Abstract
Although erythroid cells and megakaryocytes arise from a common progenitor, their terminal maturation follows very different paths; erythroid cells undergo cell-cycle exit and enucleation, whereas megakaryocytes continue to progress through the cell cycle but skip late stages of mitosis to become polyploid cells. In our efforts to identify genes that participate in this process, we discovered that survivin, a member of the inhibitor of apoptosis family that also has an essential role in cytokinesis, is differentially expressed during erythroid versus megakaryocyte development. Erythroid cells express survivin throughout their maturation, whereas megakaryocytes express ≈4-fold lower levels of survivin mRNA and no detectable protein. To investigate the role of survivin in these lineages, we overexpressed or knocked down survivin from mouse bone marrow cells and then examined erythroid and megakaryocyte development. These studies revealed that overexpression of survivin antagonized megakaryocyte growth, maturation, and polyploidization but had no effect on erythroid development. This block in polyploidization was accompanied by increased expression of p21 and decreased expression of megakaryocyte genes such as von Willebrand factor and β1-tubulin. In contrast, a reduction in survivin expression interfered with the formation of erythroid cells but not megakaryocytes. Last, consistent with the requirement for survivin in the survival of proliferating cells, survivin-deficient hematopoietic progenitors failed to give rise to either erythroid or megakaryocytic colonies. Together, these studies show that whereas survivin expression is essential for megakaryocyte and erythroid progenitors, its down-regulation is required for terminal differentiation of megakaryocytes.
Keywords: erythropoiesis, hematopoiesis, megakaryopoiesis
Survivin is a 16.5-kDa protein with a single baculovirus inhibitor of apoptosis (IAP) repeat (BIR) domain and a coiled-coil region in its C terminus (1). Because of the presence of the BIR domain, survivin has been placed in the IAP family. Indeed, several reports have shown that overexpression of survivin is associated with inhibition of cell death. However, the mechanism of the antiapoptotic function of survivin is unclear. Although some studies have demonstrated a direct interaction with caspases 3 and 7, other studies failed to detect an association between these proteins (2, 3). It is possible that the antiapoptotic effect of survivin may be mediated by indirect association with other proapoptotic or antiapoptotic molecules (4–7). Altogether, the role of survivin as a bona fide inhibitor of apoptosis remains controversial. In contrast, survivin unequivocally has an essential, evolutionarily conserved role in mitosis. Survivin expression is generally cell-cycle-regulated, with expression peaking during G2/M, when survivin functions as an essential chromosome passenger protein to regulate cytokinesis (8, 9). Survivin is essential for the viability of proliferating cells, because knocking out or interfering with its activity results in abnormal cytokinesis, polyploidization, and eventual cell death (10, 11). Murine gene-targeting studies have confirmed that survivin is an essential protein, because homozygous knockout embryos displayed gross cellular degeneration, lacked an inner cell mass, and failed to progress beyond embryonic day 4.5. Furthermore, the cells within the mutant embryos exhibited disrupted microtubule formation and polyploidization (8).
Although survivin is generally cell-cycle-regulated, there are a few examples in which survivin is expressed throughout the cell cycle. For example, survivin is expressed at low levels in fresh umbilical-cord blood and bone-marrow-derived quiescent CD34+ cells, and it is rapidly up-regulated after incubation with a cytokine mixture consisting of thrombopoietin, stem cell factor (SCF), and flt3 ligand (12). Interestingly, although survivin expression was highest in the G2/M phases of the cell cycle, it was also detected throughout the cell cycle (12). With respect to other hematopoietic cells, survivin also has an important role in the survival of terminally differentiated neutrophils under inflammatory conditions (13) and in the development and homeostasis of T cells (14, 15). Recently, it has been demonstrated that mature antigen responding CD4 T cells require sustained survivin expression to maintain T cell proliferation. Also, this sustained survivin expression was induced by OX40 cosignaling independent of mitotic progression (16).
Because several articles have shown that the experimental reduction of survivin leads to polyploidization, we sought to determine whether megakaryocytes, the only hematopoietic cell that undergoes repeated rounds of DNA synthesis without cell division, would express survivin. Megakaryocytes and red blood cells share a common progenitor, the megakaryocyte–erythroid progenitor, but their respective terminally differentiated cells have very different functions and express a very different set of genes. Moreover, there are major differences in cell-cycle progression and nuclear maturation; erythroid cells undergo cell-cycle arrest, nuclear condensation, and enucleation, whereas megakaryocytes proceed through endomitosis. Here, we show that although survivin is expressed in erythroid cells during their maturation, as late as the orthochromatic stage of differentiation, murine megakaryocytes express ≈4-fold lower levels of survivin mRNA and no detectable survivin protein. This difference in expression is likely to be physiologically relevant because we discovered that experimental modulation of survivin differentially affects the outgrowth of these two cells. Overexpression of survivin in murine bone marrow progenitors led to a decreased production of megakaryocytes and a block in their terminal maturation and polyploidization. In contrast, a reduction in survivin expression by RNA interference (RNAi) or heterozygous deletion of the survivin gene caused a decrease in the outgrowth of erythroid cells but had no effect on megakaryocytes. Furthermore, consistent with the requirement for survivin in proliferating cells, survivin-deficient bone marrow progenitors failed to give rise to erythroid or megakaryocytic colonies in vitro. Thus, although survivin is required at the progenitor stage of both lineages, its down-regulation is an essential component of megakaryocyte maturation. These results have implications in other areas of biology; we predict that other types of polyploid cells, including trophoblast giant cells and hepatocytes, require down-regulation of survivin for their maturation.
