). However, in bacteria, the corresponding CR-II/CR-III domain is not essential for catalysis, rather it plays a role in substrate discrimination (17–21). In contrast to P RNA, MRP RNA does not appear to have any conserved sequence motifs in domain 2.
RNase P and MRP genes, as many other non-coding RNAs, are poorly annotated in genome projects. As a result, for many species we lack sequences of P and MRP RNAs. In part, this is because no effective methods have been developed for their identification from genomic sequences. In particular, MRP RNA has been poorly documented, and we only know sequences from yeasts, a few vertebrates and Arabidopsis. To remedy this situation, we have made an extensive inventory of both P and MRP RNAs. We present here a number of putative novel genes resulting from an analysis of available genome sequence data. To do this, we have made use of methods that we previously used to identify SRP RNA genes that are based on pattern searches and covariance models (22,23). We have also developed a method based on the conserved features of the CR-I and CR-V regions. The novel P and MRP RNA sequences should significantly improve our understanding of the structure and evolution of RNase P and MRP.
MATERIALS AND METHODS
Sources of genomic sequences
Genomic sequences were obtained from NCBI (www.ncbi.nlm.nih.gov/entrez/, ftp.ncbi.nih.gov/genomes/), EMBL (www.ebi.ac.uk) and ENSEMBL (www.ensembl.org). We also made use of TraceDB (ftp.ncbi.nlm.nih.gov/pub/TraceDB) with shotgun reads from genome sequencing projects, including different strains of Drosophila. Plasmodium sequences were from PlasmoDB at www.plasmodb.org.
We have received permission from TIGR to use genome data of Trypanosoma cruzi, Entamoeba histolytica, Theileria parva, Toxoplasma gondii and Trichomonas vaginalis.
Cryptosporidium hominis and Cryptosporidium parvum genome sequences were from http://cryptodb.org/. Leishmania species were from the Sanger centre (www.sanger.ac.uk). Chlamydomonas reinhardtii sequences were obtained at genome.jgi-psf.org/chlre2/chlre2.home.html and Giardia lamblia at jbpc.mbl.edu/Giardia-HTML/index2.html. For Saccharomyces P and MRP identification, we made also use of the Saccharomyces database (http://www.yeastgenome.org/).
Software
Multiple alignments of RNA primary sequences were made using ClustalW (24) and in some cases formatted using BOXSHADE (http://www.isrec.isb-sib.ch/ftp-server/boxshade/3.3.1/). RNA secondary structure predictions were carried out by MFOLD (25) or RNAALIFOLD (26). Sequence logos were created using software from Weblogo (27).
Prediction of P and MRP RNA genes using pattern matching and covariance models
RNA genes were predicted as described previously (22,23) by applying a combination of pattern searches using rnabob (www.genetics.wustl.edu/eddy/software/#rnabob) and cmsearch of the Infernal package (28,29). The rnabob searches made use of descriptors based on consensus features of the P and MRP RNAs. Descriptors included the CR-I, CR-IV and CR-V motifs as well as base-pairing rules consistent with the helix P2.
Examples of patterns used for P RNA are for CR-I GNAANNUCNGNG and for CR-V ACNNNAUNCNGANNA. For MRP RNA, GAAANUCNCCG was used for CR-I and ACNNNANGGNGCUNA was used for CR-V. When a P or MRP RNA gene was not found using these patterns new searches were carried out where mismatches were allowed.
Prediction of P and MRP RNA using hidden Markov models (HMMs) of P4 motifs
The CR-I and CR-V motifs from available P and MRP RNA sequences were extracted. Whenever a phylogenetic group was clearly overrepresented in the dataset we removed members from that family. The list of organisms and sequences remaining in the dataset is shown in the Web Supplement at http://bio.lundberg.gu.se/p_mrp/. The CR-I and CR-V sequences from P and MRP RNA were used to make a multiple alignment with ClustalW and that alignment was in turn used as input to hmmbuild (default parameters) of the hmmer package (http://hmmer.wustl.edu). After calibration with hmmcalibrate (default parameters), the two models were combined into one file and used by hmmpfam to search genomic sequences. The result of hmmpfam was analyzed with a script to identify regions in the genomic sequences that contained the CR-I and CR-V motifs and where the distance between the two motifs was <3000 bases. The resulting regions were sorted on the basis of the mean of the CR-I and CR-V expect values.
RESULTS AND DISCUSSION
The RNA components of RNases P and MRP have a conserved structural design as shown in Figure 1. Domain 1 contains the ubiquitous P1, P2 and P3 helices as well as the CR-I, CR-IV and CR-V regions. The P4 helix is formed by pairing of elements from the CR-I and CR-V regions. Domain 2 in RNase P has conserved elements CR-II and CR-III, but such elements are apparently absent in MRP RNA.
To design a computational approach to identify P and MRP RNA genes in genomic sequences, the conserved elements may be used to make a pattern search as described previously for SRP RNA (22,23) and P RNA (30). Covariance models (22,31) of the two RNAs could also be used to search candidates identified in a pattern search or, as in Rfam, candidates resulting from a BLAST search to query all known members of the RNA family (29,32). We, however, found that pattern searches applied to P and MRP RNAs often produced a large number of false-positive hits. Furthermore, when matched against the covariance models in Rfam, true hits sometimes gave rise to very low score values. In particular, this was true for MRP RNA. This was a result of a bias in the sequences available to produce the Rfam covariance model, but also because this RNA is highly variable between species.
