Abstract
In this paper we report that the inclusion of heat-resistant RecA protein from a thermophilic bacteria, Thermus thermophilus, and its cofactor (ATP) in PCR effectively eliminates non-specific PCR products. The effect of RecA protein, which catalyzes pairing between homologous DNA molecules with great fidelity in genetic recombination, is due to its promotion of precise priming in PCR (i.e. priming at sites where the primer sequence is completely complementary to that of the target sequence). In addition, the RecA protein substantially reduces the primer concentration required for PCR. These experimental results have led to the realization of multiplex PCR, which involves PCR for multiple sites in the same reaction mixture. We were able to successfully perform multiplex PCR with over a dozen reactions without affecting the amplification pattern of the PCR products.
INTRODUCTION
PCR has been used extensively as a potent tool to amplify specific DNA sequences from genomic DNA. Though its usefulness is widely accepted, several technical problems still remain to be solved. Perhaps, the most significant is the appearance of non-specifically amplified products, which often complicates the analysis of PCR products. To circumvent this, where feasible, appropriate priming sites may be selected whose complementary sequences are least likely to produce illegitimate (non-specific) priming in the DNA molecules. However, the optimized priming sites to reduce non-specific PCR products may not be preferred when particular or exact priming sites are required. Furthermore, in practice, even with the most optimized primer sequences, non-specific PCR products cannot be completely eliminated, particularly in instances where complex whole genomic DNA is used as the template.
We attempted to eliminate non-specific PCR products by employing heat-stable RecA protein. RecA is known to be involved in DNA recombination in prokaryotes by catalyzing the pairing of homologous DNA sequences (1–4). Non-specific PCR products are most likely to be derived from false priming at sites where primers recognize similar but not completely complementary sequences. Thus, we speculated that the presence of RecA protein could greatly stimulate pairing between completely matched primers and targeted sequences; therefore, reduce false priming and lead to the elimination of non-specific PCR products.
Recently, a heat-stable RecA protein from a thermophilic bacteria, Thermus thermophilus (Tth), has been isolated and fully characterized (5–8). This prompted us to examine whether the presence of RecA in PCR effectively eliminated non-specific PCR products, thus making it possible to amplify the sequences between any sites of interest without needing to consider the optimal priming sites for each PCR. Furthermore, the elimination of non-specific PCR products has the potential to create a means to perform multiplex PCR (PCR for multiple sites in a single test tube), which has not been perfected to date.
In this article we show that the T.thermophilus RecA protein effectively eliminates non-specific PCR products. We also report that in the presence of the thermostable RecA protein, multiplex PCR, involving more than one dozen simultaneous PCRs, can be conducted in a single test tube.
MATERIALS AND METHODS
Expression and purification of TthRecA protein
The T.thermophilus RecA gene encodes a protein consisting of 263 amino acids. The gene was cloned in an expression vector with a T7 promoter (pET3b; Novagen), pET3b-RecA, was overexpressed in Escherichia coli BL21(DE3)/pLysS cells (Novagen). Purification of the RecA protein is carried out as described with a minor modification. The cells were grow to an OD550 of 0.5 and were treated with isopropyl β-D-thiogalactopyranoside at a final concentration of 1 mM for 4 h. The cells were then harvested, resuspended in a buffer containing 50 mM Tris–HCl (pH 8.0), 5 mM EDTA, 8.4 mM β-mercaptoethanol, 25% (w/v) sucrose and 1 M KCl, and disrupted by sonication. After removing debris by centrifugation (46 000 g for 90 min), the supernatant was heated for 60 min at 65°C and centrifuged (46 000 g for 15 min) to remove the precipitate. The supernatant was dialyzed against a buffer containing 20 mM potassium phosphate (pH 6.5), 1 mM EDTA, 5 mM β-mercaptoethanol and 10% (v/v) glycerol, and then applied to a phosphocellulose (P11 Cellulose; Whatman, Japan) column that had been equilibrated by using the same buffer. The RecA protein was eluted with a linear gradient of KCl (0–1.5 M). Fractions containing the RecA protein, detected by SDS–PAGE, were pooled and dialyzed against a buffer consisting of 50 mM Tris–HCl (pH 7.5), 0.1 mM EDTA, 5 mM β-mercaptoethanol, 1.5 M KCl and 10% (v/v) glycerol. After the addition of equal amounts of glycerol, the sample was stored at −20°C until use. The concentration of the RecA protein was determined using a molar absorption coefficient of 14 600 M−1 cm−1 at 277 nm.
PCR
Primers used for PCR were purchased from Sigma–Aldrich, Japan. PCR was performed using DNA polymerase from T.aquaticus (Taq) or T.thermophilus (Tth) according to the manufacturer's instructions [rTaq or ExTaq DNA polymerase plus ‘hot start’ antibody (Takara-bio)]. The same results were obtained by the other polymerase systems: LA Taq polymerase (Takara-bio), Tth polymerase (Applied-boisystems); Expand High Fidelity, Expand High FidelityPLUS and Expand Long Template polymerase (Roche-diagnostics); TITANIUM Taq polymerase (Becton-Dickinson-Clontech); and Taq polymerase (Promega). Essentially, the same results were obtained by employing a basic reaction mixture, which contained 10 mM Tris–HCl, pH 8.3, 50 mM KCl and 1.5 mM MgCl2. Primer concentrations used for each experiment are described in the figure legends. PCR thermal cycles were used as follows: 30 s at 94°C for DNA denaturation, followed by 35 cycles for 10 s at 94°C, 30 s at 55°C and 90 s at 68°C. A final elongation step was performed for 3 min at 68°C. Multiplex PCR was performed under the same conditions described above using the DNA polymerase (rTaq or ExTaq DNA polymerase plus ‘hot start’ antibody or rTth DNA polymerase).