Materials and Methods
Cell Culture and Differentiation Assays. Human K562 cells were cultured in RPMI medium 1640 supplemented with 10% FCS. Erythroid and megakaryocytic differentiation of K562 cells was induced by addition of 1 μM cytosine arabinoside (araC) and 10 nM phorbol 12-myristate 13-acetate (PMA), respectively. Primary bone marrow cells were obtained from femurs and tibiae of 8- to 10-week-old C57BL/6 mice and expanded for 4 days in a serum-free expansion medium containing SCF, IL-3, and a low dose (2 units/ml) of erythropoietin (EPO). Cells were then differentiated by using higher doses (10 units/ml) of EPO for 4 days (17). Differentiation of human CD34+ cells to erythroid cells was performed as described (18). For analysis of ploidy, GFP+/CD41+ cells were sorted by FACS and permeabilized by 70% ethanol, and the DNA content was analyzed by flow cytometry after staining with propidium iodide. Approval for the use of animals in this study was granted by the University of Chicago Institutional Animal Care and Use Committee.
Quantitative RT-PCR (qRT-PCR). RNA from BSA-gradient-purified murine erythroid cells or megakaryocytes (19), expanded ex vivo, were extracted by using TRIzol reagent (Invitrogen). For the experiments shown in Fig. 2, RNA was extracted from FACS-purified CD41+ or Ter119+ cells by using TRIzol reagent. Relative quantitation of real-time PCR product was performed as described (20). Semiquantitative RT-PCR was performed by using standard methods, with serial 5-fold dilutions of input cDNA included in the PCRs. Primer sequences are available on request.
Fig. 2.
Effect of survivin overexpression on megakaryocytic and erythroid differentiation. (a) Human survivin cDNA was cloned into the MIGR1 retroviral vector. (b) Survivin expression in bone marrow cells infected with the MIGR1 or the MIGR1-survivin retrovirus. (c and d) Erythroid and megakaryocytic differentiation as measured by Ter119 and CD41 positivity, respectively, in primary bone marrow cells infected with control (MIGR1) or survivin (MIGR1-survivin)-expressing retroviruses. CD41 and Ter119 expression were analyzed in GFP+ cells. Data from representative (c) and mean ± SD of three experiments (d) are shown. SSC, side scatter. *, P = 0.004 for difference in CD41 expression and 0.008 for difference in Ter119 expression (Student's t test). (e) Cell-cycle analysis of the CD41+ GFP+ subpopulation of primary bone marrow cultures infected with either MIGR1 or the MIGR1-survivin (MIGR1-SUR) retrovirus after 2 days of differentiation. (f) qRT-PCR analysis of transcripts expressed in sorted CD41+ GFP+ cells infected with either MIGR1 or the MIGR1-survivin retrovirus. VWF, von Willebrand factor.
Survivin Overexpression and Knockdown Liquid Cultures. The human survivin cDNA was cloned into the MIGR1 vector, whereas the mouse survivin short hairpin RNA (shRNA; 386-CAATTGAGCAGCTGGCTGCC-407) and the control shRNA (human survivin; 12-GAATCGCGGGACCCGTTGGCAGAGGTGGC-40) were introduced downstream of the U6 promoter by using a PCR strategy (21) and cloned into the MSCV vector with a phosphoglycerate kinase (PGK)-Puro-internal ribosomal entry site (IRES)-GFP cassette. Primary bone marrow cells were infected by spinoculation as described (20) on days 2 and 3 of expansion. Differentiation was initiated the next day, and cells were analyzed by FACS for differentiation after an additional 3 or 4 days.
Colony-Forming Assays. Primary mouse bone marrow cells were collected from survivin+/+, survivinfl/+, or survivinfl/fl mice (15) and enriched for progenitors with the Easy Sep negative-selection mouse hematopoietic progenitor-enrichment kit (Stem Cell Technologies, Vancouver). The cells were then infected by spinoculation with retroviruses expressing either Cre and GFP (MSCV-Cre-IRES-GFP) or GFP alone (MSCV-IRES-GFP) on days 2 and 3 of expansion. The next day, GFP+ cells were collected by FACS and plated into methylcellulose. For the growth of mature burst-forming units (BFU)-E colonies, 20,000 GFP+ cells were seeded into MethoCult 3234 (Stem Cell Technologies), supplemented with 10 units/ml EPO. Mature BFU-E colonies were enumerated after 4 days. For megakaryocyte colony assays, 40,000 GFP+ cells were mixed with MegaCult-C (Stem Cell Technologies), containing Tpo, IL-3, IL-6, and IL-11 and plated onto two double-chamber culture slides, and colonies were enumerated after 8 days. For the colony assays shown in Fig. 3, C57BL/6 bone marrow cells were treated as described above, except that they were infected with retroviruses harboring either the human survivin cDNA (MIGR1-survivin) or the vector alone (MIGR1).
