Abstract
Copper is an essential micronutrient that plays a central role for a broad range of biological processes. Although there is compelling evidence that the intracellular milieu does not contain any free copper ions, the rapid kinetics of copper uptake and release suggests the presence of a labile intracellular copper pool. To elucidate the subcellular localization of this pool, we have synthesized and characterized a membrane-permeable, copper-selective fluorescent sensor (CTAP-1). Upon addition of Cu(I), the sensor exhibits a 4.6-fold emission enhancement and reaches a quantum yield of 14%. The sensor exhibits excellent selectivity toward Cu(I), and its emission response is not compromised by the presence of millimolar concentrations of Ca(II) or Mg(II) ions. Variable temperature dynamic NMR studies revealed a rapid Cu(I) self-exchange equilibrium with a low activation barrier of ΔG‡ = 44 kJ·mol–1 and kobs ∼ 105 s–1 at room temperature. Mouse fibroblast cells (3T3) incubated with the sensor produced a copper-dependent perinuclear staining pattern, which colocalizes with the subcellular locations of mitochondria and the Golgi apparatus. To evaluate and confirm the sensor's copper-selectivity, we determined the subcellular topography of copper by synchrotron-based x-ray fluorescence microscopy. Furthermore, microprobe x-ray absorption measurements at various subcellular locations showed a near-edge feature that is characteristic for low-coordinate monovalent copper but does not resemble the published spectra for metallothionein or glutathione. The presented data provide a coherent picture with strong evidence for a kinetically labile copper pool, which is predominantly localized in the mitochondria and the Golgi apparatus.
Keywords: photoinduced electron transfer, metal exchange kinetics, dynamic NMR, microprobe x-ray absorption near-edge spectroscopy
Copper is an essential trace element for all currently known life forms. It is required as a cofactor for many fundamental biological processes (1); however, it also catalyzes the production of highly reactive oxygen species, leading to oxidative damage of lipids, proteins, DNA, and other biomolecules (2). To protect the cell from copper toxicity, nature developed sophisticated machinery for uptake, storage, and trafficking of this element (3–6). Recently, it became increasingly evident that defects in these regulatory processes cause serious diseases such as the Menkes syndrome, Wilson's disease, and certain neurodegenerative diseases including amyotropic lateral sclerosis (ALS) and Alzheimer's disease (7–10). Given the high concentration of cytosolic copper-binding proteins and ligands such as superoxide dismutase, metallothionein, or glutathione, the thermodynamically estimated level of solvated copper ions in the cytoplasm lies well below a single ion per cell (11). At the same time, a significant fraction of cellular copper exchanges surprisingly quickly with the extracellular medium. For example, the intracellular copper concentration increases within <1 h by 4- to 20-fold upon incubation in growth medium supplemented with a micromolar concentration of copper and decreases with a similar rate upon washing in basal medium (12). Hence, the rapid kinetics of copper uptake and release may suggest the presence of a kinetically labile, intracellular copper pool; however, at present, the subcellular location and nature of this pool and the fundamental question of how copper is stored as part of the copper regulatory machinery are elusive. Cytosolic metallothionein has been suggested as a possible buffer ligand for intracellular copper storage (13, 14). Despite its high copper affinity, this protein could provide a sufficiently rapid exchange kinetics by means of an associative mechanism as already demonstrated for intermolecular exchange of zinc (15). Cytosolic glutathione might be similarly involved in coordination of excess copper. Alternatively, secretory vesicles containing copper ATPases have been recently shown to undergo copper-dependent trafficking (16) and might function as storage containers of intracellular copper. Such compartmentalization would spatially separate the copper ions from the cytoplasm and prevent potentially damaging redox reactions with other biomolecules.
Fluorescent sensors that can permeate the plasma membrane have proven to be powerful and nondestructive tools for the study of intracellular metal ion distributions of calcium (17), magnesium (18), or zinc (19, 20), yet rigorous analytical techniques for sensitive in vivo measurements of intracellular copper levels are lacking (for a novel, protein-based approach using a Cu(I)-selective transcriptional factor (CueR) as sensor, see ref. 21). Here, we report the development, characterization, and evaluation of a membrane-permeable copper-selective fluorescent sensor for imaging of kinetically labile copper pools. Because the reducing environment of the cytosol is expected to stabilize monovalent copper (6), the sensor has been specifically designed for this oxidation state. In addition, a series of synchrotron-based x-ray fluorescence microprobe (microXRF) experiments were performed to determine the intracellular copper topography and thus to evaluate the performance of the sensor. The data are consistent with the presence of a labile pool of low-coordinate, monovalent copper, which appears to be predominantly localized in the mitochondria and the Golgi apparatus.
Experimental Methods
Synthesis. We synthesized 4-(1,4,7,10-tetrathia-13-aza-cyclopentadec-13-yl)-benzene (2) as described in ref. 22. Detailed procedures for the synthesis of CTAP-1 including spectral data and analytical characterizations are provided in Supporting Text, which is published as supporting information on the PNAS web site.