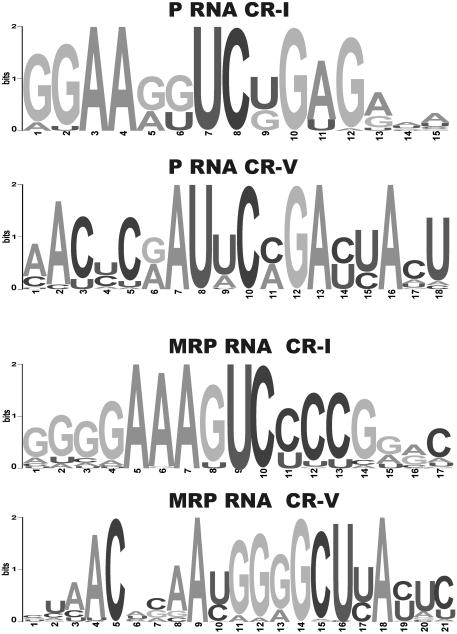
We, therefore, developed a complementary approach to identify P and MRP RNA genes. It is based on the observation that the CR-I and CR-V motifs are strongly conserved, both in P and MRP RNAs. The degree of conservation is shown in the sequence logos in Figure 2. We created an HMM model of available CR-I and CR-V sequences using the hmmer package (http://hmmer.wustl.edu) and used that model to search genomic sequences. The output from this search was analyzed to identify hits where CR-I and CR-V with low expect values occurred in close proximity to each other (see Materials and Methods). For all the RNAs presented in this work, the expect values were clearly below background values, typically two orders of magnitude. Advantages of this method are that large genomes may be searched quickly (100 Mb in a few minutes) and that the search in a highly specific manner identifies the P and MRP RNA genes.
Figure 2.
Sequence logos of the CR-I and CR-V regions of P and MRP RNA. Logos were created using multiple alignments of available P and MRP sequences and software from Weblogo (27). For multiple alignments see Web Supplement at http://bio.lundberg.gu.se/p_mrp/. P RNA CR-I and CR-V and MRP RNA CR-I and CR-V correspond to positions 1–12, 2–16, 4–13 and 4–22, respectively, in the sequence logos shown.
The candidates identified in the search based on HMM profiles were further analyzed to check that other conserved features of the RNA were present. Thus, we required base pairing between the CR-I and CR-V motifs, and we required the presence of CR-IV as well as the helices P1, P2 and P3. In the case of P RNA, the CR-II and CR-III motifs should also be present. All candidates from the HMM profile search with expect values significantly below background values did indeed meet these criteria.
The folding of the molecule was predicted using MFOLD (25). This algorithm does not take pseudoknots, such as the P4 helix into consideration. We, therefore, used MFOLD with constraints that did not allow the CR-I and CR-V to basepair. Furthermore, the CR-IV motif as well as the P RNA CR-II and CR-III motifs were not allowed to pair. With these constraints, MFOLD typically predicted P1, P2, P3 and eP19 helices associated with previously known P and MRP structures (Figure 1). However, because the domain 2 is highly variable, particularly for MRP RNA, we cannot be confident that MFOLD will predict the biologically relevant structure in this part. In the case of P RNA, we made use of CR-II, CR-III, P10/P/11 and P12 for structural prediction of domain 2, and for this reason the prediction of P RNA folding in this region is often more reliable than for MRP RNA. In cases where a good match to a covariance model of Rfam (29,32) was observed, MFOLD was used with constraints based on this match. The prediction of secondary structure is more reliable when several sequences are available from closely related species. In such cases, we also used RNAALIFOLD (26), using as input a multiple alignment produced by ClustalW (24). It should be kept in mind that many of the structures that we present at this stage are predicted by programs, such as MFOLD and RNAALIFOLD, and are not to expected to be as exact as careful inspection of sequences and alignments including covariation analysis.
To predict P and MRP RNA genes, we analyzed a range of organisms where a substantial amount of genomic sequence data is available (Table 1). In the vast majority of organisms, we found a P or MRP candidate. All sequences as well as secondary structure models for most sequences are shown in the Web Supplement to this paper at http://bio.lundberg.gu.se/p_mrp/. Mainly novel sequences are listed in Table 1. As sources for previously known P and MRP RNA sequences, we used the RNase P database (33), Rfam (29), NONCODE (34) and Entrez queries in GenBank (35).
Table 1.
Novel P and MRP RNAs
| P RNA | MRP RNA | |
|---|---|---|
| Diplomonads | ||
| Giardia lamblia | +* | |
| Parabasalidea | ||
| Trichomonas vaginalis | +(2)* | + |
| Dictyosteliida | ||
| Dictyostelium discoideum | +* | + |
| Entamoebidae | ||
| Entamoeba histolytica | + | |
| Alveolata | ||
| Theileria annulata | + | + |
| Babesia bovis | + | + |
| Plasmodium falciparum | + | + |
| Plasmodium berghei | + | + |
| Plasmodium chabaudi | + | |
| Plasmodium knowlesi | + | + |
| Plasmodium yoelii | + | + |
| Plasmodium vivax | + | + |
| Plasmodium reichenowi | + | |
| Plasmodium gallinaceum | + | + |
| Eimeria tenella | + | + |
| Toxoplasma gondii | + | + |
| Oxytricha trifallax | + | |
| Tetrahymena thermophila | + | + |
| Cryptosporidium parvum | + | + |
| Cryptosporidium hominis | + | + |
| Heterokonta | ||
| Phytophthora ramorum | + | |
| Phytophthora sojae | + | |
| Thalassiosira pseudonana | + | |
| Red algae | ||
| Cyanidioschyzon merolae | + | |
| Green algae | ||
| Chlamydomonas reinhardtii | + | |
| Volvox carteri | + | |
| Plants | ||
| Arabidopsis thaliana | +(2) | |
| Oryza sativa | + | |
| Insects | ||
| Drosophila melanogaster | +(P)* | + |
| Drosophila mojavensis | + | + |
| Drosophila virilis | + | |
| Drosophila ananassae | + | + |
| Drosophila erecta | + | + |
| Drosophila simulans | + | + |
| Drosophila yakuba | + | + |
| Drosophila willistoni | + | + |
| Drosophila pseudoobscura | + | + |
| Anopheles gambiae | + | + |
