Abstract
Although most globins, including the N-terminal domains within chimeric proteins such as flavohemoglobins and globin-coupled sensors, exhibit a 3/3 helical sandwich structure, many bacterial, plant, and ciliate globins have a 2/2 helical sandwich structure. We carried out a comprehensive survey of globins in the genomes from the three kingdoms of life. Bayesian phylogenetic trees based on manually aligned sequences indicate the possibility of past horizontal globin gene transfers from bacteria to eukaryotes. blastp searches revealed the presence of 3/3 single-domain globins related to the globin domains of the bacterial and fungal flavohemoglobins in many bacteria, a red alga, and a diatom. Iterated psi-blast searches based on groups of globin sequences found that only the single-domain globins and flavohemoglobins recognize the eukaryote 3/3 globins, including vertebrate neuroglobins, α- and β-globins, and cytoglobins. The 2/2 globins recognize the flavohemoglobins, as do the globin coupled sensors and the closely related single-domain protoglobins. However, the 2/2 globins and the globin-coupled sensors do not recognize each other. Thus, all globins appear to be distributed among three lineages: (i) the 3/3 plant and metazoan globins, single-domain globins, and flavohemoglobins; (ii) the bacterial 3/3 globin-coupled sensors and protoglobins; and (iii) the bacterial, plant, and ciliate 2/2 globins. The three lineages may have evolved from an ancestral 3/3 or 2/2 globin. Furthermore, it appears likely that the predominant functions of globins are enzymatic and that oxygen transport is a specialized development that accompanied the evolution of metazoans.
Keywords: evolution, sequences
Thirty years ago, our knowledge of globin sequences was limited to vertebrate α- and β-globins, myoglobins (Mbs), and the symbiotic Hbs of legume plants. The ensuing years brought the discovery of 3/3 nonsymbiotic plant Hbs (NsHbs) in plants, symbiotic Hbs in plants other than legumes, and chimeric flavohemoglobins (FHbs), which are comprised of an N-terminal globin linked to an FAD reductase domain, in bacteria and yeasts (1–8). Concurrently, globins shorter than normal (<130 aa), the “truncated” Hbs, were observed in protozoa and bacteria (9, 10), and globins longer than normal (>150 aa), which aligned with the truncated Hbs, were found in green alga (11) and plants (12). The crystal structures of these globins (13–15) exhibited a novel globin fold comprised of a 2/2 helical sandwich secondary structure. The recent rapid accumulation of genomic information has resulted in the discoveries of new types of globins, including the 3/3 neuroglobins (Ngbs) and 3/3 cytoglobins (Cygbs) believed to occur in all vertebrates (16), and of globins in organisms in which their presence had not been suspected, such as nematodes (17), an insect (18), and a urochordate (19). Furthermore, genomic data brought to light the existence of 3/3 single-domain globins (SDgbs), which aligned with the globin domain of the FHbs (20). A class of gene-regulating, chimeric heme proteins, the globin-coupled sensors (GCSs), was discovered in several bacterial groups and an archaean (21). The related single-domain protoglobins (Pgbs) found in archea and bacteria have been proposed to represent the “ancestral” globin (22). Here, we report the results of a comprehensive survey of globins.
Materials and Methods
Globin Sequences. Putative globins were identified in the genomes of 36 eukaryotes, including Cyanidioschyzon merolae (The C. merolae Genome Project, http://merolae.biol.s.u-tokyo.ac.jp), Thalassiosira pseudonana (Doe Joint Genome Institute database, http://genome.jgi-psf.org/thaps1/thaps1.home.html), and Chlamydomonas reinhardtii (The Chalmy Center, www.chlamy.org); 22 archea; and 129 bacteria (Tables 1 and 2, which are published as supporting information on the PNAS web site) by using a library of hidden Markov models (23) (Medical Research Council, http://supfam.mrc-lmb.cam.ac.uk/SUPERFAMILY) and with blastp and tblastn (Version 9.2.2) searches (24) (National Center for Biotechnology Information, www.ncbi.nlm.nih.gov/blast). Borderline globin sequences were evaluated with fugue (University of Cambridge, Cambridge, U.K.) (61).