Steady-State Fluorescence Spectroscopy. All sample solutions were filtered through 0.2-μm Teflon membrane filters to remove interfering dust particles or fibers. Fluorescence emission spectra were recorded with a fluorimeter (path length, 1 cm; cell volume, 3.0 ml; PTI, Lawrenceville, NJ). The fluorescence spectra have been corrected for the spectral response of the detection system (emission correction file provided by instrument manufacturer) and for the spectral irradiance of the excitation channel (via calibrated photodiode). Quantum yields were determined by using quinine sulfate dihydrate in 0.5 M H2SO4 as fluorescence standard [Φf = 0.54 ± 0.05 (23)]. To a 5 μM solution of CTAP-1 in 10 mM Pipes (pH 7.20, 25°C) were stepwise added 0.1 M eq aliquots of [Cu(I)(CH3CN)4]PF6 (5 mM stock solution in CH3CN). After each addition, the solution was equilibrated with stirring until no further increase of the emission intensity at 480 nm (excitation at 365 nm) was observed (typically 10 min). To minimize photobleaching by the incident light, the emission intensity was checked by short scans (2 sec) in intervals of 20 sec. Upon equilibration, the emission spectrum was acquired over the entire wavelength range (0.2 sec integration time per 1 nm step size). In a control experiment addition of an equivalent amount of neat acetonitrile had no effect on the emission intensity. Additional binding studies to determine the Cu(I) stability constant of CTAP-1 are provided in Supporting Text.
Variable-Temperature NMR Spectroscopy. A series of 1H NMR spectra were acquired for a solution of ligand 2 (3 mg) in CD3CN (700 μl) at temperature settings ranging from –40 to + 20°C in 10°C intervals. At each temperature, the sample was allowed to equilibrate for at least 20 min. The measurements were repeated for solutions containing ligand 2 and 0.5 or 1.0 molar equivalent (molar eq) [Cu(I)(CH3CN)4]PF6, respectively. The temperature calibration of the variable temperature control unit was verified on the basis of the chemical shift changes of a methanol sample. Full line shape analysis by using the gnmr software package (24) (gnmr software calculates the full bandshape of signals that are broadened as a result of chemical exchange) provided the exchange rates with kobs = rate/c(Lfree) + rate/c(CuL).
Cell Culture. Mouse NIH 3T3 cells were cultivated in DMEM supplemented with 5% calf serum and 200 mM l-glutamine. Copper uptake studies were performed in the same medium but supplemented with 150 μM copper(II) chloride. For the staining experiments cells were incubated with 10 μM CTAP-1 (in DMEM) for 50 min at 37°C, washed with PBS, and fixed with 3.7% paraformaldehyde for 30 min before mounting on slides with ProLong (Molecular Probes). For immunofluorescence colocalization, NIH 3T3 cells were fixed (3.7% paraformaldehyde, 30 min), permeabilized (0.2% Triton X-100, 2 min), and incubated with either mouse anti-GS28 IgG1 (Stressgen Bioreagents, Victoria, Canada) or mouse anti-OxPhos complex V IgG1 (Molecular Probes) primary antibodies (1:500 dilution, 1 h), respectively, and goat anti-mouse IgG Alexa Fluor 546 (Molecular Probes) as a secondary antibody following a standard immunofluorescence protocol. After the last washing step, the cells were further incubated with 10 μM CTAP-1 (in PBS, filtered) for 50 min at room temperature. The cells were imaged with a Zeiss Axiovert fluorescence microscope equipped with a standard filter set (DAPI, FITC, RITC).
microXRF. Scanning x-ray fluorescence (XRF) microscopy was performed at beamline 2-ID-D of the Advanced Photon Source at the Argonne National Laboratory (25). Incident x-rays of 10 keV (1 eV = 1.602 × 10–19 J) energy were chosen to excite elements from P to Zn. A Fresnel zone plate focused the x-ray beam to a spot size of 0.2 × 0.2 μm2 on the specimen, which was raster-scanned (26). XRF from the specimen was captured with an energy dispersive Ge detector. Spectral analysis of the fluorescence spectrum of each raster pixel then provided spatial images for each element (27). At the hard x-ray regime, biological cells do not cause significant absorption or beam spreading, hence no specimen thinning is required, and the fluorescence image represents a two-dimensional projection of the volumetric distribution for each element. For sample preparation, cells were allowed to attach to the formvar layer of a gold electron microscopy (EM) grid. Upon treatment with CTAP-1 as described above, cells were washed in PBS, fixed in methanol/acetone (50/50) at –20°C for 2 min, and then air dried.
Microprobe X-Ray Absorption Near-Edge Structure (microXANES). Once an interesting area was identified in the XRF image, microXANES (28) was performed by scanning the energy of the incident x-ray across the Cu K edge (≈8.979 keV). The zone plate and the specimen remained stationary during the measurement, which ensured that the illuminated area was fixed during the energy scan, although slight defocusing increased the spot size to ≈0.25 × 0.25 μm2.