| Aedes aegypti | + | |
| Apis mellifera | + | + |
| Tribolium castaneum | + | |
| Urochordata | ||
| Ciona savignyi | + | + |
| Cephalochordata | ||
| Branchiostoma floridae | +(2) | |
| Echinodermata | ||
| Strongylocentrotus purpuratus | + | |
| Nematodes | ||
| Brugia malayi | + | + |
| Caenorhabditis briggsae | + | |
| Caenorhabditis remanei | + | |
| Vertebrates | ||
| Danio rerio | +(P) | + |
| Fugu rubripes | + | + |
| Tetraodon negroviridis | + | + |
| Loxodonta africana | + | |
| Monodelphis domestica | + | |
| Canis familiaris | + | + |
| Macaca mulatta | (P) | + |
| Fungi, Ascomycota and Saccharomycetes | ||
| Saccharomyces paradoxus | + | + |
| Saccharomyces mikatae | + | + |
| Saccharomyces kudriavzevii | + | + |
| Candida glabrata | + (P) (L) | + |
| Kluyveromyces waltii | + | + |
| Eremothecium gossypii | + | + |
| Debaryomyces hanseni | (P) | + |
| Candida albicans | + | + (L) |
| Candida dubliniensis | + | + (L) |
| Candida tropicalis | + | + |
| Yarrowia lipolytica | + | + |
| Pichia guilliermondi | + (P) | + |
| Pichia angusta | + | |
| Fungi, Ascomycota and Pezizomycotina | ||
| Aspergillus nidulans | + | + |
| Aspergillus fumigatus | + | + |
| Coccidioides immitis | + | + |
| Histoplasma capsulatum | + | + |
| Uncinocarpus reesii | + | + |
| Neurospora crassa | +(2) | + |
| Podospora anserina | + | + |
| Magnaporthe grisea | + | |
| Fusarium graminearum (G.zeae) | + | + |
| Trichoderma reesei | + | + |
| Botrytis cinerea | + | |
| Fungi and Basidiomycota | ||
| Coprinus cinereus | + | + |
| Phanerochaete chrysosporium | +(L) | + |
| Laccaria bicolor | + | + |
| Microsporidia | ||
| Nosema (Antonospora) locustae | + | + |
| Encephalitozoon cuniculi | + | + |
RNase P and MRP RNAs identified in this work are indicated (+). In certain organisms, we found two RNA gene candidates [+(2)]. Sequences that are previously known from the RNase P database (33) or Rfam are marked with ‘(P)’. The sequences indicated by an asterisk were recently independently identified and experimentally verified by Marquez et al. (36). In some cases, an unusually long (L) RNA sequence was observed. See text for further details.
In organisms where we failed to identify a gene we cannot exclude that the gene will be found once the genome is fully sequenced. However, in the case of plants, none of the genomes reveals a P RNA candidate. It, therefore, seems likely that if there is a P RNA gene in that group encoded by the nuclear genome it must be very different from previously known genes of this kind. Similarly, in the case of the Trypanosomatids Trypanosoma brucei, T.cruzi, Leishmania major and Leishmania infantum, we have so far failed to identify a P or MRP RNA. This would suggest that these genes, if they exist, are different in this phylogenetic group.
As a rule, the P and MRP RNA genes of a particular organism appear in different locations in the genome. However, in Theileria annulata, Babesia bovis, Eimeria tenella and T.gondii the two genes appear in tandem with a spacer of ∼50 nt. Interestingly, the P and MRP RNA sequences in B.bovis and T.gondii are more similar as compared with P/MRP RNA pairs in other organisms (for details see Web Supplement). These observations further emphasize the close evolutionary relationship of P and MRP RNA.
In some of the RNAs identified in this work, the domain 2 is very large as compared with that of previously described P or MRP RNAs. Similarly, some of the helices in domain 1 are in some cases much longer than normal. We cannot exclude that some of these sequences are part of introns. However, with the possible exception of the very large Phanerochaete P RNA and Candida MRP RNAs referred to below, we find this unlikely. In the following, we will assume that P and MRP RNA genes do not have introns and that the genomic sequence is colinear with the mature RNA sequence.
Although we have not tested whether all candidates predicted in this work are indeed expressed, we believe that the RNAs identified in this work are true P or MRP RNAs. This is based on the finding that they all have the consensus properties of such RNAs and the probability that such sequences would occur by chance only is negligible.
Furthermore, the predicted Dictyostelium P and MRP RNAs referred to below were shown to be expressed (Fredrik Söderbom, personal communication). In addition, at a final stage in the preparation of this paper several novel P RNAs were reported by Marquez et al. (36). P RNA sequences that we found from Giardia, Dictyostelium and Trichomonas were experimentally verified by these authors through PCR from genomic DNA. These results make us more confident that our methods to predict P and MRP RNA genes are reliable.
RNase P RNA
An RNase P gene was found in all the organisms as indicated in Table 1. The P RNA sequences vary considerably in length, from T.annulata of 216 nt to much larger RNAs as in Plasmodium knowlesi (696 nt).
In a majority of cases, P RNA seems to be encoded by a single copy gene. However, in T.vaginalis we found two gene candidates, one of which was previously identified (GenBank accession no. AY627758) (36). The two RNAs differ in only four positions. Two of the changes involve a compensatory base change in P3. Also in Neurospora crassa we found two P RNA sequences (for these examples see below and http://bio.lundberg.gu.se/p_mrp).