Gel electrophoresis
PCR products were electrophoresed in 1.2% agarose gel for 1 h (4 V/cm) in a Tris–acetate/EDTA buffer [40 mM Tris–acetate (pH 8.3) and 1 mM EDTA] with a sample loading buffer [0.05% bromophenol blue and 3% Ficoll; Type 400 (Pharmacia)]. After electrophoresis, the DNA bands were stained with ethidium bromide and visualized on a Fluoro Imager (Fluoro Imager 595; Molecular Dynamics).
Real-time PCR
Real-time PCR and assays were performed using an ABI PRISM 7700 and an RT–PCR kit reagent (SYBR premix ExTaq Perfect Real Time plus ‘hot start’ antibody; Takara-bio). Template DNA (100 ng) was added to the reaction mixture to a final volume of 100 μl) containing 1× Takara-bio SYBR premix ExTaq master mixture and primers (0.2 μM each). The mixture was split into three 30 μl PCRs and they were subjected to thermo cycling (10 s at 94°C for 1 cycle, followed by 40 cycles of 5 s at 94°C and 30 s at 60°C). The values of the triplicated samples were averaged and shown.
RESULTS
Effect of RecA protein on PCR-based DNA amplification
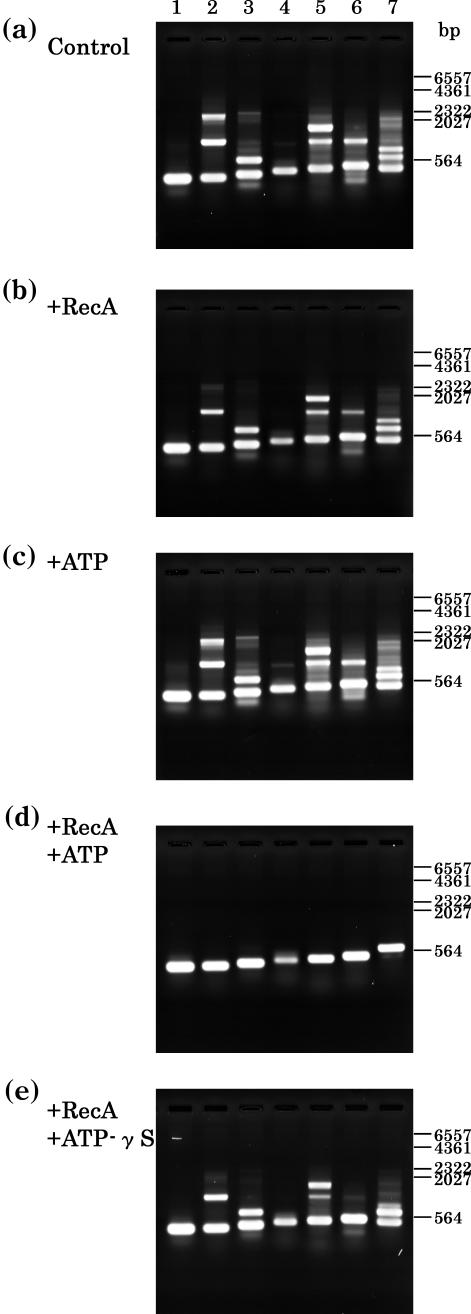
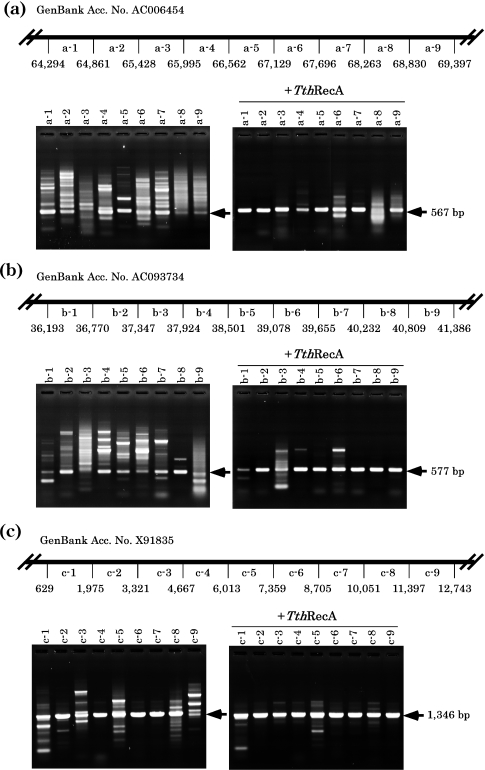
In order to examine whether the presence of RecA protein could minimize false priming and thus reduce non-specific PCR products, we compared the patterns of PCR products obtained in the presence (or absence) of heat-stable TthRecA. As shown in Figure 1, the presence of TthRecA with its cofactor, ATP in PCR essentially eliminated the appearance of non-specific PCR products for seven randomly selected sites in human genomic DNA (Figure 1d). Unhydrolyzable ATP analogs such as ATP-γS did not substitute for ATP (Figure 1e). The requirement of ATP strongly suggests that the effect of TthRecA is associated with the enzymatic activity of the protein (Discussion). To further validate the effect of the RecA protein, three regions (Figure 2a, 567 bp; Figure 2b, 577 bp; Figure 2c, 1346 bp) in human genomic DNA were equally subdivided into nine 567, 577 and 1346 bp sites, respectively (each designated a-1 through a-9, b-1 through b-9 and c-1 through c-9, Figure 2a–c). Using primers (20 bp) complementary to the terminal sequence of each subdivided site, PCR was performed in the presence of TthRecA. As shown in the figure, the presence of TthRecA eliminated non-specific PCR products in most of the 27 PCR sites examined, as evidenced by the greatly simplified patterns of the amplified products.
