Abstract
The regular spacing of somites during vertebrate embryogenesis involves a dynamic gradient of FGF signaling that controls the timing of maturation of cells in the presomitic mesoderm (PSM). How the FGF signal is transduced by PSM cells is unclear. Here, we first show that the FGF gradient is translated into graded activation of the extracellular signal-regulated kinase (ERK)/mitogen-activated protein kinase (MAPK) pathway along the PSM in the chicken embryo. Using in ovo electroporation of PSM cells, we demonstrate that constitutive activation of ERK signaling in the PSM blocks segmentation by preventing maturation of PSM cells, thus phenocopying the overexpression of FGF8. Conversely, inhibition of ERK phosphorylation mimics a loss of function of FGF signaling in the PSM. Interestingly, video microscopy analysis of cell movements shows that ERK regulates the motility of PSM cells, suggesting that the decrease of cell movements along the PSM enables mesenchymal PSM cells to undergo proper segmentation. Together, our data demonstrate that ERK is the effector of the gradient of FGF in the PSM that controls the segmentation process.
Keywords: extracellular signal-regulated kinase, fibroblast growth factor, somite
Somitogenesis leads to the subdivision of the paraxial mesoderm into transient epithelial metameric units, called somites (1). Somite formation begins anteriorly and proceeds posteriorly, passing like a wave through the paraxial mesoderm as successive groups of cells segregate from the PSM at regular intervals, in concert with extension of the body axis (2). Somite formation involves an oscillator (the segmentation clock) that drives cyclic gene expression. This periodic signal is converted into the repeated array of somite boundaries by a spacing mechanism relying on a traveling threshold of FGF8 and Wnt3a that regresses with body axis extension (3). FGF8 mRNA and protein are distributed in a graded fashion in the caudal presomitic mesoderm (PSM) of vertebrate embryos (4–6), and overexpression of FGF8 in the PSM cells prevents them from differentiating (5). These data led to the idea that high concentrations of FGF8 are required to actively maintain newly formed PSM cells in an immature state. Thus, because of the progressive decrease of FGF8 expression during maturation of the PSM, cells reach a threshold of FGF signaling at a given level in the PSM, called the “determination front”, where they activate their segmentation program. This determination front marks a molecular transition for PSM cells, as shown by the down-regulation of posterior genes such as brachyury and the activation of new sets of genes such as paraxis in the anterior PSM (5). In chick and fish embryos, FGFR1 is the only FGF receptor to be expressed in the PSM, and in mouse, the FGFR1 knockout disrupts somite formation, suggesting that FGFR1 mediates FGF8 activity in somitogenesis (4, 5, 7, 8).
FGF signaling activates a variety of downstream effectors, such has mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinase (ERK), MAPK/p38, phosphatidylinositol 3-kinase (PI3K), or phospholipase Cγ (PLCγ) (9), but which of these pathways regulates the maturation of PSM cells in response to FGF8 is currently unknown. In zebrafish, the distribution of the activated diphosphorylated form of MAPK/ERK (dpERK) overlaps with that of fgf8 in the caudal PSM (4). Moreover, treatment of zebrafish embryos with the FGFR1 inhibitor SU5402 blocks ERK activation in the PSM, suggesting that ERK might be one of the downstream effector of FGF signaling in this tissue (4). However, the exact function of ERK during segmentation in fish was not investigated, and, because SU5402 blocks all pathways downstream of FGFR1, which of them mediates the maturation of PSM cells remains unknown. In mouse embryos, no graded distribution of dpERK could be detected in the tail bud (6, 10), suggesting that alternative pathway(s) might be activated in amniote somitogenesis. Here, we have investigated the intracellular effectors of FGF signaling during avian segmentation. We provide evidence for a gradient of ERK activation established in response to FGF signaling. We show that the maturation of the PSM is directly regulated by ERK activity. Furthermore, we show that the MAPK/ERK pathway imposes a caudo-rostral gradient of cell motility. The gradual decrease of cell movements along the PSM may enable mesenchymal PSM cells to organize themselves properly at the onset of the segmentation program.