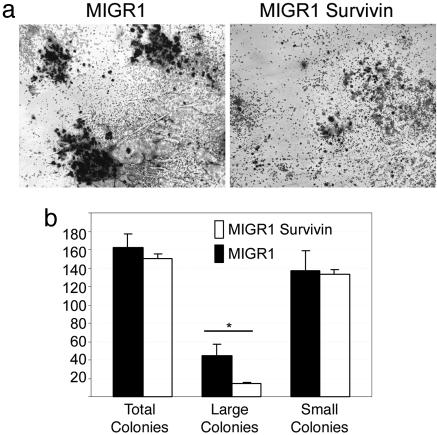
Fig. 3.
Effect of survivin overexpression on CFU-MK formation. Lineage-depleted bone marrow cells were infected with control (MIGR1) or survivin (MIGR1-survivin)-expressing retroviruses as shown in Fig. 2. Sorted GFP+ cells were plated in methylcellulose media to promote megakaryocyte colony formation. (a) Colonies were evaluated for total numbers and morphology after staining for acetylcholinesterase. (b) The number of total colonies, as well as the proportion of small and large colonies, were compared between cells infected with control (MIGR1) or survivin (MIGR1-survivin)-expressing retroviruses. Median ± SE from three experiments is shown. *, P < 0.05 (Mann–Whitney U test).
Cell Staining, Antibodies, and Flow Cytometry. Surface staining for human CD41, human CD42, and mouse Ter119 (BD Pharmingen) was performed by using phycoerythrin (PE)-conjugated antibodies in Ca2+-free, Mg2+-free PBS with 2% serum. Surface staining for mouse CD41 or CD61 was analyzed by using a purified anti-mouse CD41 or CD61 antibody (BD Pharmingen), followed by staining with PE or PE-Cy5-conjugated secondary antibody (Jackson ImmunoResearch). Cytoplasmic staining for survivin expression was performed as described (14), with a polyclonal anti-survivin antibody (AF886; R & D Systems) after fixing the cells in 2% paraformaldehyde, followed by permeabilization with Perm/Wash buffer (BD Biosciences). All flow cytometry was performed on a FACScan (BD Biosciences), and data were analyzed by using flojo software. The anti-survivin mouse mAb (6E4; Cell Signaling Technology, Beverly, MA) was used for immunofluorescence staining. Western blot analyses to detect survivin in K562 cells were performed by using the polyclonal and monoclonal anti-survivin antibodies, and analyses to detect survivin in primary cells were performed with the polyclonal anti-survivin antibody.
Results
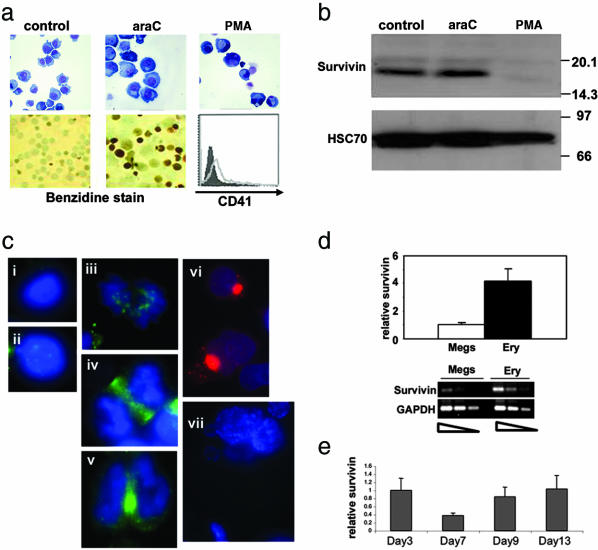
To determine whether megakaryopoiesis is associated with a decrease in survivin expression, we induced megakaryocytic differentiation in the bipotential human cell line K562 with the phorbol ester PMA. At 96 h after PMA treatment, the cells exhibited basophilic cytoplasm, indented nuclei, and staining for both CD41 and CD42, consistent with megakaryocytic differentiation (Fig. 1a and data not shown). As expected, there was a marked reduction in survivin expression in this population of cells (Fig. 1b). Furthermore, we could not detect survivin in primary polyploid megakaryocytes generated from murine fetal livers (Fig. 1c, vii), consistent with ref. 22. In contrast, K562 cells treated with araC, which undergo erythroid differentiation (as evidenced by benzidine staining; Fig. 1a), displayed persistent survivin expression (Fig. 1b). Primary human erythroid progenitors at the basophilic stage of differentiation exhibited survivin localization consistent with a role as a chromosomal passenger protein (8) (Fig. 1c i–v), whereas cells at the terminal stage of differentiation (orthochromatic) uniformly displayed a focal, cytoplasmic localization that is distinct from that observed in cycling cells (Fig. 1c vi). Note that primary human erythroid cells continued to express survivin at the orthochromatic stage of differentiation, when most cells are exiting the mitotic cell cycle (23). We also performed real-time qRT-PCR on RNA isolated from purified populations of murine erythroid cells and megakaryocytes (Fig. 1d). Survivin mRNA was detected in both lineages, but its expression was reduced ≈4-fold in megakaryocytes as compared with red blood cells. This difference was similar to that observed in K562 cells treated with either PMA or araC (data not shown). By using qRT-PCR, we also discovered that survivin mRNA was present in primary human erythroid cells during their in vitro differentiation from CD34+ cells through the orthochromatic stage of maturation (Fig. 1e). The persistent expression of survivin RNA and protein during late stages of erythroid maturation suggests that survivin may also have an important role in the terminal differentiation of this lineage.