Results and Discussion
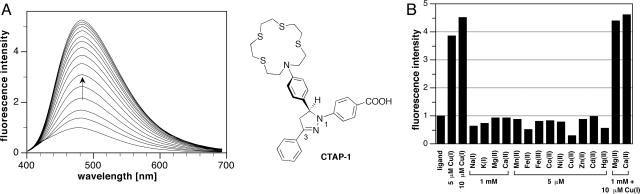
Fluorescent Probe Design. To visualize kinetically labile copper with subcellular resolution, a fluorescent sensor that undergoes bright fluorescence enhancements is required; however, monovalent copper typically acts as an efficient fluorescence quencher (29) and therefore poses a significant design challenge. As recently demonstrated by Rurack et al. (30), electronic decoupling combined with a rigid sensor architecture is an effective strategy to achieve large fluorescence enhancements, even for typical quenchers such as Hg(II), Ag(I), or Cu(II). The fluorescence intensity of these sensors is modulated by means of quenching through photoinduced electron transfer (PET), which, upon binding of the analyte, becomes energetically unfavorable and results in a fluorescence increase. The quenching efficiency is essentially determined by the relative rate of radiative deactivation vs. electron transfer. According to Marcus' theory, the latter directly correlates with the driving force –ΔGPET of the PET process, which can be estimated based on the oxidation potential E1/2(D+/D) of the donor, typically a Lewis base involved in analyte coordination, the excited state energy ΔG00 of the fluorophore, and the ground-state reduction potential E1/2(A/A–) of the fluorophore acting as electron acceptor (31). Upon coordination of the metal cation, the oxidation potential of the Lewis base increases, which in turn leads to a decrease of –ΔGPET and the ET rate, and thus an increase of the fluorescence intensity. In principle, this approach should also be applicable for sensing of Cu(I); however, depending on the ligand environment, this cation also may act as electron donor for the PET process, thus significantly limiting the decrease of –ΔGPET and thus the emission enhancement upon Cu(I) binding. Because of the smaller size of the available free energy window Δ(–ΔGPET), the design of a Cu(I)-selective sensor requires careful tuning of –ΔGPET by means of adjusting the reduction potential of the fluorophore and its excited-state energy. Although for most fluorophore platforms the two parameters are interdependent, we recently found that 1,3-diarylpyrazolines permit selective adjustment of both parameters (32). This fluorophore platform is not only attractive due to this unique property, it also provides a high degree of synthetic flexibility, a large molar absorptivity, high quantum yield, and low toxicity in biological applications (32). Combination of a tetrathiaza crown ether as copper-selective receptor with a photophysically tailored, disubstituted pyrazoline fluorophore yielded the water-soluble sensor CTAP-1 (Fig. 1) (CTAP, copper-responsive triarylpyrazoline). The sulfur-rich ligand environment is expected to yield a sufficiently positive oxidation potential for the corresponding Cu(I) complex (33), thus not only maximizing the free energy change window but also protecting coordinated copper from disproportionation.
Fig. 1.
Metal-dependent fluorescence response of CTAP-1. (A) (Left) Fluorescence emission spectra (λex 365 nm) of CTAP-1 (5 μM in 10 mM Pipes, pH 7.20, 25°C) as a function of added Cu(I) [0.1 molar eq aliquots of 5 mM Cu(CH3CN)4PF6 in CH3CN]. (Right) Structure of CTAP-1. (B) Emission response of CTAP-1 at 480 nm (λex 365 nm) as a function of various added metal cations (5 μM ligand/10 mM Pipes, pH 7.20).
Photophysical Properties and Cu(I)-Binding Studies. An aqueous solution of the water-soluble sensor CTAP-1 (5 μM in 10 mM Pipes, pH 7.2, 25°C) was titrated with 0.1 molar eq aliquots of a 5 mM stock solution of [Cu(I)(CH3CN)4]PF6 in acetonitrile. In the absence of Cu(I), the fluorescence emission was almost completely quenched with a quantum yield of 3% (Fig. 1 A). After addition of 1 molar eq Cu(I), the fluorescence intensity at 485 nm increased 4.6-fold and reached a quantum yield of 14%. Furthermore, the emission intensity increased linearly up to 0.8 molar eq, suggesting a fractional saturation (ratio between the complex concentration and total metal concentration) near unity at each titration point. Because reliable binding constants can only be obtained from titration data with fractional saturation of <80% (34), the data set is not suitable to extract the Cu(I) stability constant of CTAP-1. The fractional saturation could principally be reduced by titrating the ligand at lower concentration. However, under these conditions the amount of Cu(I) that is not bound to CTAP-1 would increase and be subject to disproportionation. It is therefore necessary to supplement the solution with a stabilizing ligand of known Cu(I) affinity. Kamau and Jordan (35) have recently demonstrated that a 0.2 M aqueous solution of CH3CN (1.2 ml in 100 ml) is sufficient to shift the disproportionation equilibrium in favor of Cu(I). Titration of CTAP-1 at 1/10 of the previous concentration in 10 mM Pipes (pH 7.2) containing 0.32 M CH3CN provided then suitable conditions in terms of fractional saturation and Cu(I) stability (see Fig. 6, which is published as supporting information on the PNAS web site). The binding isotherm is consistent with a 1:1 complex stoichiometry, and nonlinear least-squares fit over the entire spectral range yielded an apparent binding affinity of logK = 6.88 ± 0.05. The stability constants for the aqueous Cu(I)–CH3CN system have been accurately determined (35) and can be used in the nonlinear least-squares data analysis to account for competitive binding by CH3CN. With this data treatment, the apparent binding affinity of CTAP-1 was estimated to be logK = 10.4 ± 0.1. The 1:1 complex stoichiometry was additionally confirmed by a 1H NMR titration experiment in neat CH3CN (see Figs. 7 and 8, which are published as supporting information on the PNAS web site).