Materials and Methods
Embryos. Fertilized chick eggs were obtained from Ozark Hatcheries (Neosho, MO) and incubated at 38°C in a humidified incubator. Embryos were staged according to the developmental table of Hamburger (11) and Hamilton (HH) and by counting somite pairs (12).
dpERK Whole-Mount Immunohistochemistry. Embryos were rapidly dissected in ice-cold PBS and immediately transferred into cold fixative to preserve endogenous phospho-ERK proteins. We used the protocol described in ref. 10 (www.mshri.on.ca/rossant/protocols.html). Briefly, embryos were fixed in 4% paraformaldehyde, dehydrated in methanol, treated with 5% H2O2, rehydrated, blocked with FBS-TBST (5% sera in TBS plus 0.1% Triton X-100), incubated with primary antibody (anti-dpERK/no. 9101, 1:50, Cell Signaling Technology, Beverly, MA) overnight (in FBS-TBST), washed six times (1 h each), incubated with biotinylated secondary antibody overnight, washed six times, incubated with streptavidin-horseradish peroxydase (HRP), and stained with diaminobenzidine (DAB). All washing and incubation steps were done at 4°C.
Bead Implantations. Heparin beads (H-5263, Sigma) were incubated in PBS, 0.1% BSA containing FGF8b (1 mg/ml) (R & D Systems) for 1 h at room temperature. PBS-soaked beads were used as controls. Beads (100- to 150-μm diameter) were selected, cut in two equal parts, and rinsed in a drop of PBS before implantation into 14- to 16-somite stage chick embryos. An incision through the ectoderm and the mesoderm was made to insert the bead between the paraxial and lateral mesoderm halfway in the PSM. Embryos were incubated for 4 h, and then fixed for whole-mount immunohistochemistry with anti-dpERK.
Embryo Culture. Chicken embryo explants at the 13- to 18-somite stage were cultured in vitro as described in ref. 13. Posterior half embryo explants were cultured for 3–6 h in a chick culture medium (5% chick serum, 2.5% FCS, and 1% bicarbonate in DMEM) containing 1–100 μM of the FGFR chemical inhibitor SU5402 (Calbiochem) or 10–100 μM of the chemical inhibitor of MAPK kinase U0126 (Calbiochem), both dissolved in DMSO. Control embryos were treated similarly with a solution containing the same concentration in DMSO. Embryo explants were fixed and processed for in situ hybridization or used for Western blot analysis.
Plasmids and in Ovo Electroporation. In ovo electroporations were performed mainly as described in ref. 5. Eggs were windowed, and the DNA solution was injected between the vitelline membrane and the epiblast in stage 4–5 Hamburger and Hamilton (HH) embryos. The primitive streak was coated with the DNA solution from the node until mid-streak. Two platinum electrodes tied together were used: one was placed directly on the streak caudal to the node, and the second was inserted into the yolk. A series of electric pulses (4 pulses, 30 volts, 50 ms) were directed by using a square wave electroporator (BTX, San Diego). Expression vectors for electroporations pCAGGS (14) for fgf8 (5), or pCIG (15) for constitutively active MAPK kinase 1 (MKK1) (MKK1ca) and dominant-negative MKK1 (MKK1dn) were purified by using an endotoxin-free maxi kit (Qiagen, Valencia, CA) and used at 0.15–2 μg/μl in a PBS solution containing 1 mM MgCl2 and 0.4 mg/ml fast green FCF (Sigma). pCIG is a bicistronic vector that drives the expression of a GFP reporter in addition to the gene of interest. pCAAGS-fgf8 embryos were coelectroporated with a pCAAGS-GFP construct. Control embryos were either electroporated with empty pCIG or coelectroporated with pCAAGS-GFP and empty pCAAGS. After electroporation, eggs were reincubated for 12, 24, or 36 h and assayed for GFP expression. Embryos were then used for Western blot analysis or fixed and processed for in situ hybridization. Some of the embryos electroporated with pCIG, pCIG-MKK1ca, or pCIG-MKK1dn were also incubated with phalloidin Alexa Fluor 546 (Molecular Probes) and Hoechst to detect the F-actin and the nucleus, respectively, by laser confocal microscopy.
Western Blotting. Western blot analysis was performed by standard procedures. The caudal part of three to four cultured or electroporated embryos was dissected out in ice-cold PBS. Samples were lysed in RIPA buffer containing 1 mM Na3VO4, 1 mM sodium pyrophosphate, and complete protease inhibitor mixture (Roche Molecular Biochemicals). Proteins were extracted and separated by SDS/PAGE on 12% acrylamide gels. The level of β-actin (A-2066, 1:10,000, Sigma) was used to normalize the dpERK (no. 9101, 1:1,000, Cell Signaling Technology) and ERK (no. 9102, 1:1,000, Cell Signaling Technology) signal intensity.