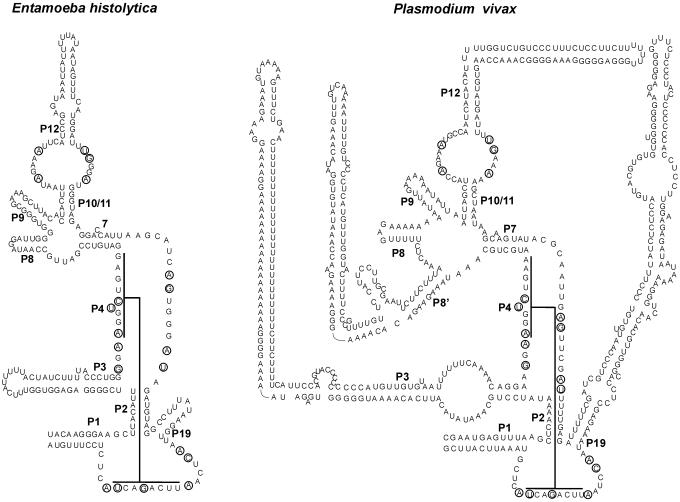
The P RNAs found here can all be folded into the consensus structure of eukaryotic P RNA. Representative structures are shown in Figure 3. (For more sequences and secondary structures see http://bio.lundberg.gu.se/p_mrp/). Eukaryotic P RNA contains helices denoted eP8 and eP9 because they seem to be the counterparts of the bacterial P8 and P9 helices. The current model of eukaryotic P RNA has an extra helix 5′ of the eP8 helix. This helix will herein be referred to as eP8′. As the three helices eP8′, eP8 and eP9 are highly variable in sequence and length it is not always possible to predict them reliably. Whenever possible we have made use of comparative structure analysis as well as covariance models obtained from Rfam to make more reliable predictions of the structure in this region.
Figure 3.
Secondary structure models of novel eukaryotic P RNAs. P RNAs shown are from Entamoeba histolytica and Plasmodium vivax. The helix numbering is as described previously by Frank et al. (44) and the extra helix present in P.vivax is labeled as P8′. As in Figure 1 interactions of CR-I and CR-V of the P4 helix are shown.
Insects and nematodes
Invertebrate P RNAs have not been described in any detail previously. Here we have identified a number of P RNAs from insects and worms and likely secondary structure models of these RNAs are presented at the Web Supplement site.
Genome sequence data for a number of Drosophila species are available (Drosophila melanogaster, Drosophila simulans, Drosophila pseudoobscura, Drosophila willistoni, Drosophila mojavensis, Drosophila erecta, Drosophila yakuba, Drosophila ananassae and Drosophila virilis) and we identified a P RNA gene in all of these. A secondary structure that is consistent with all these Drosophila sequences are shown at the Web Supplement site. This model is also consistent with P RNA sequences from Apis mellifera (honeybee), Anopheles gambiae (malaria mosquito) and Aedes aegypti (yellow fever mosquito). However, only the Drosophila RNAs have an eP8′ helix.
Previously, a P RNA sequence was identified in Caenorhabditis elegans (Rfam database). Here, we also identified homologs in Caenorhabditis briggsae and Brugia malayi.
Fungi and microsporidia
A number of yeast P RNAs from organisms closely related to Saccharomyces were identified. We also found a P RNA homolog in Eremothecium gossypii.
The P RNAs from Neurospora and Aspergillus all have a relatively long eP8′ helix. The proposed structure for one of the two Neurospora sequences that we found is shown at the Web Supplement site. In Aspergillus, the P3 helix has been extended as compared with other fungi. We also identified an RNA in Trichoderma reseii that has an extra helix in the eP8/eP8′ region.
In the Basidiomycota group, we found a P RNA in Coprinus cinereus, Phanerochaete and Laccaria. Interestingly, the Phanerochaete RNA is 1143 nt long and seems to have a large insertion in the eP15 region. However, the secondary structure of the insertion is very difficult to predict, as there are no closely related sequences to make a comparative approach possible. It is also interesting to note that in Candida glabrata a very long P RNA sequence has been identified (37).
The microsporidia Nosema (Antonospora) locustae and Encephalitozoon cuniculi P RNAs identified here conform to the consensus structure of P RNAs but they are relatively small. In particular, the helix 3 is very small and is lacking the internal loop characteristic of that helix in most other P RNAs. The ‘minimal’ structure of microsporidia RNAs will be discussed further below in the context of MRP RNA.
Other eukaryotic groups
Among the protists we identified a P RNA gene from E.histolytica (265 nt, Figure 3), T.gondii, E.tenella and Cryptosporidium. We also identified P RNA of G.lamblia (248 nt), T.vaginalis (251 nt), Dictyostelium (421 nt), T.annulata (216 nt) and B.bovis (230 nt) that were recently reported (36). To our knowledge, the RNA from T.annulata is the smallest P RNA found so far.
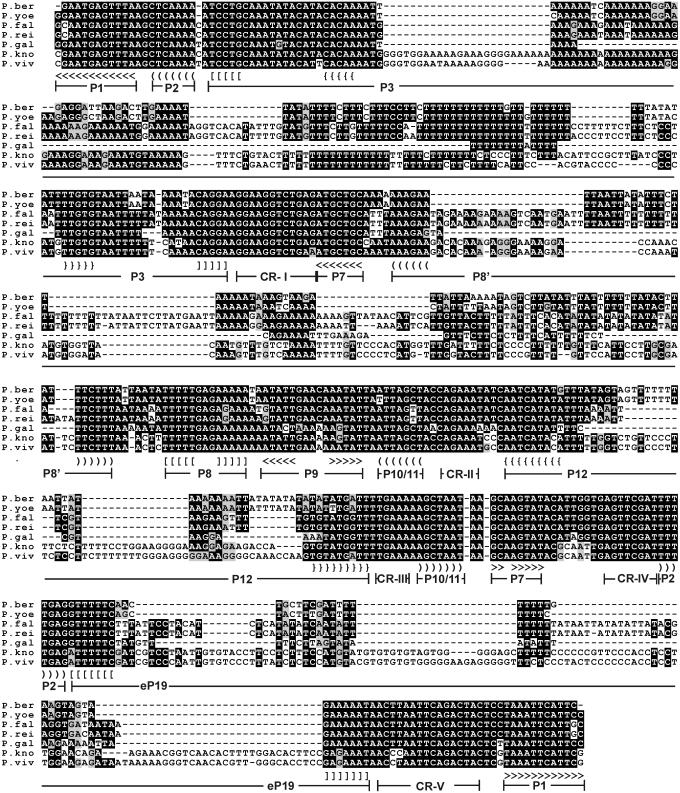
RNase P genes were also identified in a range of Plasmodium species that have been sequenced: Plasmodium gallinaceum, Plasmodium yoelii, Plasmodium berghei, Plasmodium falciparum, Plasmodium reichenowi, Plasmodium vivax (Figure 3) and Plasmodium knowlesi (38). They are larger than the other protist P RNAs and show a large variation in size with 387, 510, 506, 618, 618, 690 and 696 nt, respectively. They are all closely related such that once we identified one sequence with the HMM profile method described above the other homologs could be identified using BLAST. A multiple alignment of the sequences is shown in Figure 4. They are all very AU-rich, reflecting the very low GC content of these organisms. All seven sequences could be folded into the structure shown in Figure 3. It is striking that in Plasmodium the helices P3, eP8′, P12 and eP19 have grown considerably. This is particularly striking in P.vivax (Figure 3) and P.knowlesi. The P3 helix has U-rich regions matched by polypurine stretches on the other strand, as though the evolution of this helix has occurred through a replication slippage mechanism. The large degree of variation in sequence of P RNA in the different Plasmodium species has no equivalent in the evolution of MRP RNA sequences (see below).