Cation Selectivity. Titration of CTAP-1 with other first-row transition metals in aqueous solution at neutral pH revealed excellent selectivity toward monovalent copper. As shown in Fig. 1B, Cu(I) is the only cation among the tested transition elements that induces a fluorescence enhancement. The emission intensity is not affected by the presence of millimolar concentrations of Ca(II) or Mg(II) (Fig. 1B, far right), an observation that is of great importance for biological applications.
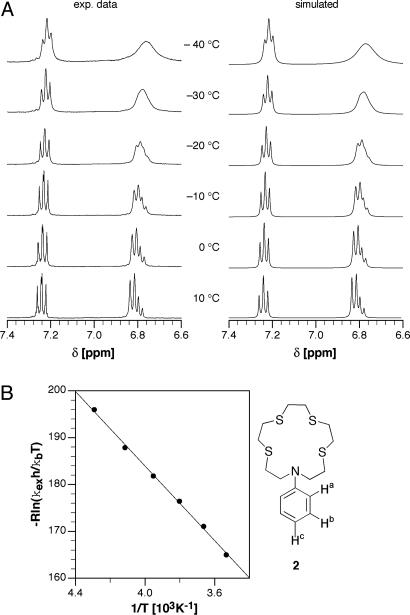
Metal Exchange Kinetics. To measure cation fluxes with temporal resolution, a sensor should provide a rapid metal exchange kinetics. Because the binding constant corresponds to the ratio of the rate constants for complexation and decomplexation, the metal exchange kinetics of ligands with high affinity is often assumed to be slow. However, this assumption is only true in the case of a dissociative exchange mechanism, whereas very fast exchange rates can be observed for associative pathways (36). To explore the Cu(I) exchange kinetics of the thiaza crown ether receptor, we performed variable temperature 1H NMR experiments with ligand 2 in the presence of 0.5 molar eq Cu(I) (Fig. 2A). Given the high Cu(I) affinity of the tetrathiaza crown ether, the concentration of free and bound ligand can be assumed to be identical under these conditions, representing a thermodynamically degenerate (self-) exchange equilibrium. At room temperature, the system is at the fast exchange limit on the NMR time scale, showing merely a single set of the averaged proton resonances. At <10°C, the resonances undergo line broadening, from which the temperature-dependent exchange rates kobs can be extracted by means of full line shape analysis (Table 1). Besides the experimental 1H NMR spectra, Fig. 2 A also depicts the simulated spectra as obtained with the gnmr software package (24).
Fig. 2.
Dynamic NMR study of the intermolecular Cu(I)-self-exchange equilibrium of ligand 2. (A) Variable-temperature 1H NMR spectra of ligand 2 in the presence of 0.5 molar eq Cu(I) (1 mM in CD3CN). (B)(Left) Eyring plot based on the exchange data given in Table 1. (Right) Structure of ligand 2.
Table 1. Temperature-dependent rate constants kobs and thermodynamic parameters for the intermolecular Cu(I)-self-exchange equilibrium of ligand 2.
| T[K] | kobs, S-1* | -R/In(kobsh/kBT) | ΔG‡, kJ·mol-1 |
|---|---|---|---|
| 283 | 43,800 | 155.6 | 44.1 |
| 273 | 22,800 | 170.8 | 43.9 |
| 263 | 11,500 | 166.1 | 43.6 |
| 253 | 5,790 | 171.5 | 43.4 |
| 243 | 2,740 | 177.4 | 43.2 |
| 233 | 1,100 | 184.6 | 43.0 |
Exchange rate based on full line shape iteration using the gnmr software package (24). Spectra of free and complexed ligand were acquired at each temperature and used as references for the exchange simulation.
An Eyring plot shows a linear correlation for the Cu(I)/ligand 2 self-exchange equilibrium and yields the activation parameters ΔH‡ = 37.8 kJ·mol–1 and ΔS‡ = –22 J·mol–1·K–1 (Fig. 2B). In principle, the metal exchange can proceed via a dissociative or associative pathway; however, the two mechanisms represent limiting cases, and both pathways may contribute more or less to the overall exchange rate (37). Given the limited accessible temperature range, the activation entropy derived from Eyring plots is inevitably associated with considerable uncertainty; nevertheless, the observed negative activation entropy indicates preferences for an associative pathway, presumably by means of formation of a “sandwich”-like transition state as observed for alkali–metal crown ether complexes in the gas phase (38). Although definite mechanistic conclusions cannot be drawn on the basis of this data set, the dynamic NMR study nevertheless demonstrates a rapid kinetics for the Cu(I)/ligand self-exchange equilibrium with a low activation barrier of ΔG‡ = 44 kJ·mol–1 and kobs ∼ 105 s–1 at room temperature.