Fig. 1.
Survivin expression during megakaryocytic and erythroid differentiation. (a) K562 cells were either untreated or cultured with araC or PMA for 96 h and assayed for erythroid and megakaryocytic differentiation by May–Grunwald–Wright–Giemsa stain (Upper), benzidine staining, and CD41 expression (Lower). (b) Survivin expression was measured by Western blot analysis of extracts from K562 cells treated as described for a. Similar results were obtained with two different anti-survivin antibodies. (c) Immunofluorescence detection of survivin expression in human CD34+ cells induced to undergo erythroid maturation. Cells at day 7 in interphase (i), prophase (ii), metaphase (iii), anaphase (iv), and telophase (v); cells at day 11 (vi); and megakaryocytes expanded from murine fetal liver (vii) are shown. Note that a different secondary antibody was used in vi as compared with the other panels. The mouse monoclonal anti-survivin antibody used in this experiment gave results that were consistent with the known localization of survivin in mitotic cells (8). (d) Quantitative assessment of survivin mRNA in purified murine megakaryocytes (Megs) and erythroid cells (Ery) cultured ex vivo. Quantitative (Upper) and semiquantitative (Lower) results are shown. (e) Levels of survivin transcripts in varying stages of erythroid maturation were assayed by qRT-PCR. Cells were collected after 3 (proliferating CD34+ cells), 7 (basophilic), 9 (polychromatophilic), or 13 (orthochromatic erythroblasts) days of culture. Survivin expression is shown relative to that detected on day 3.
To investigate whether forced expression of survivin could influence the expansion and/or differentiation of hematopoietic progenitors, we infected primary mouse bone marrow cells with retroviruses harboring either the human survivin cDNA fused to an IRES-GFP cassette or the IRES-GFP cassette alone (Fig. 2a). After infection, cells were grown in a single liquid culture under conditions that favored the development of erythroid cells and megakaryocytes. These culture conditions allowed us to investigate the requirement for survivin in the development of these specific populations. Infected cells were distinguished by the presence of GFP expression, and the differentiation toward erythroid cells or megakaryocytes was monitored by the expression of lineage-specific surface antigens. Overexpression of survivin in the MIGR1-survivin-infected cells was verified by Western blot analysis (Fig. 2b). In a representative experiment, bone marrow populations that expressed GFP alone gave rise to 66% Ter119+ erythroid cells and 13% CD41+ megakaryocytes (Fig. 2c). Expression of survivin led to an increase in erythroid differentiation (to 79% Ter119+ cells), with a concomitant decrease in the extent of megakaryocytic differentiation (to 7% CD41+ cells). Note that the same phenotype was observed when megakaryocytes were detected by staining for a different megakaryocyte marker, CD61, and that the cells that stained for CD41 or CD61 were consistently larger than nonstained cells (data not shown); these assays confirm their identity as megakaryocytes. Because ectopic survivin expression led to a significant decrease in megakaryocyte outgrowth (Fig. 2d;14 ± 1% versus 8 ± 1%, P = 0.004), down-regulation of survivin is likely to be an important step in megakaryopoiesis. Conversely, overexpression of survivin led to an increase in the number of Ter119+ cells (Fig. 2d; 57 ± 8% versus 68 ± 10%, P = 0.008). Of note, analysis of survivin expression in purified Ter119+ and CD41+ cells by qRT-PCR verified that survivin was overexpressed in the terminally differentiated cells by 4- to 7-fold (data not shown). Together, these results suggest that elevated levels of survivin favor the expansion of erythroid cells over megakaryocytes.
Because survivin overexpression might be predicted to interfere with polyploidization, we next compared the DNA content of CD41+ cells generated in the presence or absence of ectopically expressed survivin. We found that there was an accumulation of CD41+ cells with a 4n DNA content and a concomitant diminution in the fraction of cells reaching a ploidy of >4n in the survivin-overexpressing population in comparison with the control MIGR1-infected cells (Fig. 2e). These observations support the hypothesis that overexpression of survivin interferes with the ability of megakaryocytes to undergo polyploidization. To investigate whether overexpression of survivin also affected other aspects of megakaryocyte maturation, we isolated mRNA from FACS-purified CD41+ GFP+ MIGR1-survivin-infected cells and from the control sorted CD41+ GFP+ MIGR1-vector-infected cells. qRT-PCR revealed that expression of late markers of megakaryocyte maturation, including β1-tubulin and, to a lesser extent, von Willebrand factor, were reduced upon overexpression of survivin (Fig. 2f). Interestingly, consistent with an aberrant cell-cycle progression, the expression of p21 was elevated in survivin-overexpressing cells (Fig. 2f). In comparison, expression of other cell-cycle regulators, including p27, cyclin B, and cyclin D1, was unaffected (Fig. 2f). Together, these results show that overexpression of survivin interferes with expansion, polyploidization, and terminal maturation of megakaryocytes.