Figure 1.
Effect of T.thermophilus RecA protein on PCR. PCR with Taq DNA polymerase (ExTaq DNA polymerase plus ‘hot start’ antibody; Takara-bio) for several randomly selected sequences (300–650 bp) in human genomic DNA was carried out in the absence or in the presence of the TthRecA protein. (a) Control, PCR under the standard conditions described in Materials and Methods. (b) Similar to (a), but with TthRecA protein (0.4 μg per 25 μl reaction mixture). (c) Similar to (a), but with ATP (400 μM). (d) Similar to (a), but with TthRecA protein (0.4 μg per 25 μl reaction mixture) and ATP (300 μM). (e) Similar to (a), but with TthRecA protein (0.4 μg per 25 μl reaction mixture) and ATP-γS (300 μM). The products were electrophoresed and stained with ethidium bromide. Molecular weight markers are indicated on the right-hand side of these panels. The oligonucleotide sequences used for the primers were as follows: 5′-ACAATGGGCTCACTCACCCA-3′ and 5′-CTAAGACCAATGGATAGCTG-3′ for lane 1 (300 bp); 5′-GCTCAGCATGGTGGTGGCAT-3′ and 5′-CCTCATACCTTCCCCCCCAT-3′ for lane 2 (319 bp); 5′-GACTACTCTAGCGACTGTCC-3′ and 5′-GACAGCCACCAGATCCAATC-3′ for lane 3 (360 bp); 5′-AACCTCACAACCTTGGCTGA-3′ and 5′-TTCACAACTTAAGATTTGGC-3′ for lane 4 (400 bp); 5′-AGGCAACTAGGATGGTGTGG-3′ and 5′-CAGGGAGCGTGTCCATAGGG-3′ for lane 5 (450 bp); 5′-CTGCTGAAAGAGATGCGGTG-3′ and 5′-AGGAAAACAGCCCAAGGGAC-3′ for lane 6 (469 bp); and 5′-ACTTTGTTCTGAGCCTCACA-3′ and 5′-GTTGCCCAATCGCCCCTCTC-3′ for lane 7 (650 bp).
Figure 2.
PCR of subdivided genomic sequences. Three regions of human genomic DNA (GenBank accession nos AC006454, AC093734 and X91835, with 5103, 5193 and 12 114 bp, respectively) were subdivided into nine 567, 577 and 1346 bp PCR sites, respectively. PCR was performed for each subdivided site using primer sets (20 bp each) corresponding to the terminal sequence of each site using the Taq DNA polymerase (ExTaq DNA polymerase plus ‘hot start’ antibody; Takara-bio). PCR was carried out in the absence and in the presence of TthRecA protein and ATP. The products were electrophoresed and stained with ethidium bromide. (a) A diagrammatic representation of the subdivided region (5103 bp in GenBank accession no AC006454) (upper panel) and the electrophoretic patterns of the PCR products (lower panel). (b) A diagrammatic representation of the subdivided region (5193 bp in GenBank accession no AC093734) (upper panel) and the electrophoretic patterns of the PCR products (lower panel). (c) A diagrammatic representation of the subdivided region (12 114 bp in GenBank accession no X91835) (upper panel) and the electrophoretic patterns of the PCR products (lower panel). Throughout (a–c), nine subdivided sites for each region are indicated as a-1 to a-9, b-1 to b-9 and c-1 to c-9. Nucleotide (nt) numbers correspond to registries in GenBank. Locations of the specific PCR products are indicated by arrows.
In online Supplementary Figure S1, we show the results of amplification in which, nine targets of human genes were amplified. It is quite clear that all the products ranging from 300 to 1350 bp were almost equally amplified with much less non-specific PCR products compared to the ones without the TthRecA protein.