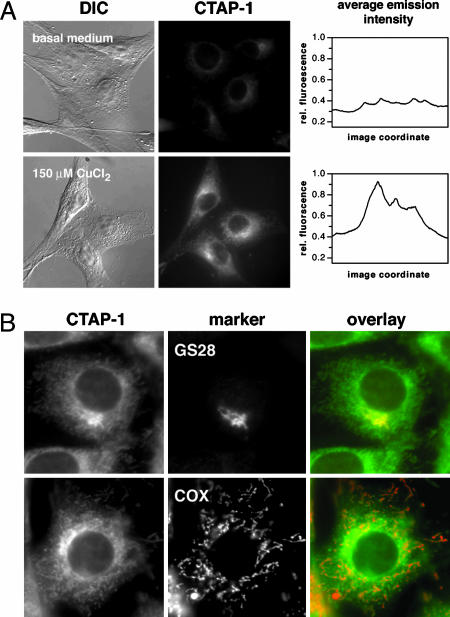
In Vivo Evaluation. Incubation of NIH 3T3 fibroblasts with 10 μM CTAP-1 produced a weak perinuclear staining pattern (Fig. 3A Upper). Supplementation of the growth medium with 150 μM CuCl2 for 12 h resulted in an ≈2- to 3-fold stronger emission intensity compared with cells cultured in basal medium (Fig. 3A Lower). The staining is particularly strong in proximity to the nuclear envelope and appears to colocalize with the Golgi apparatus. To quantitatively assess differences in emission intensities between the two growth conditions, the depicted fluorescence micrographs were acquired with a high-resolution 12-bit charge-coupled device camera using precisely identical exposure times and microscope settings. For better comparison, the profile of the average emission intensity across the x-coordinate of each fluorescence micrograph is shown in Fig. 3A Right. Incubation in medium supplemented with 150 μM bathocuproin sulfonate, a copper-selective membrane-impermeable extracellular chelator, led to a relatively small decrease of the fluorescence intensity compared with basal conditions (data not shown). These results correlate well with data from the literature acquired by means of metabolic labeling with 64Cu or by atomic absorption spectroscopy, showing an ≈3- to 5-fold increase at 100 μM CuCl2 and 30% decrease upon incubation with 200 μM bathocuproin sulfonate (16).
Fig. 3.
Fluorescence micrographs showing the intracellular staining pattern of NIH 3T3 cells incubated with 10 μM CTAP-1 for 50 min. (A)(Left and Center) Influence of the extracellular copper concentration. (Upper) Cells were grown in basal medium. (Lower) Medium supplemented with 150 μM CuCl2 for 8 h. (Right) The graphs depict the average intensity profile along the x-coordinate of the fluorescence micrographs (DIC, differential interference contrast image). (B) Immunofluorescence colocalization of the CTAP-1 staining pattern (Left) with two cellular marker antibodies (Center). (Upper) Anti-GS28 to visualize the Golgi apparatus. (Lower) Anti-OxPhos Complex V to visualize mitochondria (for details, see Experimental Methods). (Right) False color overlay (CTAP-1, green; antibody, red; areas of colocalization appear in orange/yellow).
To identify organelles or compartments that might be associated with the observed staining pattern, we performed immunofluorescence colocalization experiments with a series of cellular markers. As shown in Fig. 3B Upper, the copper-dependent staining pattern of CTAP-1 matches the subcellular localization of the Golgi apparatus; however, additional, weaker stained areas can be detected throughout the cytoplasm. Visualization of the intracellular mitochondria distribution revealed a high degree of colocalization with this weaker pattern (Fig. 3B Lower). Because the net charge of CTAP-1 at physiological pH is either neutral or negative, staining artifacts through accumulation within the negatively charged environment of the mitochondrial matrix are less probable. Other staining artifacts could be introduced through the mildly acidic luminal pH of the Golgi apparatus [pHG = 6.4 ± 0.3 (39)], potentially leading to protonation-induced fluorescence enhancements (32); however, the emission intensity of an aqueous solution of CTAP-1 (0.1 M KCl ionic background) decreases upon acidification from pH 7.2 to 4.5 (see Fig. 9, which is published as supporting information on the PNAS web site), indicating that the observed perinuclear staining pattern is not due to protonation of CTAP-1. In an additional control experiment, we tested whether the intracellular fluorescence of CTAP-1 observed under high-copper growth conditions is reversed upon addition of an exogenous Cu(I) chelator. For this purpose, NIH 3T3 fibroblast cells were cultured first for 12 h in medium supplemented with 150 μM of Cu(II) chloride and then treated with 2 μM CTAP-1 in the presence or absence of 10 mM 3,6-dithia-1,8-octanediol (DTO) as exogenous chelator (for experimental details, see Supporting Text). Quantitative image analysis of the fluorescence micrographs revealed that the average intracellular fluorescence intensity of the control cells was ≈50% higher (40 cells; P < 0.0001, Student's t test) compared with DTO-treated cells (see Fig. 10, which is published as supporting information on the PNAS web site). Furthermore, a set of cell-free fluorescence experiments in buffer solution demonstrated that DTO is able to competitively remove Cu(I) from CTAP-1. Addition of 2 mM DTO was sufficient to reverse the Cu(I)-induced emission increase of CTAP-1 but had no effect on the emission intensity in the absence of Cu(I) (see Fig. 11, which is published as supporting information on the PNAS web site).
In summary, the data suggest that under copper-supplemented growth conditions, the fibroblast cells accumulate copper predominantly in the Golgi region and mitochondria. The latter observation is corroborated through studies by Winge and coworkers (40), who recently demonstrated the existence of a nonproteinaceous copper pool in the mitochondrial matrix of yeast cells.