To further characterize the effects on hematopoiesis seen in our liquid-culture experiments, we next performed erythroid and megakaryocyte colony assays. BFU-E colony formation, with respect to both number and size of colonies, was not significantly affected by overexpression of survivin (data not shown). In contrast, overexpression of survivin had a marked effect on megakaryocyte colonies. Although the total number of colonies was not altered, overexpression of survivin gave rise to abnormal colonies that were small and poorly formed (Fig. 3a). The number of large colonies, defined as those harboring ≥50 megakaryocytes, was decreased ≈3-fold in outgrowths from progenitors overexpressing survivin (Fig. 3b). These findings demonstrate that overexpression of survivin did not affect the commitment of cells to the megakaryocyte lineage.
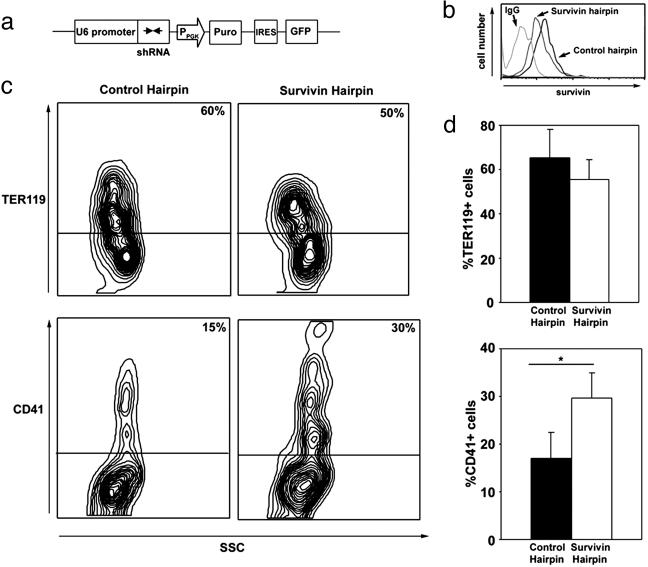
If the regulation of survivin contributed to differential expansion of these two lineages, then reducing survivin expression in bone marrow progenitors might lead to an increase in megakaryocytes. To determine the effect of survivin down-regulation, we PCR-generated an shRNA against the C terminus of the mouse survivin cDNA (survivin hairpin 1), which is downstream of the U6snRNA promoter (21), and we then incorporated the U6shRNA cassette into the MSCV-PIG retroviral vector, which contains a PGK-Puro-IRES-GFP expression cassette (Fig. 4a). As a control, we used a hairpin that targeted the 5′ noncoding region of the human survivin cDNA (control hairpin). Primary murine bone marrow cells were infected with either the mouse or human survivin shRNA-expressing retrovirus and then differentiated in liquid culture. Flow cytometry for intracellular survivin confirmed the reduced survivin expression in bone marrow cells infected with mouse survivin hairpin (Fig. 4b). Consistent with the antiapoptotic role of survivin, we discovered that the bone marrow culture infected with the survivin shRNA, but not the population infected with human shRNA or the survivin cDNA, exhibited a moderate decrease in the percentage of cells expressing GFP over the course of differentiation, with a concomitant increase in the proportion of GFP+ cells that expressed annexin V (data not shown). Subsequent analysis of hematopoietic differentiation revealed a reduction in the number of Ter119+ cells generated from bone marrow harboring the survivin hairpin as compared with the control hairpin (from 60% to 50%; Fig. 4c). Concomitant with the reduction of Ter119+ erythroid cells, there was an increase in CD41+ megakaryocytes in cells harboring the survivin hairpin (from 15% to 30%; Fig. 4c). These data constitute a statistically significant increase in megakaryopoiesis upon reduced survivin expression (17 ± 5% versus 30 ± 6%, P = 0.01; Fig. 4d). Thus, down-regulation of survivin favored the expansion of megakaryocytes over erythroid cells.
Fig. 4.
Effect of survivin down-regulation on megakaryocytic and erythroid differentiation. (a) U6shRNAi cassette containing the shRNAi against mouse survivin (survivin hairpin) or human survivin (control hairpin) were cloned into the MSCV-PIG vector. PGK, phosphoglycerate kinase. (b) Intracellular survivin expression in GFP+ cells populations was measured by flow cytometry. (c and d) Erythroid and megakaryocytic differentiation as measured by Ter119 and CD41 positivity, respectively, in primary bone marrow cells infected with the control or survivin hairpin. Data from a representative experiment (c) and the mean ± SD of three experiments (d) are shown. *, P < 0.01.
Our RNAi studies suggested that megakaryocytes and erythroid cells exhibit differential requirements for survivin. To determine unambiguously whether survivin is required for development of only one or both of these lineages, we performed hematopoietic colony assays with bone marrow progenitors from survivin conditionally targeted mice (15). First, we harvested bone marrow from 8- to 10-week-old survivin+/+, survivinfl/+, and survivinfl/fl mice; infected these cells with retroviruses that expressed either Cre and GFP or GFP alone; sorted for GFP+ cells; and then performed in vitro colony-forming assays. Heterozygous loss of survivin resulted in >50% reduction in survivin mRNA expression (Fig. 5a), with only the deleted and wild-type alleles detectable by PCR (Fig. 5b). The introduction of Cre into survivinfl/fl progenitors resulted in a decrease in survivin mRNA to <20% of that in GFP-infected control cells (Fig. 5a). This residual expression of survivin was most likely caused by incomplete excision of the floxed allele because the floxed and deleted alleles both were detected by PCR (Fig. 5b).