PCR with primers carrying mismatched bases
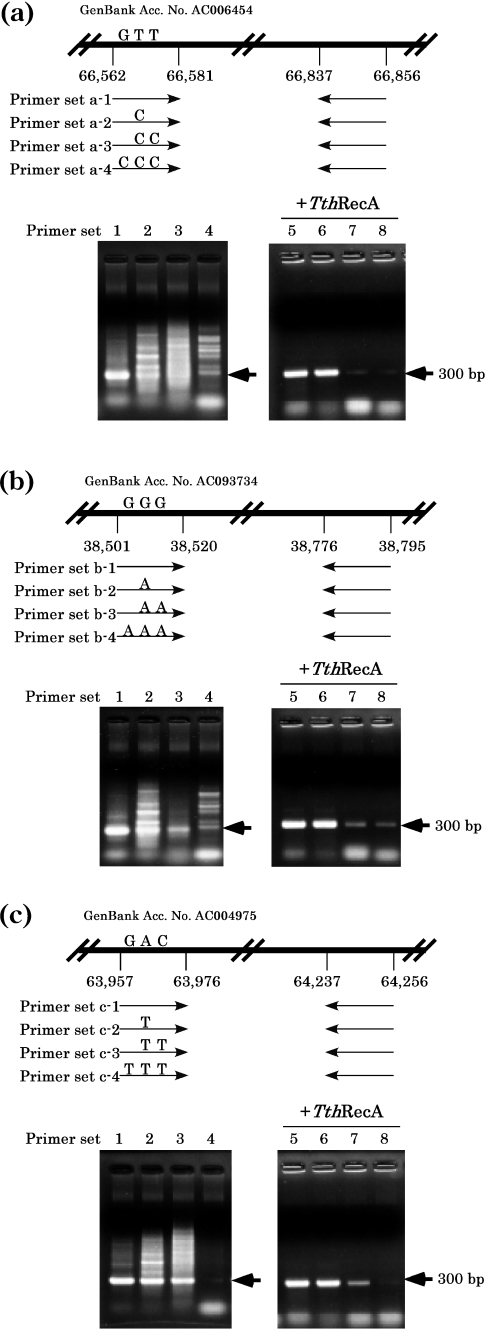
We investigated whether the effect of TthRecA on reducing non-specific PCR products was due to the activity associated with RecA catalyzing the pairing of homologous DNA sequences. To examine this, PCR was performed with three different sets of primers (Figure 3a–c) in which one to three bases (among 20 bases) that mismatched the sequence of the priming sites were introduced in one of the primers. In the presence of TthRecA, the amount of amplified PCR products decreased drastically when PCR primers with more than two mismatched bases were employed (Figure 3a–c, right panels).
Figure 3.
PCR with primers carrying mismatched bases. PCR was performed at two human genomic sites with primers (20 bp), one of which (forward primer) carried one, two or three mismatched bases in the middle of the primer, in the absence (left) or presence (right) of TthRecA protein and ATP using the Taq DNA polymerase (rTaq DNA polymerase plus ‘hot start’ antibody). (a) Upper panel: a diagrammatic representation of the location of the PCR site (20 bp between nt 66 562 and nt 66 581 in GenBank accession no AC006454) and of the primers. Lower panel: lanes 1 and 5, PCR products using primers without mismatched bases (primer set a-1); lanes 2 and 6, PCR products using primers (primer set a-2 with one mismatched base at nt 66 566, T to C); lanes 3 and 7, PCR products using primers (primer set a-3 with two mismatched base at nt 66 566 and nt 66 571, both T to C); and lanes 4 and 8, PCR products using primers (primer set a-4 with three mismatched base at nt 66 566 and nt 66 571, T to C and nt 66 576, G to C). The oligonucleotide sequences used for the forward primers (mismatched bases are underlined) are as follows: primer set a-1, 5′-CATGGCACCTGCTCTGAGAC-3′; primer set a-2, 5′-CATGGCACCCGCTCTGAGAC-3′; primer set a-3, 5′-CATGGCACCCGCTCCGAGAC-3′; and primer set a-4, 5′-CATGCCACCCGCTCCGAGAC-3′. (b) Upper panel: a diagrammatic representation of the location of the PCR site (20 bp between nt 38 501 and nt 38 520 in GenBank accession no. AC0937734) and of the primers. Lower panel: lanes 1 and 5, PCR products using primers without mismatched bases (primer set b-1); lanes 2 and 6, PCR products using primers (primer set b-2 with one mismatched base at nt 38 505, G to A); lanes 3 and 7, PCR products using primers (primer set b-3 with two mismatched base at nt 38 505 and nt 38 510, both G to A); and lanes 4 and 8, PCR products using primers (primer set b-4 with three mismatched base at nt 38 505, nt 38 510 and nt 38 515, all G to A). The oligonucleotide sequences used for the forward primers are as follows: primer set b-1, 5′-ATCTGTGTGGTTCGGCTCTG-3′; primer set b-2, 5′-ATCTGTGTGATTCGGCTCTG-3′; primer set b-3, 5′-ATCTGTGTGATTCGACTCTG-3′; and primer set b-4, 5′-ATCTATGTGATTCGACTCTG-3′. (c) Upper panel: a diagrammatic representation of the location of the PCR site (20 bp between nt 63 957 and nt 63 976 in GenBank accession no. AC004975) and of the primers. Lower panel: lanes 1 and 5, PCR products using primers without mismatched bases (primer set c-1); lanes 2 and 6, PCR products using primers (primer set c-2 with one mismatched base at nt 63 961, A to T); lanes 3 and 7, PCR products using primers (primer set c-3 with two mismatched base at nt 63 961 and nt 63 966, A to T and C to T); and lanes 4 and 8, PCR products using primers (primer set c-4 with three mismatched base at nt 63 961, nt 63 966 and nt 63 971, A to T, C to T and G to T). The oligonucleotide sequences used for the forward primers are as follows: primer set c-1, 5′-GCAGGCACCAAGAACTACTG-3′; primer set c-2, 5′-GCAGGCACCTAGAACTACTG-3′; primer set c-3, 5′-GCAGGCACCTAGAATTACTG-3′; and primer set c-4, 5′-GCAGTCACCTAGAATTACTG-3′. The sequences for the backward primers are 5′-TCACCTCCCAGCCTGGCCCA-3′ for (a), 5′-AGGGAGATGTTCTCATAAAT-3′ and 5′-CTGTAAGTGGCAGACATTAC-3′ for (b). Nucleotide numbers correspond to registries in GenBank. Locations of the specific PCR products are indicated by arrows.