Figure 4.
Multiple alignment of Plasmodium P RNAs. Organisms shown are P.berghei, P.yoelii, P.falciparum, P.reichenowi, P.gallinaceum, P.knowlesi and P.vivax. Alignment is based on a ClustalW alignment (45) using gap extension penalty = 0 and was manually edited in the P8′ region to achieve a better structural alignment. Consensus elements are highlighted with BOXSHADE. Positions of helices and conserved regions referred to in Figure 1 are indicated. Angular, curved, square and curly brackets indicate base pairing. Not all paired regions in P3, P8′, P12 and eP19 are shown.
We have so far not been able to identify a P RNA in heterokonta, plants and red algae, phylogenetic groups where we found an MRP RNA (Table 1, see below).
RNase MRP RNA
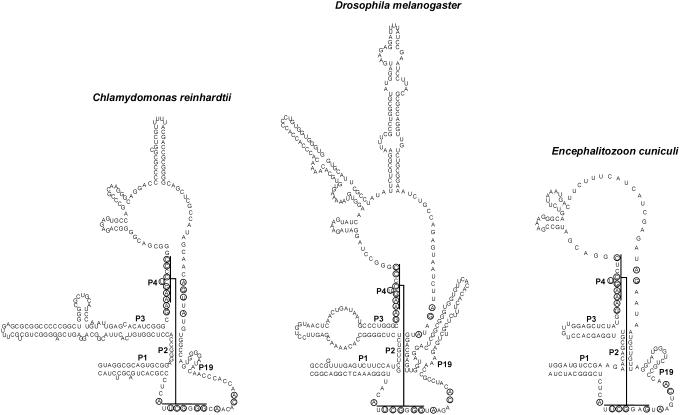
In the majority of organisms analyzed we found an MRP RNA gene as indicated in Table 1. Secondary structure models of representative examples of novel MRP RNAs are shown in Figure 5. A large degree of variation in domain 2 is observed. The smallest MRP RNAs found so far are those of N.locustae (160 nt) and E.cuniculi (165 nt). Some of the fungi RNAs are very large in comparison, e.g. Magnaporthe grisea homolog (567 nt).
Figure 5.
Secondary structure models of selected novel eukaryotic MRP RNAs. RNAs shown are from Chlamydomonas reinhardtii, D.melanogaster and Encephalitozoon cuniculi. Secondary structure of D.melanogaster was inferred from a comparative approach using available Drosophila sequences as described in the text.
As with P RNA, we typically identified only one MRP RNA gene in each organism. One exception is Arabidopsis where we found two genes.
Plants and red algae
An MRP RNA homolog was previously found in Arabidopsis thaliana (39). As mentioned above, we found an additional homolog in the same species (http://bio.lundberg.gu.se/p_mrp/). We also identified an MRP RNA gene in Oryza sativa, in the multicellular green algae Volvox carteri, in the unicellular green algae C.reinhardtii (Figure 5) and in the red algae Cyanidioschyzon merolae. These findings indicate that MRP RNA is ubiquitous in the plant group. All these sequences are consistent with the secondary structure shown for C.reinhardtii (Figure 5). It is interesting to note that no P RNAs were identified in the plants and red algae.
Heterokonta
In the heterokonta group, we found MRP RNA genes in Phytophthora (Phytophthora ramorum and Phytophthora sojae) and Thalassiosira pseudonana. The proposed secondary structure of the Phytophthora sequence is very similar to the vertebrate and plant MRP RNAs.
Vertebrates
Previously, MRP sequences have been described for man, mouse, rat, cow and Xenopus. Here, we have also identified MRP in Fugu rubripes, Danio rerio and Tetraodon negroviridis.
Insects and nematodes
As with P RNA, we found MRP RNA genes from different Drosophila species. We also identified homologs in A.mellifera and A.gambiae. We have not been able to detect an MRP RNA in C.elegans or in C.briggsae. However, an MRP RNA was identified in another nematode, B.malayi (http://bio.lundberg.gu.se/p_mrp/). The RNAs of Brugia as well as those of the insects adopt a structure very similar to vertebrate MRP RNAs. However, the Drosophila structure is unusual in that it seems to have an extended helix eP9 (Figure 5).
A comparison of the MRP RNAs from plants, red algae, heterokonta, vertebrates, insects and nematodes shows that they all may be folded into very similar structures as examplified by C.reinhardtii and D.melanogaster in Figure 5. As discussed below the fungi are different, particularly in domain 2.