Whole-Mount in Situ Hybridization and Immunohistochemistry on Section. Whole-mount in situ hybridizations were performed as described in ref. 16. The chick Brachyury, Sprouty2, meso2, and paraxis have been described (17–20). After imaging, some of the stained embryos were embedded for cryosection in gelatin-sucrose and cut at 10 μm. GFP was detected on section with a mAb anti-GFP (no. 1814460, 1:200, Roche Molecular Biochemicals) by diaminobenzidine staining.
Time-Lapse Analysis. We used a simplified new culture system to perform time-lapse analysis of embryos electroporated with PCIG, PCIG-MKK1ca and PCIG-MKK1dn. After electroporation, embryos were reincubated for 12 h and then transferred onto an agarose-albumen culture plate, dorsal-side up. The plate was then placed upside-down on a heated stage of an epifluorescence microscope. Images were taken at 2-min intervals with GFP filter for 3–6 h by using the openlab (Improvision, Lexington, MA) software. The cell tracking and measurements were performed by using image pro plus (Media Cybernetics, Silver Spring, MD) software. GFP-positive embryos displaying sufficient mosaicism to allow single-cell tracking were selected. For measurements, a minimum of three embryos per construct were analyzed. For each embryo, at least 10 cells in the posterior PSM and 10 cells in the anterior PSM were randomly picked. Cells were tracked only along the XY plan, and the calculated velocity might therefore be underestimated. Data were produced with excel (Microsoft), and statistical significance (P < 0.05) was determined by using a paired Student's t test.
Results and Discussion
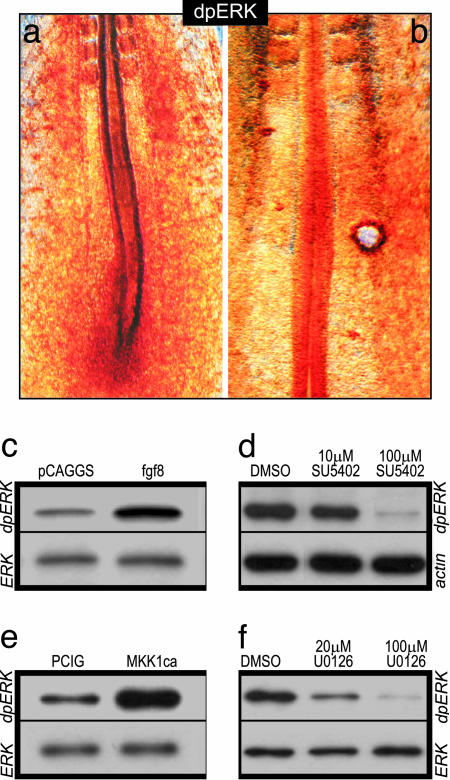
ERK Is Activated by FGF8 Signaling in the Chick PSM. The FGF-signaling gradient in the PSM was shown to control the timing of cell maturation along this tissue. Caudal high level of FGF signaling blocks the response to differentiation signals of the newly produced PSM cells, thus maintaining them in an immature state (2, 4, 5, 21). FGF signaling is known to activate several intracellular effectors, including MAPK/ERK and phosphatidylinositol 3-kinase (PI3K) (9), but which of these pathways regulates the maturation of PSM cells in response to FGF8 is currently unclear. Whereas in zebrafish the function of ERK in mediating the control of PSM maturation has not been investigated (4), in mouse, the FGF8 gradient was shown to correlate with graded phosphorylation of AKT, the kinase downstream of PI3K (22), suggesting that an AKT-dependent pathway might mediate FGF signaling in this species (6). In the present study, we used the chicken embryo as a model to analyze which pathway(s) regulates the maturation of PSM cells in response to FGF8. We first investigated the pattern of ERK and AKT activation in the chick PSM using anti-dpERK and anti-pAKT antibodies. By contrast to the mouse studies, we did not observe any spatial correlation between pAKT and Fgf8 mRNA distribution both by Western blot and immunohistochemistry in 2-day-old embryos (data not shown). However, we observed a clear caudo-rostral gradient of dpERK expression in the PSM, with a higher expression in the primitive streak/tail bud, which progressively diminished anteriorly (Fig. 1a). Given these results, we focused our investigations on the role of ERK signaling during somitogenesis. We first tested whether FGF8 can activate ERK in the chick PSM. We grafted heparin-acrylic beads soaked in FGF8b between the PSM and the lateral mesoderm of 10- to 20-somite stage embryos (5); the grafts were done in the mid-PSM, and embryos were fixed 4 h after the operation. Whole-mount immunohistochemistry with the anti-dpERK antibody showed that ERK is activated around the FGF8 bead (Fig. 1b). This activation was more important anterior to the bead, possibly because of the presence of endogenous inhibitors of FGF signaling in the posterior PSM, such as the Sprouty proteins (23). We also analyzed ERK activation by Western blot after in ovo electroporation of the PSM with an fgf8-expressing vector. A strong signal of ERK phosphorylation in the posterior part of electroporated embryos can be detected after Fgf8 overexpression (Fig. 1c). Therefore, FGF8 signaling can trigger ERK activation in the chick PSM. We next addressed whether FGF signaling was necessary for the establishment of the caudorostral gradient of activated ERK in the chick PSM. As assessed by Western blot, SU5402 treatment, which blocks FGFR1 phosphorylation, prevented ERK phosphorylation in the posterior part of cultured embryos (Fig. 1d). Together, these data indicate that FGF signaling is necessary and sufficient to activate the ERK pathway in the chick PSM.