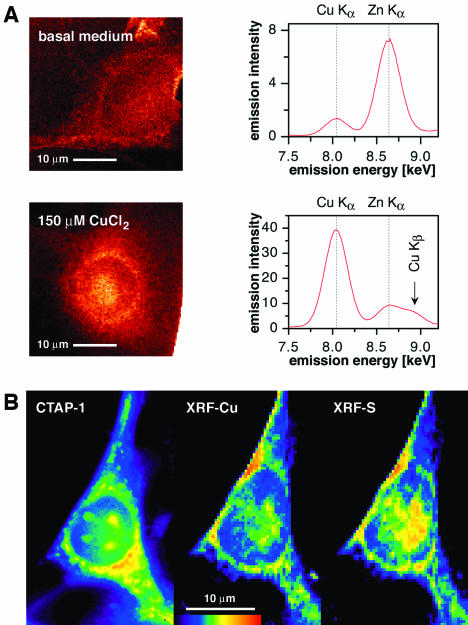
microXRF. To study the subcellular distribution of copper with an independent technique, we performed a series of microXRF experiments at the Advanced Photon Source of the Argonne National Laboratory. The instrument at the 2-ID-D beamline has a detection limit of ≈10–19 mol·μm–2 for manganese, iron, copper, or zinc and a spatial resolution of 200 nm, rendering it well-suited for quantitative elemental mapping of single cells with subcellular resolution (27). Cells, which were cultured in medium supplemented with 150 μM CuCl2, yielded an ≈20-fold increased Cu Kα signal at 8.0 keV compared with cells grown in basal medium (Fig. 4A). The perinuclear intensity distribution of the XRF copper map strongly resembles the staining pattern observed with CTAP-1. Interestingly, the ratio between the average emission intensity of copper and zinc appears reversed; however, the total amount of zinc varies only little between the two samples.
Fig. 4.
Imaging of the subcellular copper distribution by microXRF. (A)(Left) XRF copper maps of NIH 3T3 cells grown in basal medium (Upper) or in medium supplemented with 150 μM CuCl2 (Lower). The emission intensity of each pixel was normalized by using the beam intensity as reference. The maps are reproduced on the following intensity scales: basal medium, 0.0002–0.0104 μg/cm2; high copper medium, 0.0017–0.1634 μg/cm2. (Right) X-ray emission spectra for copper and zinc averaged over the entire area of the corresponding cell (normalized to beam intensity). (B) False-color micrographs of a single NIH 3T3 fibroblast cell grown in medium supplemented with 150 μM CuCl2 and treated with 10 μM CTAP-1 for 50 min. (Left) Epifluorescence image acquired with a DAPI filter set. (Center) XRF copper map. (Right) XRF sulfur map. The relative intensities increase in the order blue, green, yellow, orange, red.
To assess the degree of colocalization between the epifluorescence staining pattern of CTAP-1 and the subcellular copper topography obtained by means of microXRF, cells were grown in medium supplemented with 150 μM CuCl2, incubated with CTAP-1, and inspected by optical and microXRF microscopy (Fig. 4B). Although drying of the cells, which is required to perform the microXRF experiments, caused some distortion of the overall structure, the micrographs indicate a high degree of colocalization. Interestingly, because the cells were fixed in methanol/acetone rather than formaldehyde, CTAP-1 also stained the nucleoli, which in agreement with the XRF map appear to contain elevated levels of copper. A comparison between the copper and sulfur topography reveals a significant colocalization, suggesting that copper might be primarily coordinated by sulfur-donor ligands.
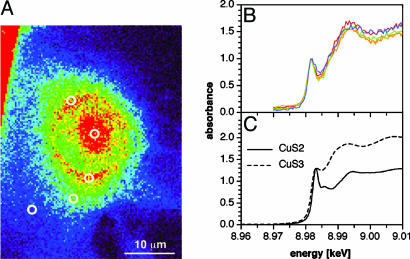
microXANES. The energy and shape of the near-edge x-ray absorption band of copper complexes is very sensitive toward variations in oxidation state and coordination environment. Whereas Cu(I) complexes exhibit a preedge feature centered around 8,984 eV characteristic for the 1s → 4p transition (41), the analogous transition of Cu(II) complexes occurs at significantly higher energy (8,986 eV). microXANES performed on a copper-loaded NIH 3T3 cell revealed at all tested subcellular locations a near-edge feature that is characteristic for monovalent copper (Fig. 5B). Given the higher edge energy of Cu(II), the observed band shape does not exclude the presence of Cu(II); nevertheless, the spectrum demonstrates that a significant portion of the total copper is present as Cu(I). Because of the complex mixture of potentially coordinating endogenous ligands, a detailed interpretation of the XANES spectra is not possible; however, qualitative comparison with two reference compounds suggests a low-coordinate linear or trigonal geometry (Fig. 5C). According to the literature, neither metallothionein nor glutathione exhibit the observed, characteristic XANES near-edge feature (42–44) and are therefore most likely not the prevalent endogenous ligands for coordination of kinetically labile copper in these pools.
Fig. 5.
NIH 3T3 fibroblast cell grown in medium supplemented with 150 μM CuCl2. (A) False-color XRF micrograph showing the copper distribution (color code as in Fig. 4B). (B) XANES spectra acquired at various locations (marked with white rings in A). (C) XANES reference spectra: CuS2 = [Cu(SC10H13)2][N(C3H7)4] (45) and CuS3 = [Cu(SC6H5)3][P(C6H5)4] (46).