Fungi and microsporidia
In the fungi group, MRP RNA was previously identified only in Saccharomyces and close relatives. We here identified MRP RNAs from several fungi groups. Interestingly, the predicted MRP RNA sequences from Candida albicans and Candida dubliniensis are extremely long (2226 and 1716 nt, respectively). Both of these RNAs are similar to other MRP RNAs in domain 1 but have a large insertion in domain 2.
We also identified MRP RNA in the Pezizomycotina group, e.g. in Aspergillus, Neurospora, Podospora and Magnaporthe. In these organisms, the domain 2 is very large, forming a long helical region. Furthermore, MRP RNA was identified in the Basidiomycota Coprinus, Laccaria and Phanerochaete. Here, the domain 2 seems to be built from three different helices but it is not clear whether they are related to the three helices of other MRP RNAs, or to the ymP5, ymP6 and ymP7 helices of Saccharomyces cerevisiae. All three Basidiomycota MRP RNAs can be folded as shown at the Web Supplement site but more sequences from this group are needed in order to reliably predict their secondary structure.
The microsporidia MRP RNAs are exceptionally small. As with P RNA, the P3 helix is very small as well as the entire domain 2. A secondary structure model for E.cuniculi is shown in Figure 5. By comparison, it is interesting to note that in the yeast MRP RNA all helices of domain 2 may be shortened significantly without effects on RNA processing (40). It would therefore seem that the microsporidia MRP RNAs represent ‘minimal’ forms of the RNA. This is consistent with the idea that microsporidia have been under a pressure to reduce their genome content, also reflected in a small average size of their proteins.
Other eukaryotic groups
A number of protist sequences were identified as indicated in Table 1 and they could all be folded into the structure characteristic of MRP RNA as shown for C.reinhardtii and D.melanogaster in Figure 5. The CR-IV consensus motif is AGNNA based on all other previously known sequences. We found here that E.tenella, T.gondii and certain Plasmodium species had a C or T in the fifth position of this motif. On the other hand, an A is found in these species one or two positions further downstream that might be functionally equivalent to the A found in the fifth position in most MRP RNAs. As with P RNA, we found a MRP candidate in many different Plasmodium species. In this way, we were able to more accurately predict the secondary structure of the Plasmodium RNAs. Compared with the P RNAs from the same Plasmodium species, the MRP sequences are much more conserved within the group.
Conserved elements of MRP RNA
As referred to above, the CR-I and CR-V motifs are the most strongly conserved in MRP RNA. As to the CR-IV motif, analysis of all available sequences shows that it has the consensus sequence ANAGNNA, where the three ‘A’s are the most strongly conserved.
Many of the MRP sequences found in this work may be folded according to a previously proposed model of mammalian MRP RNA (3), and the structures shown in Figure 5 include three helices in domain 2. We noted that the stem–loop structure in domain 2 that follows CR-I is conserved. In most eukaryotes, this loop has five bases with the consensus sequence ‘GARAR’, but in some phylogenetic groups it is a tetraloop with the consensus GARA (Web Supplement and Figure 5). Only in very few organisms, such as the Basidiomycota, we were not able to identify a corresponding motif. S.cerevisiae and Saccharomyces pombe also have a similar motif. However, a helix in this region conflicts with current models of yeast MRP, as there is evidence that this region of the RNA is either single-stranded (15) or involved in a P7-like stem (41).
Sequence conservation in the P and MRP RNA P3 helices
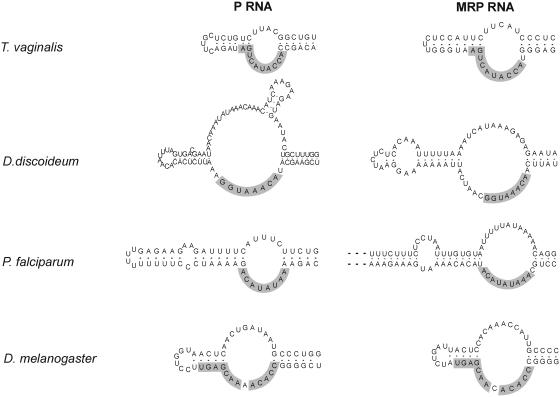
P and MRP RNA sequences from one organism typically share sequence elements in the P3 region, most often in the lower strand of the internal loop of P3. This was first observed for a number of yeast species and human (15). It has also been shown by experiments with the yeast RNAs that the P3 domains are functionally interchangeable (42). It is hypothesized that the P and MRP RNAs tend to coevolve in the P3 region because they share one or more protein subunits that interact with this region. The yeast Pop1p protein is one protein implicated in the binding to P3 (43).
Analysis of the novel RNAs identified in this work reveals notable cases of sequence conservation in P3 as observed for yeast and human. Examples for Trichomonas, Drosophila, Dictyostelium and Plasmodium are shown in Figure 6. As with yeast and human, conserved sequences are mainly observed in the lower strand of the interior loop. The only cases where we were not able to observe this kind of sequence similarity was for the microsporidia N.locustae and E.cuniculi. On the other hand, these RNAs have a shorter P3 helix and lack the interior loop. One may, therefore, speculate that these organisms are also different in terms of their RNase P and MRP protein subunits.
Figure 6.
Sequence conservation between P and MRP RNA P3 helices. Sequences identical in P and MRP RNA are shaded.
CONCLUSIONS
We used an efficient method to identify P and MRP RNAs, which is based on the conserved properties of the P4 helix of these RNAs. All candidates found also have other properties of these RNAs, such as the CR-IV motif and P1, P2 and P3 helices. It is therefore highly likely that the sequences that we present as a result of this work are bona fide P and MRP RNA genes.
We report for P and MRP RNA more than 100 novel sequences. By comparison, only 22 MRP RNA sequences are reported in Rfam and out of these 17 are of the Ascomycota branch. For P RNA, 56 are reported in Rfam, out of which 21 are from Ascomycota. Therefore, this investigation significantly increases the number of available P and MRP RNA sequences.