Taken together, we conclude that through stimulating legitimate pairing (i.e. pairing between primers and complementary sequence on template DNA) TthRecA greatly increases the specificity of PCR, eliminating the appearance of most, though not all, non-specific PCR products.
The effect of primer concentration
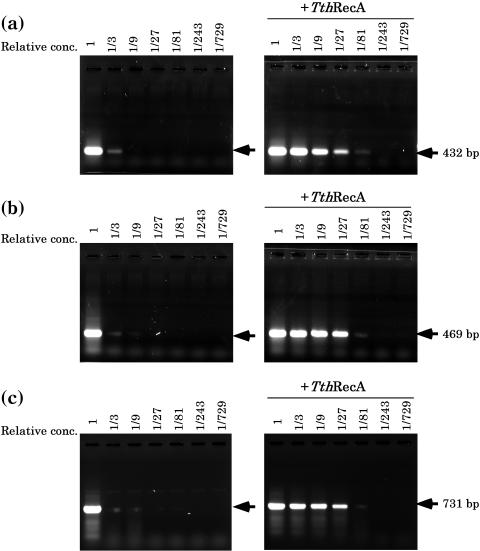
Since RecA protein enzymatically catalyzes pairing between homologous sequences, we expected that the TthRecA protein-assisted PCR would proceed with much lesser concentration of primers than that required for conventional PCR (without the TthRecA protein). Figure 4 shows that while the amount of PCR products decreased drastically as primer concentration was reduced to one-third in the control experiment (without TthRecA) (Figure 4a–c, left panels), in the presence of TthRecA substantial amounts of PCR products were still obtained even when the primer concentration was reduced to 1/27 of the original concentration (Figure 4a–c, right panels).
Figure 4.
The effect of primer concentration on PCR products. PCR was performed at different concentrations of primers at three human genomic DNA sites [(a) beta globin gene, (b) blue corn gene and (c) beta globin gene] in the absence or in the presence of TthRecA protein (and ATP). Throughout (a–c), PCR products at serially diluted primer concentrations (the original concentration: 0.2 μM each primer sets) are shown. Locations of the specific amplification products are indicted by arrows. The oligonucleotide sequences used for the primer sets are 5′-GGCAGCTTTCATGGGCACTG-3′ and 5′-GACAGGGCTGGACTGACATT-3′ for (a); 5′-CTGCTGAAAGAGATGCGGTG-3′ and 5′-AGGAAAACAGCCCAAGGGAC-3′ for (b); 5′-CTTTTTGTTCCCCCAGACAC-3′ and 5′-GCACTGGCTTAGGAGTTGGA-3′ for (c).
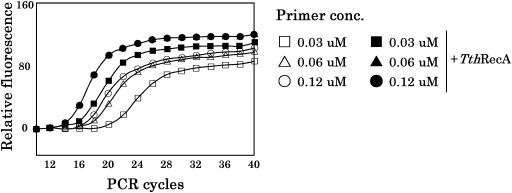
The results were further validated by real-time PCR in which the polymerization process was followed as a function of the number of PCR cycles. The results are shown in Figure 5. It is clear that TthRecA greatly stimulates polymerization particularly at low primer concentrations, indicating that in the presence of TthRecA, pairing of primers to complementary sequence occurs with high efficiency, consequently requiring considerably low primer concentrations than conventional PCR.
Figure 5.
Real-time PCR in the presence of TthRecA protein (and ATP). PCR was carried out in the absence and in the presence of TthRecA protein (and ATP) at different concentrations of primers. We employed three primer concentrations (0.03, 0.06 and 0.12 μM) and followed the appearance of PCR products (measured by the intensity of fluorescence) at each PCR cycle. In the figure, squares, triangles and circles represent primer concentrations of 0.03, 0.06 and 0.12 μM, respectively. Open symbols correspond to the control samples (without TthRecA protein and ATP) and closed symbols correspond to samples that contain TthRecA protein and ATP. The oligonucleotide sequences used for the primers are 5′-ACAATGGGCTCACTCACCCA-3′ and 5′-CTAAGACCAATGGATAGCTG-3′ (300 bp).