Concluding Remarks
Copper-selective fluorescence sensing combined with XRF imaging microscopy provided a coherent picture regarding the subcellular localization of copper in the studied NIH 3T3 cell line. The copper-dependent intracellular staining pattern observed with CTAP-1 and XRF microscopy strongly supports the existence of the hypothesized labile copper pool, which appears to be localized in mitochondria and the Golgi apparatus. microXANES experiments confirmed the predominance of low-coordinate, monovalent copper throughout the cell and are inconsistent with predominant coordination to metallothionein or glutathione. At present, the nature of the endogenous ligand is elusive; however, Winge and coworkers (40) recently provided the first evidence that kinetically labile, mitochondrial copper is coordinated to a nonproteinaceous, low-molecular-weight ligand carrying a negative net complex charge. It is critical to note that the fluorescent sensor cannot provide actual quantitative concentration information. Because copper must be assumed to be coordinated to a yet unknown endogenous ligand, the sensor is inevitably engaged in a competitive metal exchange equilibrium, whose position depends not only on the relative binding affinities but also the local concentrations of the sensor and endogenous ligand. Although XRF microscopy can provide quantitative information about the total copper concentration regardless of its kinetic availability, copper-selective fluorescence sensing complements this technique in a synergistic fashion, thus representing a powerful partnership for the investigation of the biological chemistry of labile copper pools and their possible role in cellular homeostasis.
Supplementary Material
Acknowledgments
We thank Jonathan D. Gitlin for his advice and James E. Penner-Hahn for providing us with XANES reference data. This work was supported by the Georgia Institute of Technology and National Institutes of Health Grants R01GM067169 and R01DK68096. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38.
Author contributions: C.J.F. designed research; L.Y., R.M., M.M.H., R.P., B.L., and C.J.F. performed research; L.Y., B.L., S.V., and C.J.F. analyzed data; and C.J.F. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PET, photoinduced electron transfer; XRF, x-ray fluorescence; microXRF, synchrotron-based microprobe XRF; XANES: x-ray absorption near-edge spectroscopy; microXANES, microprobe XANES; molar eq, molar equivalent.
References
- 1.Tapiero, H., Townsend, D. M. & Tew, K. D. (2003) Biomed. Pharmacol. 57, 386–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Halliwell, B. & Gutteridge, J. M. (1984) Biochem. J. 219, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Field, L. S., Luk, E. & Culotta, V. C. (2002) J. Bioenerg. Biomembr. 34, 373–379. [DOI] [PubMed] [Google Scholar]
- 4.Huffman, D. L. & O'Halloran, T. V. (2001) Annu. Rev. Biochem. 70, 677–701. [DOI] [PubMed] [Google Scholar]
- 5.Arnesano, F., Banci, L., Bertini, I. & Ciofi-Baffoni, S. (2004) Eur. J. Inorg. Chem., 1583–1593.
- 6.Puig, S. & Thiele, D. J. (2002) Curr. Opin. Chem. Biol. 6, 171–180. [DOI] [PubMed] [Google Scholar]
- 7.Gitlin, J. D. (2003) Gastroenterology 125, 1868–1877. [DOI] [PubMed] [Google Scholar]
- 8.Mercer, J. F. B. (2001) Trends Mol. Med. 7, 64–69. [DOI] [PubMed] [Google Scholar]
- 9.Multhaup, G., Schlicksupp, A., Hesse, L., Beher, D., Ruppert, T., Masters, C. L. & Beyreuther, K. (1996) Science 271, 1406–1409. [DOI] [PubMed] [Google Scholar]
- 10.Waggoner, D. J., Bartnikas, T. B. & Gitlin, J. D. (1999) Neurobiol. Dis. 6, 221–230. [DOI] [PubMed] [Google Scholar]
- 11.Rae, T., Schmidt, P., Pufahl, R., Culotta, V. & O'Halloran, T. (1999) Science 284, 805–808. [DOI] [PubMed] [Google Scholar]
- 12.Herd, S. M., Camakaris, J., Christofferson, R., Wookey, P. & Danks, D. M. (1987) Biochem. J. 247, 341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bertinato, J. & L'Abbe, M. R. (2004) J. Nutr. Biochem. 15, 316–322. [DOI] [PubMed] [Google Scholar]
- 14.Palmiter, R. D. (1998) Proc. Natl. Acad. Sci. USA 95, 8428–8430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maret, W., Larsen, K. S. & Vallee, B. L. (1997) Proc. Natl. Acad. Sci. USA 94, 2233–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamza, I., Prohaska, J. & Gitlin, J. D. (2003) Proc. Natl. Acad. Sci. USA 100, 1215–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tepikin, A., ed. (2001) Calcium Signaling: A Practical Approach (Oxford Univ. Press, Oxford).