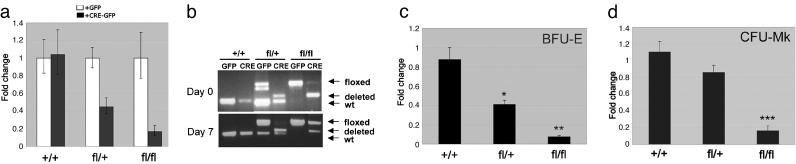
Fig. 5.
Survivin is required for BFU-E and CFU-MK formation. (a) Level of survivin mRNA was quantified by qRT-PCR in the different GFP+ populations before plating. (b) Excision was monitored by PCR in DNA from cells collected at days 0 and 7 of methylcellulose culture. wt, wild type. (c and d) BFU-E and CFU-MK assays. Data are depicted as the changes in the number of colonies upon expression of Cre in comparison with expression of GFP alone. Mean ± SD of three or four experiments are shown. *, P < 0.02; **, P < 0.01; ***, P < 0.0002. Note that the difference between CFU-Mk+/+ to +/fl was not significant (Student's t test).
Data from the colony assays revealed that erythroid progenitors are more sensitive to the levels of survivin than those of the megakaryocyte lineage. Heterozygous survivinfl/+ progenitors expressing Cre gave rise to <50% BFU-Es in comparison with those expressing GFP alone (Fig. 5c). In marked contrast, megakaryocyte colony formation was unaffected by the heterozygous loss of survivin (Fig. 5d). In comparison, complete excision of survivin affected the formation of colonies of both lineages. The survivinfl/fl progenitors that were infected with Cre failed to generate significant numbers of BFU-Es or colony-forming units (CFU)-Mks. The few colonies that were formed in these experiments corresponded to those that escaped complete excision by Cre, as determined by PCR of DNA isolated from the residual survivinfl/fl MSCV-Cre-GFP+ colonies (Fig. 5b Lower). Thus, we conclude that survivin is essential for the proliferation and/or survival of erythroid–megakaryocyte progenitors but that erythroid cells have a second requirement for high levels of survivin in a cell downstream of the common progenitor.
Discussion
Survivin is a protein with multiple functions, including an essential role in cytokinesis and a possible role as an inhibitor of apoptosis. Here, we demonstrate a differential requirement for survivin during erythroid and megakaryocytic maturation. Our data suggest that survivin is involved in at least three different stages of erythroid and megakaryocytic development. First, we infer that there is an essential requirement for survivin in the proliferation of the common progenitor, because survivin-deficient bone marrow failed to give rise to either BFU-E or CFU-Mk colonies. Second, we show that there is an additional requirement for high-level survivin expression in erythroid progenitors downstream of the megakaryocyte–erythroid progenitor. Erythroid colony formation was reduced significantly when survivin expression was decreased by 50%, whereas that of the megakaryocyte lineage was unaffected. Furthermore, in liquid culture, a reduction in survivin expression led to a preferential expansion of megakaryocytes. Last, we demonstrate that survivin expression needs to be reduced during the maturation of megakaryocytes; overexpression of survivin caused a block in megakaryocyte polyploidization and a reduced expression of megakaryocyte-specific genes. Furthermore, the size of megakaryocyte colonies was significantly reduced when survivin was overexpressed. Although overexpression of survivin has not been associated with inhibition of proliferation or cell death, our data suggest that its presence in a cell that is programmed to undergo endomitosis is detrimental.
Maturing megakaryocytes enter an endomitotic phase in which cells proceed through prophase and metaphase but exit anaphase prematurely (24). Recent evidence suggests that endoreplication is likely to be a consequence of a unique regulation of chromosome passenger proteins, such as BubR1 (25), Aurora B, and survivin (22). The kinase Aurora B is recruited to the kinetochores by survivin to regulate mitosis and cytokinesis (3). Although it is expressed in murine megakaryocytes during prophase, it is absent or mislocalized during late anaphase, when Aurora B and survivin usually become localized to the midzone in dividing cells (22). Interestingly, overexpression of Aurora B in cell lines treated with phorbol esters prevented polyploidization (26) and transgenic mice that overexpress Aurora B in megakaryocytes show evidence of abnormal maturation, with an enhanced number of megakaryocytes with increased proliferative potential and, in some cases, a mild decrease in ploidy level (22). These findings support the conclusion that Aurora B needs to be degraded, or mislocalized, at late stages of the endomitotic cell cycle in megakaryocytes.
Although we found that survivin mRNA is present in murine megakaryocytes, we failed to detect survivin at the protein level when assayed by both Western blot analysis and immunofluorescence. These observations suggest that survivin is likely to be regulated at a posttranscriptional level in megakaryocytes. These results are consistent with those of Ravid and colleagues (22), who reported that survivin could not be detected in murine megakaryocytes at any stage of polyploidization. In contradiction to those findings, a recent report (27) has shown that survivin is expressed at both the mRNA and protein level in human megakaryocytes. A possible explanation for this discrepancy may be the fact that human megakaryocytes reach a much lower ploidy when cultured ex vivo, as compared with their murine counterparts.