Position-Specific Iterated blast (psi-blast). psi-blast searches were carried out by using sequences representative of various groups of globins as queries (www.ncbi.nlm.nih.gov/blast). The first round was a standard blastp search employing the blosum62 substitution matrix and default settings with a threshold of 0.005. A position-specific scoring matrix constructed from selected aligned sequences created the profile used as a query in the second iteration. This step was repeated in subsequent iterations until all of the known members of a given group of globins were identified above the threshold.
Alignment of Sequences and Molecular Phylogeny. Manual alignment, employing the Mb-fold pattern of predominantly hydrophobic residues at 37 solvent-inaccessible positions, was performed as described in ref. 25. For details, see Supporting Materials and Methods, which is published as supporting information on the PNAS web site. Bayesian analysis was carried out by using mrbayes 2.01 and the Jones–Taylor–Thornton transition matrix (26). Four chains were run simultaneously for 106 generations, and trees were sampled every 100 generations, producing a total of 104 trees. paup 4.0b10 was used for viewing and editing. modeltest 3.5 was used to determine the evolutionary model that best fit the small subunit rRNA sequences.
Results
Algal Globins. We identified one (CMR319C, 185 aa), two (Scaff_137, 158 aa, and Scaff_18, 160 aa) and three (C160981, C160982, and C157690) globins in the genomes of C. merolae, T. pseudonana, and C. reinhardtii, respectively. blastp searches (Tables 3 and 4, which are published as supporting information on the PNAS web site) showed that CMR319C and Scaff_137 recognized the bacterial SDgbs, vertebrate Ngbs, and bacterial and eukaryote FHbs, whereas Scaff_18 and the C. reinhardtii globins recognized only 2/2 Hbs.
Other Globins. A total of 21 SDgbs occur in 19 bacteria, of which 5 have FHbs as well (Table 3). Self-consistent lists of each of the three classes of bacterial 2/2 Hbs (10), ≈22 2/2 Hb1s, 59 2/2 Hb2s, and 25 2/2 Hb3s were obtained by psi-blast searches (Fig. 3 and Table 5, which are published as supporting information on the PNAS web site). A total of 42 GCSs and six Pgbs occur in microbial genomes (Table 6, which is published as supporting information on the PNAS web site). The alignment of globin sequences is shown in Fig. 3.
psi-blast Searches. The results in Table 3 reveal that the bacterial SDgbs and FHbs recognized vertebrate Ngbs first, followed by vertebrate, plant, and various metazoan Hbs. The results in Tables 4 and 6 demonstrate that (i) the individual groups of 2/2 Hbs did not recognize plant 3/3 symbiotic Hbs and NsHbs; (ii) all 2/2 Hbs recognized [expect value (E) ≈ e-4] bacterial and eukaryotic FHbs; and (iii) the GCSs recognized the Pgbs and the bacterial and eukaryotic FHbs (E ≈ e-4), whereas the Pgbs recognized only GCSs.
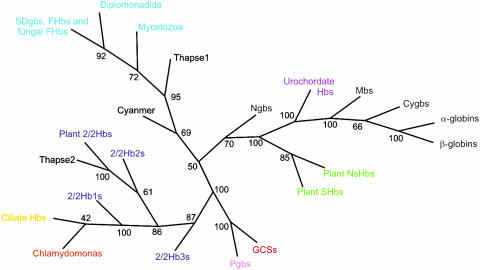
Molecular Phylogeny. The Bayesian trees of SDgbs, FHb globin domains, FHb complete sequences, and the small subunit rRNA sequences of FHb-containing organisms are respectively provided in Figs. 4–7, which are published as supporting information on the PNAS web site. A Bayesian tree of 2/2 Hbs, including one from the euryarcheote Haloarcula marismortui (27), is provided in Fig. 8, which is published as supporting information on the PNAS web site. A Bayesian phylogenetic tree, which is based on 150 sequences representing various groups of globins, is shown in Fig. 1. A detailed version of the tree and the alignment are given in Figs. 9 and 10, which are published as supporting information on the PNAS web site, respectively.
Fig. 1.
A consensus phylogenetic tree of major globin groups from the three kingdoms of life. The detailed version and the alignment are provided in Figs. 9 and 10, respectively.