The RNA sequences now available will be helpful in understanding the structure and evolution of these RNAs. On the one hand, a comparison of all P and MRP sequences shows that all novel sequences may be folded into structures with the P1, P2, P3 and P4 helices characteristic of previously studied P and MRP RNAs. On the other hand, there is a large degree of variation in certain parts of the RNA. One striking example is the Plasmodium P RNA, where the helices P3, eP8′, P10-12 and eP19 are very different within the Plasmodium group. MRP RNAs show a large variation in domain 2. For example, Neurospora, Aspergillus and Magnaporthe have an extensive helical structure in this part whereas the microsporidia RNAs are extremely simple. We do not know the biological significance of this variation. It might be related to the protein subunits of the ribonucleoprotein complexes and for this reason it will be interesting to inventory the RNase P and MRP protein subunits in all the different organisms where we have found an RNA component.
In a majority of organisms, we have been able to identify both P and MRP RNA genes. Exceptions are the Trypanosomatids where we have not been able to find a P or MRP RNA. Furthermore, we have failed to identify P RNA in the plant and red algae groups. If P RNAs exist in this latter group they must be very different from previously known P and MRP RNAs.
Finally, we have observed that certain protists have their P and MRP RNA genes located in tandem. The P and MRP RNA sequences seem to be more similar in these organisms as compared with organisms where the genes are located separately. These findings are interesting from an evolutionary perspective and suggest that the RNAs might be even more closely related than previously anticipated.
Acknowledgments
P.P. was supported by the Swedish Research School of Genomics and Bioinformatics and M.A.R. by an EU grant QLK3-CT-200-00082. Funding to pay the Open Access publication charges for this article was provided by Magnus Bergvalls Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Xiao S., Scott F., Fierke C.A., Engelke D.R. Eukaryotic ribonuclease P: a plurality of ribonucleoprotein enzymes. Annu. Rev. Biochem. 2002;71:165–189. doi: 10.1146/annurev.biochem.71.110601.135352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lygerou Z., Mitchell P., Petfalski E., Seraphin B., Tollervey D. The POP1 gene encodes a protein component common to the RNase MRP and RNase P ribonucleoproteins. Genes Dev. 1994;8:1423–1433. doi: 10.1101/gad.8.12.1423. [DOI] [PubMed] [Google Scholar]
- 3.Schmitt M.E., Clayton D.A. Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 1993;13:7935–7941. doi: 10.1128/mcb.13.12.7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lygerou Z., Allmang C., Tollervey D., Seraphin B. Accurate processing of a eukaryotic precursor ribosomal RNA by ribonuclease MRP in vitro. Science. 1996;272:268–270. doi: 10.1126/science.272.5259.268. [DOI] [PubMed] [Google Scholar]
- 5.Chu S., Archer R.H., Zengel J.M., Lindahl L. The RNA of RNase MRP is required for normal processing of ribosomal RNA. Proc. Natl Acad. Sci. USA. 1994;91:659–663. doi: 10.1073/pnas.91.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gill T., Cai T., Aulds J., Wierzbicki S., Schmitt M.E. RNase MRP cleaves the CLB2 mRNA to promote cell cycle progression: novel method of mRNA degradation. Mol. Cell. Biol. 2004;24:945–953. doi: 10.1128/MCB.24.3.945-953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai T., Aulds J., Gill T., Cerio M., Schmitt M.E. The Saccharomyces cerevisiae RNase mitochondrial RNA processing is critical for cell cycle progression at the end of mitosis. Genetics. 2002;161:1029–1042. doi: 10.1093/genetics/161.3.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ridanpaa M., Ward L.M., Rockas S., Sarkioja M., Makela H., Susic M., Glorieux F.H., Cole W.G., Makitie O. Genetic changes in the RNA components of RNase MRP and RNase P in Schmid metaphyseal chondrodysplasia. J. Med. Genet. 2003;40:741–746. doi: 10.1136/jmg.40.10.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forster A.C., Altman S. Similar cage-shaped structures for the RNA components of all ribonuclease P and ribonuclease MRP enzymes. Cell. 1990;62:407–409. doi: 10.1016/0092-8674(90)90003-w. [DOI] [PubMed] [Google Scholar]
- 10.Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 11.Pace N.R., Brown J.W. Evolutionary perspective on the structure and function of ribonuclease P, a ribozyme. J. Bacteriol. 1995;177:1919–1928. doi: 10.1128/jb.177.8.1919-1928.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krasilnikov A.S., Xiao Y., Pan T., Mondragon A. Basis for structural diversity in homologous RNAs. Science. 2004;306:104–107. doi: 10.1126/science.1101489. [DOI] [PubMed] [Google Scholar]
- 13.Krasilnikov A.S., Yang X., Pan T., Mondragon A. Crystal structure of the specificity domain of ribonuclease P. Nature. 2003;421:760–764. doi: 10.1038/nature01386. [DOI] [PubMed] [Google Scholar]
- 14.Chen J.L., Nolan J.M., Harris M.E., Pace N.R. Comparative photocross-linking analysis of the tertiary structures of Escherichia coli and Bacillus subtilis RNase P RNAs. EMBO J. 1998;17:1515–1525. doi: 10.1093/emboj/17.5.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X., Frank D.N., Pace N., Zengel J.M., Lindahl L. Phylogenetic analysis of the structure of RNase MRP RNA in yeasts. RNA. 2002;8:740–751. doi: 10.1017/s1355838202022082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pagan-Ramos E., Lee Y., Engelke D.R. A conserved RNA motif involved in divalent cation utilization by nuclear RNase P. RNA. 1996;2:1100–1109. [PMC free article] [PubMed] [Google Scholar]