Although many groups have shown that loss of survivin leads to aberrant cell division, our data demonstrate a physiologically relevant setting for the polyploidization that accompanies survivin down-regulation. We found that overexpression of survivin interfered with polyploidization and development of primary murine megakaryocytes. As part of this block, the expression of p21 was elevated in survivin-overexpressing cells (Fig. 2f). This change is noteworthy because overexpression of p21 in megakaryocytes has been reported to cause a marked inhibition of polyploidization (28), and knocking out p21 has been linked to an increased state of polyploidization (29, 30). These results have implications in other areas of biology; we predict that other types of polyploid cells, including trophoblast giant cells and hepatocytes, require down-regulation of survivin for their maturation. Indeed, a recent study has shown that overexpression of survivin interferes with the polyploidization of vascular smooth muscle cells (31).
Also, we have found that a partial reduction of survivin expression did not affect megakaryocyte growth. In liquid culture, this decrease favored their expansion over that of erythroid cells. Consistent with these results, reduced expression of the mitotic checkpoint protein BubR1 also differentially affected erythroid and megakaryocytic development. BubR1 heterozygous deficient mice displayed increased megakaryopoiesis coupled with decreased erythropoiesis, and many BubR1 heterozygous mice displayed marked anemia (25). Wang et al. (25) concluded that the primary consequence of the reduced expression of BubR1 was an increase in megakaryopoiesis. They then speculated that a weakened spindle checkpoint, which would lead to frequent chromosomal missegregation, would be better tolerated in a cell that is poised to undergo polyploidization. In contrast to this autonomous effect on megakaryocytes, the reduction in erythropoiesis was attributed to the “progenitor steal” effect, which was described as the depletion of the bipotential progenitor by the enhanced differentiation of megakaryocytes at the expense of erythroid cells. Our data suggest that an alternative explanation may be that BubR1 acts in concert with survivin to have an additional, essential role in the erythroid lineage.
Survivin is one of the most highly expressed genes in a wide spectrum of tumors (2). Because survivin is generally not expressed in adult tissues, it has been viewed as an excellent target for cancer therapy. However, recent reports (12–16) have shown that survivin has an essential role in variety of hematopoietic cells. Our studies reveal that survivin is also essential for erythropoiesis and suggest that inhibiting survivin may also interfere with the continued production and/or the survival of red blood cells.
Acknowledgments
We thank A. Winoto (University of California, Berkeley) for the survivin floxed mice; H. Nakshatri (Indiana University, Indianapolis) for the human survivin cDNA; K. Macleod (University of Chicago, Chicago) for the MSCV-Cre plasmid; S. Lowe (Cold Spring Harbor Laboratories, Cold Spring Harbor, NY) for the MSCV PIG vector; G. Mundaschau, A. Muntean, and J. Ulaszek for technical assistance; and C. Leung, S. Orkin, M. Weiss, G. Blobel, and B. Kee for critical reading of the manuscript. This work was supported in part by a Junior Faculty Award from the American Society of Hematology and National Institutes of Health Grants DK61464 and CA101774 (to J.D.C.).
Author contributions: S.G., Y.X., G.K., A.W., and J.D.C. designed research; S.G., Y.X., and G.K. performed research; and S.G., A.W., and J.D.C. analyzed data and wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: araC, cytosine arabinoside; PMA, phorbol 12-myristate 13-acetate; IRES, internal ribosomal entry site; shRNA, short hairpin RNA; BFU, burst-forming units; CFU, colony-forming units; EPO, erythropoietin; SCF, stem cell factor; RNAi, RNA interference; qRT-PCR, quantitative RT-PCR.