Discussion
psi-blast Searches. We have used psi-blast searches with successive iterations based on the sequences of a given group of globins to detect distant evolutionary relationships to other groups of globins (Tables 3–6). The matches between the query sequence and each of the sequences in a database are assessed by the E value, which is also a function of the query length. Thus, E = 0.01 signifies that the probability of finding by chance another match with the query sequence having the same score is 1 in 100. We define recognition to be a hit with E < 0.005, the default threshold, and with the pairwise alignment fulfilling the following two criteria: proper alignment of the F8 His residues and of helices BC through G. The E values, rounded off to the nearest integer, are provided for the first member of a globin group hit (recognized) by the query sequence(s).
Three Globin Lineages in Two Structural Classes. The phylogenetic tree of 21 SDgbs in 19 bacteria (Fig. 4) illustrates their broad distribution among bacterial groups. Similar SDgbs were found in the red alga C. merolae (28) and the diatom T. pseudonana (29). The crystal structures of the SDgbs from Vitreoscilla stercoraria (30) and Pseudomonas aeruginosa (Protein Data Bank ID code 1TU9) and of the FHbs from Escherichia coli (31) and Ralstonia (Alcaligenes) eutrophus (32) show them to have a 3/3 helical sandwich structure. We have used psi-blast searches with successive iterations based on the sequences of all of the bacterial SDgbs and FHbs to detect similarities to other globin sequences, because it is known to be a powerful tool for the detection of remote homology in sequence (33, 34). The results (Table 3) show that the bacterial SDgbs recognize vertebrate Ngbs, Cygbs, α- and β-globins, plant 3/3 symbiotic Hbs, NsHbs, and many invertebrate globins: urochordate, echinoderm, annelid, crustacean, insect, mollusc, and nematode. Furthermore, the bacterial and eukaryote FHbs recognize the same groups as do the SDgbs, except for the Cygbs and the mollusc globins. Although the vertebrate Ngbs recognize all of the groups of globins, the highest recognition, i.e., the lowest E values, were with the bacterial SDgbs. The crystal structures of invertebrate globins have the same 3/3 helical sandwich structure as the vertebrate globins (35, 36). Hence, the bacterial and eukaryote SDgbs and FHbs, together with the metazoan globins, form the dominant lineage of extant globins belonging to the 3/3 structural class.
psi-blast searches using 2/2 Hb sequences as queries (Tables 4 and 5), show that (i) the plant and ciliate 2/2 Hbs recognize only the bacterial 2/2 Hbs from the same group, 2/2 Hb2s and 2/2 Hb1s, respectively, and (ii) the bacterial 2/2 Hbs and all 2/2 Hbs recognize (E ≈ e-4) bacterial and eukaryotic FHbs. Consequently, the 2/2 Hbs of bacteria, ciliates, plants, and algae represent another globin lineage, one that belongs to another structural class because of their different secondary structure (13–15). Fig. 11, which is published as supporting information on the PNAS web site, shows representative 3/3 and 2/2 structures.
The GCSs and the related Pgbs found in a couple of archaea and a variety of bacteria recognize each other (Table 6) and are clearly related (20, 21). The GCSs, but not the Pgbs, recognize (E ≈ e-4) the bacterial and eukaryotic FHbs. Thus, the GCSs and Pgbs represent another globin lineage belonging to the 3/3 structural class based on the only known crystal structure, that of Bacillus subtilis GCS (37).
The condensed Bayesian tree of representative globins shown in Fig. 1 (the full version is provided in Fig. 9), is consonant with our proposal of three globin lineages, with the SDgb/FHb lineage encompassing the extant 3/3 metazoan and plant globins. Surprisingly, the tree indicates a closer relationship between the 2/2 Hbs and the GCSs/Pgbs than would be expected from the lack of mutual recognition in psi-blast searches. This discrepancy may be explained by the greater homogeneity of the latter two groups relative to the SDgbs/FHbs and the fact that Bayesian phylogeny does not rely on profile similarities but compares the entire sequences.