Multiplex PCR amplification
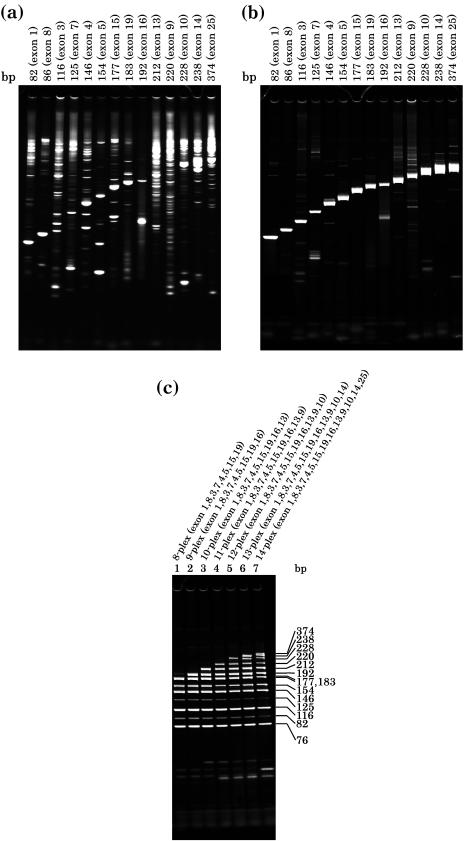
Encouraged by the elimination of non-specific PCR products and the particularly low concentrations of primer required for PCR in the presence of TthRecA, we examined the effect of the protein in multiplex PCR. Despite its potential as a valuable research technique, multiplex PCR has not been widely used, mainly because of the presence of non-specific PCR products and the requirement for relatively high primer concentrations to satisfy each PCR, which may inhibit the procedure as the number of PCRs increases. We show the results of multiplex PCR for human apoB gene exons ranging from 76 to 374 bp in which up to 14 parallel PCRs corresponding to each exon were conducted in the same reaction mixture. As predicted, in the presence of TthRecA, individual PCRs for each exon (the exons presented in the figure were rearranged according to their size, from 76 to 374 bp) show substantial improvement in the specificity of polymerization, which virtually eliminated non-specific PCR products for most of the exons (Figure 6b), compared to those performed without TthRecA (Figure 6a). In Figure 6c, we show the results of multiplex PCR, in which PCR for 8–14 exons of the gene was run in parallel in the presence of TthRecA. The result shows that multiplex PCR for as many as 14 sites can be done without significantly affecting the polymerization pattern of each PCR. Essentially, the same effect of TthRecA was observed in the multiplex PCR with less than eight simultaneous reactions (data not shown). The results demonstrate that by employing the thermostable TthRecA protein multiplex PCR is now possible.
Figure 6.
Multiplex PCR. PCR for the 14 exons of the human apoB gene was carried out in the absence (a) or in the presence (b) of TthRecA protein and ATP. All 14 exons of the gene were subjected to PCR. In the figure, the PCR products for each exon is shown after having been rearranged according to the size of each exon, the smallest (exon 1, 82 bp) far left and the largest (exon 25, 374 bp) far right. Throughout (a and b), all samples were amplified with apoB exon primers at the same primer pair concentrations (0.1 μM per pair). (c) Multiplex PCR for exons 8–14 of the apoB gene was performed and the PCR products for each multiplex PCR are shown. The exons subjected to each multiplex PCR are also shown on the top of the figure. Samples were amplified with apoB 8-plex to 14-plex primers at the following primer pair concentrations: lanes 1–4, 0.1 μM per pair; lanes 5–7, 0.05 μM per pairs. Aliquots of 4 μl volume were electrophoresed through 12.5% acrylamide gel in Tris–borate/EDTA buffer, and stained with SYBR Green (SYBR Green I; Novagen). The signals were detected on a Fluoro Imager (Fluoro Imager 595; Molecular Dynamics). Product sizes (82–374 bp) are indicated on the right-hand side of (c).
DISCUSSION
In conventional PCR, non-specific PCR products are apparently derived from illegitimate (false) pairing of PCR primers with similar, but not identical sequences, which are present in DNA molecules, particularly genomic DNA from higher organisms. In this paper, we have shown that the addition of heat-stable RecA protein from a thermophilic bacteria, T.thermophilus, and its cofactor (ATP) (9) to PCR mixtures effectively eliminates non-specific PCR products, thus making it possible to amplify almost any sequence of interest without having to consider the selection of optimal primer sites. Based on this finding, we were able to amplify each exon of a specific gene from a complex genomic DNA sample. We showed that the efficiency of PCR amplification was drastically reduced when mismatched bases to the targeted sequence were introduced in the primers. From the results presented here, it is apparent that TthRecA eliminates non-specific PCR products through its enzymatic activity, and presumably this is achieved through the precise positioning of primers with their complementary sequences.
RecA protein and its homologs are believed to play a central role in genetic recombination by catalyzing the pairing of homologous DNA molecules with high fidelity (10–13). In this process, RecA protein first forms a complex with ATP and single-stranded DNA and the complex searches for and pairs with a homologous sequence in the target single-stranded or double-stranded DNA molecules to form an intermolecular double-stranded structure (known as D-loop structure when target DNA is double-stranded in recombination in the cells) (14,15). The structure is subsequently processed by actions of DNA polymerases and other enzymes to complete recombination. It is known that hydrolysis of ATP is required for recycling the pairing process whereas just for the interaction between the two target DNA molecules, ATP can be replaced by unhydrolyzable ATP analogs such as ATP-γS. Since the effect of RecA protein was observed only with ATP, not with ATP-γS, it is most probably the elimination of non-specific PCR products by RecA protein is a result of repeated cycles of pairing DNA sequences between PCR primers and their target DNA molecules, which minimizes mismatches between them (16–19).