References
- 1.Ambrosini, G., Adida, C. & Altieri, D. C. (1997) Nat. Med. 3, 917-921. [DOI] [PubMed] [Google Scholar]
- 2.Altieri, D. C. (2003) Oncogene 22, 8581-8589. [DOI] [PubMed] [Google Scholar]
- 3.Reed, J. C. & Bischoff, J. R. (2000) Cell 102, 545-548. [DOI] [PubMed] [Google Scholar]
- 4.Marusawa, H., Matsuzawa, S., Welsh, K., Zou, H., Armstrong, R., Tamm, I. & Reed, J. C. (2003) EMBO J. 22, 2729-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song, Z., Yao, X. & Wu, M. (2003) J. Biol. Chem. 278, 23130-23140. [DOI] [PubMed] [Google Scholar]
- 6.Dohi, T., Okada, K., Xia, F., Wilford, C. E., Samuel, T., Welsh, K., Marusawa, H., Zou, H., Armstrong, R., Matsuzawa, S., et al. (2004) J. Biol. Chem. 279, 34087-34090. [DOI] [PubMed] [Google Scholar]
- 7.Sun, C., Nettesheim, D., Liu, Z. & Olejniczak, E. T. (2005) Biochemistry 44, 11-17. [DOI] [PubMed] [Google Scholar]
- 8.Uren, A. G., Wong, L., Pakusch, M., Fowler, K. J., Burrows, F. J., Vaux, D. L. & Choo, K. H. (2000) Curr. Biol. 10, 1319-1328. [DOI] [PubMed] [Google Scholar]
- 9.Wheatley, S. P., Carvalho, A., Vagnarelli, P. & Earnshaw, W. C. (2001) Curr. Biol. 11, 886-890. [DOI] [PubMed] [Google Scholar]
- 10.Li, F., Ambrosini, G., Chu, E. Y., Plescia, J., Tognin, S., Marchisio, P. C. & Altieri, D. C. (1998) Nature 396, 580-584. [DOI] [PubMed] [Google Scholar]
- 11.Li, F., Ackermann, E. J., Bennett, C. F., Rothermel, A. L., Plescia, J., Tognin, S., Villa, A., Marchisio, P. C. & Altieri, D. C. (1999) Nat. Cell Biol. 1, 461-466. [DOI] [PubMed] [Google Scholar]
- 12.Fukuda, S. & Pelus, L. M. (2001) Blood 98, 2091-2100. [DOI] [PubMed] [Google Scholar]
- 13.Altznauer, F., Martinelli, S., Yousefi, S., Thurig, C., Schmid, I., Conway, E. M., Schoni, M. H., Vogt, P., Mueller, C., Fey, M. F., et al. (2004) J. Exp. Med. 199, 1343-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okada, H., Bakal, C., Shahinian, A., Elia, A., Wakeham, A., Suh, W. K., Duncan, G. S., Ciofani, M., Rottapel, R., Zuniga-Pflucker, J. C. & Mak, T. W. (2004) J. Exp. Med. 199, 399-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xing, Z., Conway, E. M., Kang, C. & Winoto, A. (2004) J. Exp. Med. 199, 69-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song, J., So, T., Cheng, M., Tang, X. & Croft, M. (2005) Immunity 22, 621-631. [DOI] [PubMed] [Google Scholar]
- 17.Kolbus, A., Pilat, S., Husak, Z., Deiner, E. M., Stengl, G., Beug, H. & Baccarini, M. (2002) J. Exp. Med. 196, 1347-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uddin, S., Ah-Kang, J., Ulaszek, J., Mahmud, D. & Wickrema, A. (2004) Proc. Natl. Acad. Sci. USA 101, 147-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drachman, J. G., Sabath, D. F., Fox, N. E. & Kaushansky, K. (1997) Blood 89, 483-492. [PubMed] [Google Scholar]
- 20.Muntean, A. G. & Crispino, J. D. (April 28, 2005) Blood, 10.1182/blood-2005-02-0551. [DOI]
- 21.Paddison, P. J., Caudy, A. A., Bernstein, E., Hannon, G. J. & Conklin, D. S. (2002) Genes Dev. 16, 948-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang, Y., Nagata, Y., Yu, G., Nguyen, H. G., Jones, M. R., Toselli, P., Jackson, C. W., Tatsuka, M., Todokoro, K. & Ravid, K. (2004) Blood 103, 3717-3726. [DOI] [PubMed] [Google Scholar]
- 23.Wojda, U., Noel, P. & Miller, J. L. (2002) Blood 99, 3005-3013. [PubMed] [Google Scholar]
- 24.Ravid, K., Lu, J., Zimmet, J. M. & Jones, M. R. (2002) J. Cell. Physiol. 190, 7-20. [DOI] [PubMed] [Google Scholar]
- 25.Wang, Q., Liu, T., Fang, Y., Xie, S., Huang, X., Mahmood, R., Ramaswamy, G., Sakamoto, K. M., Darzynkiewicz, Z., Xu, M. & Dai, W. (2004) Blood 103, 1278-1285. [DOI] [PubMed] [Google Scholar]
- 26.Kawasaki, A., Matsumura, I., Miyagawa, J., Ezoe, S., Tanaka, H., Terada, Y., Tatsuka, M., Machii, T., Miyazaki, H., Furukawa, Y. & Kanakura, Y. (2001) J. Cell Biol. 152, 275-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geddis, A. E. & Kaushansky, K. (2004) Blood 104, 1017-1024. [DOI] [PubMed] [Google Scholar]
- 28.Baccini, V., Roy, L., Vitrat, N., Chagraoui, H., Sabri, S., Le Couedic, J. P., Debili, N., Wendling, F. & Vainchenker, W. (2001) Blood 98, 3274-3282. [DOI] [PubMed] [Google Scholar]
- 29.Mantel, C., Braun, S. E., Reid, S., Henegariu, O., Liu, L., Hangoc, G. & Broxmeyer, H. E. (1999) Blood 93, 1390-1398. [PubMed] [Google Scholar]
- 30.Fukuda, S., Mantel, C. R. & Pelus, L. M. (2004) Blood 103, 120-127. [DOI] [PubMed] [Google Scholar]
- 31.Nagata, Y., Jones, M. R., Nguyen, H. G., McCrann, D. J., St. Hilaire, C., Schreiber, B. M., Hashimoto, A., Inagaki, M., Earnshaw, W. C., Todokoro, K. & Ravid, K. (2005) Exp. Cell Res. 305, 277-291. [DOI] [PubMed] [Google Scholar]