Evidence for Horizontal Gene Transfer (HGT) of FHbs and 2/2 Hbs. Three indications of possible HGT occur in the phylogenetic tree of SDgbs and FHbs (Fig. 5, branches 1–3). The nestings of several yeast and the two Dictyostelium FHbs within a clade of Firmicutes, β- and ε-Proteobacteria, and the location of Giardia FHb within γ-Proteobacteria are supported by 100% probabilities. The occurrence of an Aspergillus FHb within several actinobacteria, α- and β-Proteobacteria, is less well supported. These indications are confirmed by the trees based on the complete FHb and small subunit rRNA sequences (Figs. 6 and 7, respectively). In the case of Giardia lamblia, HGT has been invoked for a dozen of its genes, including FHb (38). The clustering of Chlamydomonas and ciliate 2/2 Hbs within the bacterial 2/2 Hb1s (90% probability) and of the plant and Thalassiosira 2/2 Hbs within bacterial 2/2 Hb2s (99% probability; observed in Fig. 8) also raise the possibility of prokaryote-to-eukaryote HGT.
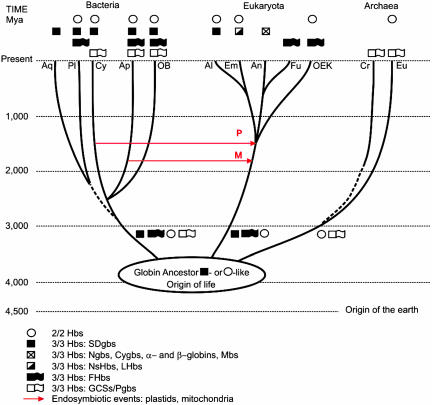
Early Globin Phylogeny. Speculations on the early phylogeny of globins should be considered in relation to the latest view of the history of life on Earth, which is depicted diagrammatically in Fig. 2 based on recent genomic analyses (39–41), with the origin of life occurring in the Hadean [≈4,100 million years ago (Mya)]. We proposed earlier, prior to the discovery of 2/2 Hbs, that all extant globins were descendants of a single-domain globin (42), which evolved from a cytochrome b-like precursor (43). Now it is more appropriate to ask whether the 3/3 and 2/2 globins evolved independently from a cytochrome b-like or other precursor(s) and which one of them represents the ancestral globin structure. During evolution, the problem of sequestering the water-insoluble heme in a hydrophobic protein cavity was solved in several ways. The similarity between the 3/3 and 2/2 structures (see Fig. 11), in contrast to the differences between them and other heme-sequestering folds (cytochrome fold, catalase, and PAS domain), argues against an independent origin of the two structures as suggested in ref. 10. The topology of our master tree shown in Fig. 1 provides no answer as to whether the 3/3 or the 2/2 fold is ancestral. Furthermore, we know from our results that 2/2 Hbs can be as long as 3/3 globins with lengths of >190 aa and that both structures are present in the three kingdoms of life. The presence of a single intron in the Paramecium 2/2 Hb gene (44) argues in favor of an ancestral 2/2 structure, which appears to provide the most parsimonious scenario for the structural evolution of globins. In contrast, a 3/3 globin ancestor similar to the SDgbs present today in all of the major bacterial groups is suggested by the following observations: the presence of SDgbs in Aquifex aeolicus and the planctomycete Rhodopirellula baltica, which represent two of the three deepest bacterial branches (45, 46), and the breadth of recognition of extant globin groups evinced by the bacterial SDgbs. Clearly, a resolution between the two scenarios is not possible at present.
Fig. 2.
A diagrammatic representation of the time scale of life based on refs. 39–41, depicting the distribution of the three globin lineages in the three kingdoms of life. The origin of Cyanobacteria (Cy) is estimated at ≈2,600 Mya, and the divergence of plants (Pl), animals (An), and fungi (Fu) is estimated at ≈1,500 Mya. The red arrows indicate the two generally accepted endosymbiotic events: between an α-proteobacterium (Ap) and a eukaryote at ≈1,800 Mya, resulting in the emergence of mitochondria (M), and between a cyanobacterium and a eukaryote at ≈1,500 Mya, resulting in the formation of plastids (P). The dotted lines indicate uncertainty in dating the split. Aq, Aquificales; OB, other bacteria; Al, Algae; Em, Embryophytes (land plants); OEk, other eukaryotes; Cr, Crenarcheota; Eu, Euryacheota.