- 17.Mobley E.M., Pan T. Design and isolation of ribozyme-substrate pairs using RNase P-based ribozymes containing altered substrate binding sites. Nucleic Acids Res. 1999;27:4298–4304. doi: 10.1093/nar/27.21.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loria A., Pan T. Domain structure of the ribozyme from eubacterial ribonuclease P. RNA. 1996;2:551–563. [PMC free article] [PubMed] [Google Scholar]
- 19.Loria A., Pan T. Modular construction for function of a ribonucleoprotein enzyme: the catalytic domain of Bacillus subtilis RNase P complexed with B.subtilis RNase P protein. Nucleic Acids Res. 2001;29:1892–1897. doi: 10.1093/nar/29.9.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Massire C., Jaeger L., Westhof E. Derivation of the three-dimensional architecture of bacterial ribonuclease P RNAs from comparative sequence analysis. J. Mol. Biol. 1998;279:773–793. doi: 10.1006/jmbi.1998.1797. [DOI] [PubMed] [Google Scholar]
- 21.Fang X., Pan T., Sosnick T.R. A thermodynamic framework and cooperativity in the tertiary folding of a Mg2+-dependent ribozyme. Biochemistry. 1999;38:16840–16846. doi: 10.1021/bi991700n. [DOI] [PubMed] [Google Scholar]
- 22.Regalia M., Rosenblad M.A., Samuelsson T. Prediction of signal recognition particle RNA genes. Nucleic Acids Res. 2002;30:3368–3377. doi: 10.1093/nar/gkf468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosenblad M.A., Zwieb C., Samuelsson T. Identification and comparative analysis of components from the signal recognition particle in protozoa and fungi. BMC Genomics. 2004;5:5. doi: 10.1186/1471-2164-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]
- 26.Hofacker I.L., Fekete M., Stadler P.F. Secondary structure prediction for aligned RNA sequences. J. Mol. Biol. 2002;319:1059–1066. doi: 10.1016/S0022-2836(02)00308-X. [DOI] [PubMed] [Google Scholar]
- 27.Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eddy S.R. A memory-efficient dynamic programming algorithm for optimal alignment of a sequence to an RNA secondary structure. BMC Bioinformatics. 2002;3:18. doi: 10.1186/1471-2105-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Griffiths-Jones S., Bateman A., Marshall M., Khanna A., Eddy S.R. Rfam: an RNA family database. Nucleic Acids Res. 2003;31:439–441. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y., Altman S. In search of RNase P RNA from microbial genomes. RNA. 2004;10:1533–1540. doi: 10.1261/rna.7970404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eddy S.R., Durbin R. RNA sequence analysis using covariance models. Nucleic Acids Res. 1994;22:2079–2088. doi: 10.1093/nar/22.11.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Griffiths-Jones S., Moxon S., Marshall M., Khanna A., Eddy S.R., Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–D124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown J.W. The Ribonuclease P Database. Nucleic Acids Res. 1999;27:314. doi: 10.1093/nar/27.1.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu C., Bai B., Skogerbo G., Cai L., Deng W., Zhang Y., Bu D., Zhao Y., Chen R. NONCODE: an integrated knowledge database of non-coding RNAs. Nucleic Acids Res. 2005;33:D112–D115. doi: 10.1093/nar/gki041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wheeler D.L., Barrett T., Benson D.A., Bryant S.H., Canese K., Church D.M., DiCuccio M., Edgar R., Federhen S., Helmberg W., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2005;33:D39–D45. doi: 10.1093/nar/gki062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marquez S.M., Harris J.K., Kelley S.T., Brown J.W., Dawson S.C., Roberts E.C., Pace N.R. Structural implications of novel diversity in eucaryal RNase P RNA. RNA. 2005;11:739–751. doi: 10.1261/rna.7211705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dujon B., Sherman D., Fischer G., Durrens P., Casaregola S., Lafontaine I., De Montigny J., Marck C., Neuveglise C., Talla E., et al. Genome evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- 38.Bahl A., Brunk B., Crabtree J., Fraunholz M.J., Gajria B., Grant G.R., Ginsburg H., Gupta D., Kissinger J.C., Labo P., et al. PlasmoDB: the Plasmodium genome resource. A database integrating experimental and computational data. Nucleic Acids Res. 2003;31:212–215. doi: 10.1093/nar/gkg081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kiss T., Marshallsay C., Filipowicz W. 7-2/MRP RNAs in plant and mammalian cells: association with higher order structures in the nucleolus. EMBO J. 1992;11:3737–3746. doi: 10.1002/j.1460-2075.1992.tb05459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li X., Zaman S., Langdon Y., Zengel J.M., Lindahl L. Identification of a functional core in the RNA component of RNase MRP of budding yeasts. Nucleic Acids Res. 2004;32:3703–3711. doi: 10.1093/nar/gkh689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walker S.C., Avis J.M. A conserved element in the yeast RNase MRP RNA subunit can participate in a long-range base-pairing interaction. J. Mol. Biol. 2004;341:375–388. doi: 10.1016/j.jmb.2004.05.076. [DOI] [PubMed] [Google Scholar]
- 42.Lindahl L., Fretz S., Epps N., Zengel J.M. Functional equivalence of hairpins in the RNA subunits of RNase MRP and RNase P in Saccharomyces cerevisiae. RNA. 2000;6:653–658. doi: 10.1017/s1355838200992574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ziehler W.A., Morris J., Scott F.H., Millikin C., Engelke D.R. An essential protein-binding domain of nuclear RNase P RNA. RNA. 2001;7:565–575. doi: 10.1017/s1355838201001996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frank D.N., Adamidi C., Ehringer M.A., Pitulle C., Pace N.R. Phylogenetic-comparative analysis of the eukaryal ribonuclease P RNA. RNA. 2000;6:1895–1904. doi: 10.1017/s1355838200001461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chenna R., Sugawara H., Koike T., Lopez R., Gibson T.J., Higgins D.G., Thompson J.D. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]