Fig. 1.
FGF signaling triggers ERK activation in the PSM. (a and b) dpERK whole-mount immunostaining in 15-somite embryos. Rostral is to the top. (a) Wild-type embryo. (b) Embryo grafted with an FGF8-soaked bead, 4 h after the graft. (c–f) Immunoblots with dpERK after electroporation with pCAGGS-fgf8 (c) or PCIG-MKK1ca (e), or after culture of posterior half embryo explants in presence of SU5402 (d) or U0126 (f). The level of phospho-ERK is compared with the level of β-actin or total ERK.
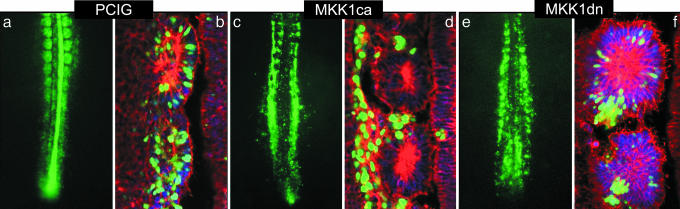
MAPK/ERK Signaling Is the Effector of the FGF Pathway Controlling Maturation of PSM Cells. Because our results indicate that ERK activation is distributed in a caudo-rostral gradient and requires FGF signaling in the chick PSM, we investigated whether this pathway mediates the FGF-dependent PSM maturation. To address this question, we first electroporated a constitutively active form of MKK1 (MKK1ca) in the nascent PSM (24). MKK1 (also called MEK1) is a MAP kinase kinase that phosphorylates and thus activates MAPK/ERK (25). Western blot analysis showed that, 1 day after electroporation, ERK phosphorylation was up-regulated in the caudal part of embryos electroporated with MKK1ca (Fig. 1e). We then analyzed the morphology of the paraxial mesoderm after such an overexpression by confocal microscopy. Frontal sections of embryos electroporated with a control vector (pCIG) expressing only the GFP reporter showed that positive cells are randomly distributed within the somitic tissue (Fig. 2 a and b). By contrast, after electroporation of MKK1ca, the transfected cells retain a mesenchymal aspect and never incorporate into epithelial somites (Fig. 2 c and d). Therefore, constitutive activation of MKK1 seems to prevent cells from activating their segmentation program. This phenotype is reminiscent of that seen after Fgf8 overexpression, except that FGF8, which is secreted, acts non-cell autonomously (5).
Fig. 2.
ERK overactivation prevents PSM cells to incorporate into somites. (a, c, and e) Distribution of GFP-positive cells of 2-day-old embryos after electroporation of pCIG (a), MKK1ca (c), or MKK1dn (e). (b, d, and f) Frontal laser confocal microscopy sections at the level of the newly formed somites of embryos shown in a, c, and e, respectively, and counterstained with Hoechst (blue) to visualize DNA and Alexa Fluor 546 phalloidin (red) to visualize F-actin (rostral to the top and lateral to the left).