The next two events in globin evolution, probably attendant on diversification and specialization of function (see below), were the covalent fusions of 3/3 SDgbs with nonglobin proteins having enzymatic or regulatory activities, resulting in FHbs and GCSs, respectively. The two events were probably independent and must also have occurred before the emergence of archaea, eukaryotes, and bacteria. Were all three lineages of globins present in each of the emerging three kingdoms of life? And when did the latter diverge? Although molecular evidence has been provided for dating the splits between archaea and eukaryotes at ≈4,000 Mya and the one between the eukaryotes and bacteria at ≈2,700 Mya (47), there is no agreement on these issues (48). If all three globin lineages were present in each of the three kingdoms of life upon their divergence, then obviously the GCSs and Pgbs were lost in the eukaryotes and the SDgbs and FHbs died out in the archaea. It is almost certain that the evolution of globins will require far more complicated scenarios than we can imagine at present. Thus, if globins were lost in any particular evolving eukaryote lineage, perhaps in the lines leading to the fungi, diplomonads, and mycetozoans, it is conceivable that they were acquired secondarily from bacteria by HGT, possibly coincident with the endosymbiotic events responsible for the origins of mitochondria and plastids (Fig. 2), a possibility that is supported by the phylogenetic trees (Figs. 5–7). Besides the foregoing two generally accepted endosymbiotic events, there is now substantial evidence of an earlier, massive HGT of ribosomal proteins between Archaea and Bacteria on the one hand and Archaea and Eukaryotes on the other (49). In addition, eukaryote initiation, termination, and elongation factors are most similar to those in the Crenarchaeota group of the Archaea, but the histone structures are common to the other main group of Archaea, the Euryarchaeota, and are not found in the Crenarchaeota or Bacteria (50). Thus, it appears that archaeal genes provided the protein-synthesizing and chromosomal-packing systems, and the bacterial genes supplied most of the metabolic and housekeeping genes. Furthermore, there is the issue of the origin of the eukaryote nucleus, which might have been the result of a much earlier union between a bacterium and an archaean (51). It is evident that the endosymbiotic events alluded to above provided ample opportunity for the shuffling of globins from all three lineages between the ancestors of the Archaea, Bacteria, and Eukaryota, irrespective of which structure was the ancestral one.
Evolution of Globin Function. We assume that the early evolution of a globin ancestor able to react with diatomic gaseous ligands was accompanied by the acquisition of several functions comprising the detection, sequestration, and detoxification of O2 and O2-derived species (e.g., NO and CO). Although atmospheric O2 levels were very low in the early history of the Earth [<0.0008 atm (1 atm = 101.3 kPa) (52)], local concentrations could have been high enough to promote the evolution of the globin ancestor. The principal role of extant 2/2 Hbs is considered to be in NO detoxification (53). Likewise, the functions of FHbs are primarily related to NO detoxification, aerobically as NO oxygenases or anaerobically as NO reductases (20, 54). Similar roles have been proposed for Vitreoscilla and Campylobacter SDgbs (55, 56). Although the function of GCSs appears to be that of an O2 sensor active in gene regulation (57), that of the few known Pgbs remains unknown.
Our main proposal is that all extant eukaryotic globins are descendants of a common ancestor 3/3 SDgb, irrespective of whether the earlier common globin ancestor had a 2/2 or a 3/3 structure, including the plant 3/3 symbiotic Hbs and NsHbs, the FHbs of lower eukaryotes, and the metazoan Hbs. Furthermore, it appears that the early functions were conserved in concert with the emergence of new functions, the latter being intimately related to the occurrence of hexa- vs. penta-coordination of the heme iron (58). The plant 3/3 symbiotic Hbs facilitate oxygen transport (58) as does Mb, which also acts as a NO scavenger (59). The plant NsHbs appear to be involved in maintaining the viability of plant cells under hypoxic conditions (60). The metazoan globins display a phenomenal diversity in structure varying from single-chain monomers to multidomain, multisubunit complexes (in annelids, molluscs, and crustaceans) and function in O2 transport and storage and protection against sulfide and have enzymatic activities ranging from oxidase and peroxidase-like to superoxide dismutase (7). The new vertebrate Ngbs and Cygbs have been proposed to function as facilitators of O2 diffusion to mitochondria and as providers of O2 to collagen prolylhydroxylases in fibroblasts, respectively (16). Because the original enzymatic functions, greatly expanded and diversified, remain the predominant functions of most extant globins, O2 and NO transport should be viewed as specialized developments that accompanied the emergence and evolution of metazoans subsequent to the rise in atmospheric O2 content to present-day levels.