Although several groups have reported that DNA-binding proteins such as SSB protein (single-stranded DNA-binding protein) reduces non-specific PCR products (20–23), there are no reports on the application of SSB protein for multiplex PCR. Under the condition we employed here, inclusion of SSB protein either from E.coli and T.thermophilus in the mixture for PCR neither reduced illegitimate priming nor primer concentrations required for PCR (Supplementary Figure S2).
In this article we also reported the effect of TthRecA in multiplex PCR. Needless to say, the demand for multiplex PCR has been increasing as laboratories want to conduct PCRs with increasingly large quantities of samples and with greater economy. Conventional PCR for the multiple sites has been tried for the analysis of mutation, deletion and polymorphism in genomic DNA (24–26), and quantification of PCR products and RNA (27–30). The problems prohibiting the success of multiplex PCR were not only to ascertain the means to eliminate non-specific PCR products, but also to determine how to reduce primer concentration required for each PCR. As discussed above, TthRecA apparently contributes to solving both these problems. We showed that in the presence of TthRecA, up to 14 exons were amplified in parallel without significantly affecting the patterns of the PCR products. Under the conditions we employed here, we can probably perform as many as 20 PCR runs in a single test tube.
The next challenge lies in developing a procedure to easily characterize each PCR product, without depending on conventional gel electrophoresis. Towards that goal, employing molecularly marked primers, we were able to detect specific PCR products by mass spectrometry after multiplex PCR with TthRecA. As more primers with a variety of molecular markers become available, RecA protein-assisted multiplex PCR should become a very effective tool to amplify and characterize any specific sequence, even when using very complex genomic DNA as the template.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
We would like to thank Dr O. Ohara for his valuable suggestions, Dr K. Kondo for their help and encouragement and Mrs S. Ishibe and Dr J. Inoue for providing the proteins to us. Funding to pay the Open Access publication charges for this article was provided by the Kazusa DNA Research Institute Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.West S.C., Cassuto E., Mursalim J., Howard-Flanders P. Recognition of duplex DNA containing single-stranded regions by RecA protein. Proc. Natl Acad. Sci. USA. 1980;77:2569–2573. doi: 10.1073/pnas.77.5.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.West S.C., Cassuto E., Howard-Flanders P. RecA protein promotes homologous-pairing and strand-exchange reactions between duplex DNA molecules. Proc. Natl Acad. Sci. USA. 1981;78:2100–2104. doi: 10.1073/pnas.78.4.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rao B.J., Dutreix M., Radding C.M. Stable three-stranded DNA made by RecA protein. Proc. Natl Acad. Sci. USA. 1991;88:2984–2988. doi: 10.1073/pnas.88.8.2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kato R., Kuramitsu S. Characterization of thermostable RecA protein and analysis of its interaction with single-stranded DNA. Eur. J. Biochem. 1999;259:592–601. doi: 10.1046/j.1432-1327.1999.00044.x. [DOI] [PubMed] [Google Scholar]
- 5.Kato R., Kuramitsu S. RecA protein from an extremely thermophilic bacterium, Thermus thermophilus HB8. J. Biochem. (Tokyo) 1993;114:926–929. doi: 10.1093/oxfordjournals.jbchem.a124278. [DOI] [PubMed] [Google Scholar]
- 6.Angov E., Camerini-Otero R.D. The recA gene from the thermophile Thermus aquaticus YT-1: cloning, expression, and characterization. J. Bacteriol. 1994;176:1405–1412. doi: 10.1128/jb.176.5.1405-1412.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wetmur J.G., Wong D.M., Ortiz B., Tong J., Reichert F., Gelfand D.H. Cloning, sequencing, and expression of RecA proteins from three distantly related thermophilic eubacteria. J. Biol. Chem. 1994;269:25928–25935. [PubMed] [Google Scholar]
- 8.Kato R., Kuramitsu S. Characterization of thermostable RecA protein and analysis of its interaction with single-stranded DNA. Eur. J. Biochem. 1999;259:592–601. doi: 10.1046/j.1432-1327.1999.00044.x. [DOI] [PubMed] [Google Scholar]
- 9.Watanabe R., Masui R., Mikawa T., Takamatsu S., Kato R., Kuramitsu S. Interaction of Escherichia coli RecA protein with ATP and its analogues. J. Biochem. (Tokyo) 1994;116:960–966. doi: 10.1093/oxfordjournals.jbchem.a124653. [DOI] [PubMed] [Google Scholar]