- 18.London, R. E. (1991) Annu. Rev. Physiol. 53, 241–258. [DOI] [PubMed] [Google Scholar]
- 19.Kikuchi, K., Komatsu, K. & Nagano, T. (2004) Curr. Opin. Chem. Biol. 8, 182–191. [DOI] [PubMed] [Google Scholar]
- 20.Lim, N. C., Freake, H. C. & Bruckner, C. (2004) Chem. Eur. J. 11, 38–49. [DOI] [PubMed] [Google Scholar]
- 21.Chen, P. & He, C. A. (2004) J. Am. Chem. Soc. 126, 728–729. [DOI] [PubMed] [Google Scholar]
- 22.Sakamoto, H., Ishikawa, J., Mizuno, T., Doi, K. & Otomo, M. (1993) Chem. Lett. 609–612.
- 23.Demas, J. N. & Crosby, G. A. (1971) J. Phys. Chem. 75, 991–1024. [Google Scholar]
- 24.Budzelaar, P. H. M. (2002) gnmr (Adept Scientific, Letchworth, U.K.).
- 25.Cai, Z., Lai, B., Yun, W., Ilinski, P., Legnini, D., Maser, J. & Rodrigues, W. (2000) Am. Inst. Phys. Proc. 507, 472–477. [Google Scholar]
- 26.Yun, W., Lai, B., Cai, Z., Maser, J., Legnini, D., Gluskin, E., Chen, Z., Krasnoperova, A. A., Vladimirsky, Y., Cerrina, F., et al. (1999) Rev. Sci. Instrum. 70, 2238–2241. [Google Scholar]
- 27.Twining, B. S., Baines, S. B., Fisher, N. S., Maser, J., Vogt, S., Jacobsen, C., Tovar-Sanchez, A. & Sanudo-Wilhelmy, S. A. (2003) Anal. Chem. 75, 3806–3816. [DOI] [PubMed] [Google Scholar]
- 28.Dillon, C. T., Kennedy, B. J., Lay, P. A., Lai, B., Cai, Z., Stampfl, A. P. J., Ilinski, P., Legnini, D., Maser, J., Rodrigues, W., et al. (2003) J. Phys. IV (French) 104, 293–296. [Google Scholar]
- 29.Rapisarda, V. A., Volentini, S. I., Farias, R. N. & Massa, E. M. (2002) Anal. Biochem. 307, 105–109. [DOI] [PubMed] [Google Scholar]
- 30.Rurack, K., Kollmannsberger, M., Resch-Genger, U. & Daub, J. (2000) J. Am. Chem. Soc. 122, 968–969. [Google Scholar]
- 31.Rehm, D. & Weller, A. (1970) Isr. J. Chem. 8, 259–271. [Google Scholar]
- 32.Fahrni, C. J., Yang, L. C. & VanDerveer, D. G. (2003) J. Am. Chem. Soc. 125, 3799–3812. [DOI] [PubMed] [Google Scholar]
- 33.Bernardo, M. M., Heeg, M. J., Schroeder, R. R., Ochrymowycz, L. A. & Rorabacher, D. B. (1992) Inorg. Chem. 31, 191–198. [Google Scholar]
- 34.Deranleau, D. A. (1969) J. Am. Chem. Soc. 91, 4044–4049. [Google Scholar]
- 35.Kamau, P. & Jordan, R. B. (2001) Inorg. Chem. 40, 3879–3883. [DOI] [PubMed] [Google Scholar]
- 36.Cheesman, B. V., Arnold, A. P. & Rabenstein, D. L. (1988) J. Am. Chem. Soc. 110, 6359–6364. [Google Scholar]
- 37.Szczygiel, P., Shamsipur, M., Hallenga, K. & Popov, A. I. (1987) J. Phys. Chem. 91, 1252–1255. [Google Scholar]
- 38.Chu, I. H., Zhang, H. & Dearden, D. V. (1993) J. Am. Chem. Soc. 115, 5736–5744. [Google Scholar]
- 39.Wu, M. M., Llopis, J., Adams, S., McCaffery, J. M., Kulomaa, M. S., Machen, T. E., Moore, H. P. H. & Tsien, R. Y. (2000) Chem. Biol. 7, 197–209. [DOI] [PubMed] [Google Scholar]
- 40.Cobine, P. A., Ojeda, L. D., Rigby, K. M. & Winge, D. R. (2004) J. Biol. Chem. 279, 14447–14455. [DOI] [PubMed] [Google Scholar]
- 41.Kau, L. S., Spirasolomon, D. J., Pennerhahn, J. E., Hodgson, K. O. & Solomon, E. I. (1987) J. Am. Chem. Soc. 109, 6433–6442. [Google Scholar]
- 42.Pickering, I. J., George, G. N., Dameron, C. T., Kurz, B., Winge, D. R. & Dance, I. G. (1993) J. Am. Chem. Soc. 115, 9498–9505. [Google Scholar]
- 43.Smith, T. A., Lerch, K. & Hodgson, K. O. (1986) Inorg. Chem. 25, 4677–4680. [Google Scholar]
- 44.Corazza, A., Harvey, I. & Sadler, P. J. (1996) Eur. J. Biochem. 236, 697–705. [DOI] [PubMed] [Google Scholar]
- 45.Koch, S. A., Fikar, R., Millar, M. & O'Sullivan, T. (1984) Inorg. Chem. 23, 121–122. [Google Scholar]
- 46.Coucouvanis, D., Murphy, C. N. & Kanodia, S. K. (1980) Inorg. Chem. 19, 2993–2998. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.