Supplementary Material
Acknowledgments
This work was supported by Fund for Scientific Research Flanders Grant G.0331.04 and European Commission Grant QLG3-CT-2002-01548. S.D. is a postdoctoral fellow of the Fund for Scientific Research Flanders.
Author contributions: S.N.V. designed research; S.N.V., D.H., X.B., J.G., R.A.-P., and M.G. performed research; D.H., X.B., J.G., S.D., R.A.-P., and M.G. analyzed data; and S.N.V., L.M., and J.R.V. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Cygb, 3/3 cytoglobin; FHb, flavohemoglobin; GCS, globin-coupled sensor; HGT, horizontal gene transfer; Mb, myoglobin; Mya, million years ago; Ngb, 3/3 neuroglobin; NsHb, 3/3 nonsymbiotic plant Hb; Pgb, protoglobin; SDgb, 3/3 single-domain globin.
References
- 1.Vinogradov, S. N., Walz, D. A., Pohajdak, B., Moens, L., Kapp, O. H., Suzuki, T. & Trotman, C. N. A. (1993) Comp. Biochem. Physiol. B 106, 1-26. [DOI] [PubMed] [Google Scholar]
- 2.Hardison, R. C. (1996) Proc. Natl. Acad. Sci. USA 93, 5675-5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arredondo-Peter, R., Hargrove, M. S., Moran, J. F., Sarath, G. & Klucas, R. V. (1998) Plant Physiol. 118, 1121-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hardison, R. C. (1998) J. Exp. Biol. 201, 1099-1117. [DOI] [PubMed] [Google Scholar]
- 5.Weber, R. E. & Vinogradov, S. N. (2001) Physiol. Rev. 81, 569-628. [DOI] [PubMed] [Google Scholar]
- 6.Hill, R. D. (1998) Can. J. Bot. 76, 707-712. [Google Scholar]
- 7.Poole, R. K. & Hughes, M. N. (2000) Mol. Microbiol. 36, 775-783. [DOI] [PubMed] [Google Scholar]
- 8.Wacjman, H. & Kiger, L. (2002) C. R. Biol. 325, 1139-1174. [DOI] [PubMed] [Google Scholar]
- 9.Wittenberg, J. B., Bolognesi, M., Wittenberg, B. A. & Guertin, M. (2002) J. Biol. Chem. 277, 871-874. [DOI] [PubMed] [Google Scholar]
- 10.Takagi, T. (1993) Curr. Biol. 3, 413-418. [Google Scholar]
- 11.Couture, M. & Guertin, M. (1996) Eur. J. Biochem. 242, 779-787. [DOI] [PubMed] [Google Scholar]
- 12.Watts, R. A., Hunt, P. W., Hvitved, A. N., Hargrove, M. S., Peacock, W. J. & Dennis, E. S. (2001) Proc. Natl. Acad. Sci. USA 98, 10119-10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pesce, A., Couture, M., Dewilde, S., Guertin, M., Yamauchi, K., Ascenzi, P., Moens, L. & Bolognesi, M. (2000) EMBO J. 19, 2424-2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milani, M., Pesce, A., Ouellet, Y., Ascenzi, P., Guertin, M. &. Bolognesi, M. (2001) EMBO J. 20, 3902-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Milani, M., Savard, P.-Y., Ouellet, H., Ascenzi, P., Guertin, M. & Bolognesi, M. (2003) Proc. Natl. Acad. Sci. USA 100, 5766-5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hankeln, T., Ebner, B., Fuchs, C., Gerlach, F., Haberkamp, M., Laufs, T. L., Roesner, A., Schmidt, M., Weich, B., Wystub, S., et al. (2005) J. Inorg. Biochem. 99, 110-119. [DOI] [PubMed] [Google Scholar]