- 10.McEntee K., Weinstock G.M., Lehman I.R. Initiation of general recombination catalyzed in vitro by the recA protein of Escherichia coli. Proc. Natl Acad. Sci. USA. 1979;76:2615–2619. doi: 10.1073/pnas.76.6.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weinstock G.M., McEntee K., Lehman I.R. ATP-dependent renaturation of DNA catalyzed by the recA protein of Escherichia coli. Proc. Natl Acad. Sci. USA. 1979;76:126–130. doi: 10.1073/pnas.76.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shibata T., DasGupta C., Cunningham R.P., Radding C.M. Purified Escherichia coli recA protein catalyzes homologous pairing of superhelical DNA and single-stranded fragments. Proc. Natl Acad. Sci. USA. 1979;76:1638–1642. doi: 10.1073/pnas.76.4.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- 14.Shibata T., Cunningham R.P., DasGupta C., Radding C.M. Homologous pairing in genetic recombination: complexes of recA protein and DNA. Proc. Natl Acad. Sci. USA. 1979;76:5100–5104. doi: 10.1073/pnas.76.10.5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szostak J.W., Orr-Weaver T.L., Rothstein R.J., Stahl F.W. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 16.Ohtani T., Shibata T., Iwabuchi M., Watabe H., Iino T., Ando T. ATP-dependent unwinding of double helix in closed circular DNA by recA protein of E.coli. Nature. 1982;299:86–89. doi: 10.1038/299086a0. [DOI] [PubMed] [Google Scholar]
- 17.Shibata T., Ohtani T., Iwabuchi M., Ando T. D-loop cycle: a circular reaction sequence which comprises formation and dissociation of D-loops and inactivation and reactivation of superhelical closed circular DNA promoted by recA protein of Escherichia coli. J. Biol. Chem. 1982;257:13981–13986. [PubMed] [Google Scholar]
- 18.Sen S., Karthikeyan G., Rao B.J. RecA realigns suboptimally paired frames of DNA repeats through a process that requires ATP hydrolysis. Biochemistry. 2000;39:10196–10206. doi: 10.1021/bi000753y. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Z., Yoon D., LaPorte J.R., Chen J. Appropriate initiation of the strand exchange reaction promoted by RecA protein requires ATP hydrolysis. J. Mol. Biol. 2001;309:29–43. doi: 10.1006/jmbi.2001.4753. [DOI] [PubMed] [Google Scholar]
- 20.Kunkel T.A., Meyer R.R., Loeb L.A. Single-strand binding protein enhances fidelity of DNA synthesis in vitro. Proc. Natl Acad. Sci. USA. 1979;76:6331–6335. doi: 10.1073/pnas.76.12.6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perales C., Cava F., Meijer W.J., Berenguer J. Enhancement of DNA, cDNA synthesis and fidelity at high temperatures by a dimeric single-stranded DNA-binding protein. Nucleic Acids Res. 2003;31:6473–6480. doi: 10.1093/nar/gkg865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olszewski M., Rebala K., Szczerkowska Z., Kur J. Application of SSB-like protein from Thermus aquaticus in multiplex PCR of human Y-STR markers identification. Mol. Cell Probes. 2005;3:203–205. doi: 10.1016/j.mcp.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 23.Rapley R. Enhancing PCR amplification and sequencing using DNA-binding proteins. Mol. Biotechnol. 1994;2:295–298. doi: 10.1007/BF02745882. [DOI] [PubMed] [Google Scholar]
- 24.Chamberlain J.S., Gibbs R.A., Ranier J.E., Nguyen P.N., Caskey C.T. Deletion screening of the Duchenne muscular dystrophy locus via multiplex DNA amplification. Nucleic Acids Res. 1988;16:11141–11156. doi: 10.1093/nar/16.23.11141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rithidech K.N., Dunn J.J., Gordon C.R. Combining multiplex and touchdown PCR to screen murine microsatellite polymorphisms. BioTechniques. 1997;23:36–45. doi: 10.2144/97231bm06. [DOI] [PubMed] [Google Scholar]
- 26.Shuber A.P., Skoletsky J., Stern R., Handelin B.L. Efficient 12-mutation testing in the CFTR gene: a general model for complex mutation analysis. Hum. Mol. Genet. 1993;2:153–158. doi: 10.1093/hmg/2.2.153. [DOI] [PubMed] [Google Scholar]
- 27.Zimmermann K., Schogl D., Plaimauer B., Mannhalter J.W. Quantitative multiple competitive PCR of HIV-1 DNA in a single reaction tube. BioTechniques. 1996;21:480–484. doi: 10.2144/96213st06. [DOI] [PubMed] [Google Scholar]
- 28.Sherlock J., Cirigliano V., Petrou M., Tutschek B., Adinolfi M. Assessment of diagnostic quantitative fluorescent multiplex polymerase chain reaction assays performed on single cells. Ann. Hum. Genet. 1998;62:9–23. doi: 10.1046/j.1469-1809.1998.6210009.x. [DOI] [PubMed] [Google Scholar]
- 29.Jin L., Richards A., Brown D.W. Development of a dual target-PCR for detection and characterization of measles virus in clinical specimens. Mol. Cell. Probes. 1996;10:191–200. doi: 10.1006/mcpr.1996.0027. [DOI] [PubMed] [Google Scholar]
- 30.Zou S., Stansfield C., Bridge J. Identification of new influenza B virus variants by multiplex reverse transcription-PCR and the heteroduplex mobility assay. J. Clin. Microbiol. 1998;36:1544–1548. doi: 10.1128/jcm.36.6.1544-1548.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.