- 17.Hoogewijs, D., Geuens, E., Dewilde, S., Moens, L., Vierstraete, A., Vinogradov, S. N. & Vanfleteren, J. R. (2005) IUBMB Life 56, 697-702. [DOI] [PubMed] [Google Scholar]
- 18.Burmester, T. & Hankeln, T. (1999) Mol. Biol. Evol. 16, 1809-1811. [DOI] [PubMed] [Google Scholar]
- 19.Ebner, B., Burmester, T. & Hankeln, T. (2003) Mol. Biol. Evol. 20, 1521-1525. [DOI] [PubMed] [Google Scholar]
- 20.Wu, G., Wainwright, L. M. & Poole, R. K. (2003) Adv. Microb. Physiol. 47, 255-310. [DOI] [PubMed] [Google Scholar]
- 21.Freitas, T. A., Hou, S. & Alam, M. (2003) FEBS Lett. 552, 99-104. [DOI] [PubMed] [Google Scholar]
- 22.Freitas, T. A., Hou, S., Dioum, E. M., Saito, J. A., Newhouse, J., Gonzalez, G., Gilles-Gonzalez, M. A. & Alam, M. (2004) Proc. Natl. Acad. Sci. USA 101, 6675-6680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gough, J., Karplus, K., Hughey, R. & Chothia, C. (2001) J. Mol. Biol. 313, 903-919. [DOI] [PubMed] [Google Scholar]
- 24.Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kapp, O. H., Moens, L., Vanfleteren, J., Trotman, C. N. A., Suzuki, T. & Vinogradov, S. N. (1995) Protein Sci. 4, 2179-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones, D. T., Taylor, W. R. & Thornton, J. M. (1992) Cabios 8, 275-282. [DOI] [PubMed] [Google Scholar]
- 27.Baliga, N. S., Bonneau, R., Facciotti, M. T., Pan, M., Glusman, G., Deutsch, E. W., Shannon, P., Chiu, Y., Weng, R. S., Gan, R. R., et al. (2004) Genome Res. 14, 2221-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsuzaki. M., Misumi, O., Shin-I, T., Maruyama, S., Takahara, M., Miyagishima, S. Y., Mori, T., Nishida, K., Yagisawa, F., Nishida, K., et al. (2004) Nature 428, 653-657. [DOI] [PubMed] [Google Scholar]
- 29.Armbrust, E. V., Berges, J. A., Bowler, C., Green, B. R., Martinez, D., Putnam, N. H., Zhou, S., Allen, A. E., Apt, K. E., Bechner, M., et al. (2004) Science 306, 79-86. [DOI] [PubMed] [Google Scholar]
- 30.Tarricone, C., Galizzi, A., Coda, A., Ascenzi, P. & Bolognesi, M. (1997) Structure (Cambridge, MA, U.S.) 5, 497-507. [DOI] [PubMed] [Google Scholar]
- 31.Ilari, A., Bonamore, A., Farina, A., Johnson, K. A. & Boffi, A. (2002) J. Biol. Chem. 277, 23725-23732. [DOI] [PubMed] [Google Scholar]
- 32.Ermler, U., Siddiqui, R. A., Cramm, R., Schroeder, D. & Friedrich, B. (1995) EMBO J. 14, 6067-6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Friedberg, I., Kaplan, T. & Margalit, H. (2000) Protein Sci. 9, 2278-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elofsson, A. (2002) Proteins 46, 330-339. [DOI] [PubMed] [Google Scholar]
- 35.Bolognesi, M., Bordo, D., Rizzi, M., Tarricone, C. & Ascenzi, P. (1997) Prog. Biophys. Mol. Biol. 68, 29-68. [DOI] [PubMed] [Google Scholar]
- 36.Dickerson, R. E. & Geis, I. (1983) Hemoglobin: Structure, Function, Evolution and Pathology (Benjamin Cummings, Menlo Park, CA).
- 37.Zhang, W. & Phillips, G. N., Jr. (2003) Structure (Cambridge, MA) 11, 1097-1110. [DOI] [PubMed] [Google Scholar]
- 38.Andersson, J. O., Sjögren, A., Davis, A., Embley, T. M. & Roger, A. J. (2003) Curr. Biol. 13, 94-104. [DOI] [PubMed] [Google Scholar]
- 39.Battistuzzi, F. U., Feijao, A. & Hedges, S. B. (2004) BMC Evol. Biol. 4, 44-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hedges, S. B. (2002) Nat. Rev. Genet. 3, 838-849. [DOI] [PubMed] [Google Scholar]
- 41.Rivera, M. C. & Lake, J. A. (2004) Nature 431, 152-155. [DOI] [PubMed] [Google Scholar]
- 42.Moens, L., Vanfleteren, J., Blaxter, M. L., van de Peer, Y., Peeters, K., Kapp, O. H., Goodman, M. & Vinogradov, S. N. (1996) Mol. Biol. Evol. 13, 324-333. [DOI] [PubMed] [Google Scholar]
- 43.Runnegar, B. (1984) J. Mol. Evol. 21, 33-41. [DOI] [PubMed] [Google Scholar]
- 44.Yamauchi, K., Ochiai, T. & Usuki, I. (1992) Biochim. Biophys. Acta. 1171, 81-87. [DOI] [PubMed] [Google Scholar]
- 45.Brochier, C. & Philippe, H. (2002) Nature 417, 244. [DOI] [PubMed] [Google Scholar]
- 46.Gribaldo, S. & Philippe, H. (2004) in Organelles, Genomes and Eukaryote Phylogeny, eds. Hirt, R. P. & Horner, D. S. (CRC, Boca Raton, FL), pp. 133-152.
- 47.Hedges, S. B., Che, S. B., Kumar, S., Wang, D.-Y., Thompson, A. S. & Watanabe, H. (2001) BMC Evol. Biol. 1, 4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Philippe, H. (2004) in Assembling the Tree of Life, eds. Cracraft, J. & Donoghue, M. J. (Oxford Univ. Press, Oxford), pp. 95-106.
- 49.Vishwanath, P., Favaretto, P., Hartman, H., Mohr, S. C. & Smith, T. F. (2004) Mol. Phylogenet. Evol. 33, 615-625. [DOI] [PubMed] [Google Scholar]
- 50.Malik, H. S. & Henikoff, S. (2003) Nat. Struct. Biol. 10, 882-891. [DOI] [PubMed] [Google Scholar]
- 51.Moreira, D., Ranjard, L. & López-Garcia, P. (2004) BioEssays 26, 1144-1145. [DOI] [PubMed] [Google Scholar]
- 52.Lenton, T. M. (2003) in Evolution on Planet Earth, eds. Rothshild, L. J. & Lister, A. M. (Academic, New York), pp. 35-54.
- 53.Milani, M., Pesce, A., Nardini, M., Ouellet, H., Ouellet, Y., Dewilde, S., Bocedi, A., Ascenzi, P., Guertin, M., Moens, L., et al. (2005) J. Inorg. Biochem. 99, 97-109. [DOI] [PubMed] [Google Scholar]
- 54.Wu, G., Wainwright, L., Membrillo-Hernandez, J. & Poole, R. K. (2004) in Respiration in Bacteria and Archea, ed. Zannoni, D. (Kluwer, Boston), Vol. 1, pp. 251-286. [Google Scholar]
- 55.Kaur, K., Pathania, R., Sharma, V., Mande, S. C. & Dikshit, K. L. (2002) Appl. Environ. Microbiol. 68, 152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Elvers, K. T., Wu, G., Gilberthorpe, N. J., Poole, R. K. & Park, S. F. (2004) J. Bacteriol. 186, 5332-5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Freitas, T. A., Saito, J. A., Hou, S. & Alam, M. (2005) J. Inorg. Biochem. 99, 23-33. [DOI] [PubMed] [Google Scholar]
- 58.Kundu, S., Trent, J. T., III, & Hargrove, M. S. (2003) Trends Plant Sci. 8, 387-393. [DOI] [PubMed] [Google Scholar]
- 59.Flögel, U., Merx, M. W., Gödecke, A., Decking, U. K. M. & Schrader, J. (2001) Proc. Natl. Acad. Sci. USA 98, 735-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Igamberdiev, A. U. & Hill, R. D. (2004) J. Exp. Bot. 55, 2473-2482. [DOI] [PubMed] [Google Scholar]
- 61.Shi, S., Blundell, T. L. & Mizugishi, K. (2001) J. Mol. Biol. 310, 243-257. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.