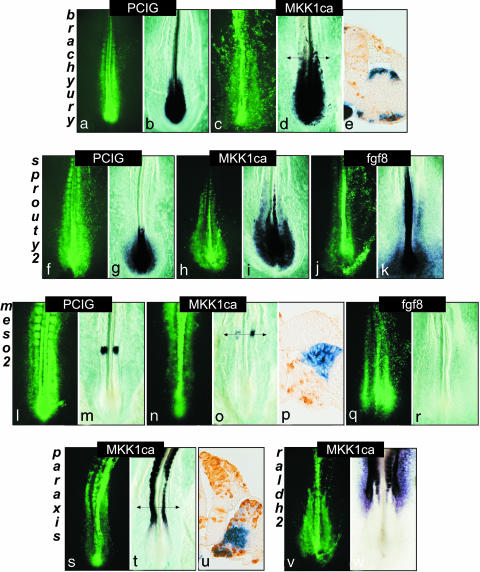
At the molecular level, we observed that Tbx6, which is normally expressed along the entire PSM, is still expressed in MKK1ca-positive cells (data not shown), suggesting that these cells maintain a PSM identity. Downstream, direct targets of FGF signaling such as Brachyury and Sprouty2 (5, 26), whose expressions are normally restricted to the caudal PSM, were found to be ectopically expressed in MKK1ca-positive cells located in the rostral PSM (Fig. 3 a–k). Meso2, the first gene that displays a segmental pattern in the chick PSM (19), was inhibited by both fgf8 and MKK1ca electroporation (Fig. 3 l–r). Paraxis, a gene involved in somite epithelialization, was also inhibited by MKK1ca (27), further confirming that ERK activity prevents cellular epithelialization (Fig. 3 s–u). Finally, the FGF-signaling gradient in the PSM has been shown to be antagonized by an opposite gradient of retinoic acid produced by the differentiated paraxial mesoderm (28, 29). Raldh2, the retinoic acid biosynthetic enzyme, is expressed in the somites and the most rostral part of the PSM. FGF8 treatment of rostral PSM explants was shown to prevent raldh2 expression (28). Here, we observed that MKK1ca-expressing cells in the rostral PSM and somites never activated raldh2 expression (Fig. 3 v and w). This finding therefore suggests that the inhibition of retinoic acid production by FGF is mediated by the MAPK/ERK pathway.
Fig. 3.
Overexpression of MKK1ca mimics FGF8 activity by maintaining PSM cells in an immature state. Embryos were either electroporated with pCIG (a, f, and l) or with MKK1ca (c, h, n, s, and v), or were coelectroporated with GFP- and FGF8-expressing vector (j and q). Panels a, c, f, h, j, l, n, p, and s show GFP expression (in green) of the corresponding whole-mount in situ hybridizations. All panels show dorsal views (rostral to the top). Panels b, d, and e were hybridized with Brachyury, and g, i, and k were hybridized with sprouty2. Panels m, o, p, and r were hybridized with meso2, t and u with paraxis, and w with raldh2. (e, p, and u) Transverse sections at the level of the arrow of the embryos shown in d, o, and t, respectively, stained with a GFP antibody (in brown) (dorsal is to the top, medial is to the left).
We then investigated the effect of blocking the MAPK/ERK pathway. We first treated embryos in culture with U0126, a chemical inhibitor known to specifically block the kinase activity of MKK1/2 (30). Western blots on extracts of posterior embryonic tissues showed that the activation of ERK was inhibited in U0126-treated embryos compared with controls (Fig. 1f). A 3-h culture period was enough to induce a down-regulation of Sprouty2 in embryos cultured with either U0126 or SU5402 (Fig. 5, which is published as supporting information on the PNAS web site, and data not shown). To further analyze the role of ERK activity during somitogenesis, we performed loss-of-function experiments by electroporating a MKK1 dominant-negative (MKK1dn) form in the chick PSM (24). After electroporation (1.5 days), cells overexpressing MKK1dn tended to cluster together in the epithelial somites, suggesting that these cells were able to undergo the mesenchymo-epithelial transition (Fig. 2 e and f). Moreover, overexpressing MKK1dn construct led to the down-regulation of posterior PSM markers such as Brachyury and sprouty2 (Fig. 5 and data not shown), in agreement with the results obtained with chemical inhibitors. However, anterior PSM markers such as paraxis were not induced prematurely in the caudal PSM (data not shown). Therefore, suppressing the FGF-mediated repression of these genes is not sufficient for their activation, suggesting that positive activating factors such as retinoic acid are likely to be required. Together, these data indicate that the maturation of PSM cells is controlled by the MAPK/ERK pathway.
Finally, we sought to determine whether the FGF-induced ERK activity gradient, in addition to positioning the determination front, might also control the segmentation clock (3). The segmentation clock drives the transcription of the “cyclic genes,” which are periodically expressed in a dynamic sequence reiterated once during each somite formation. Their dynamic behavior and their genetic regulation change in the anterior PSM at the level of the determination front where the progression of the wave of cyclic gene expression slows down and ultimately stops where the next somite will form (31). To test whether the changes in cyclic gene behavior are regulated by the FGF/ERK signaling, we treated embryos in culture with SU5402 to block FGF signaling or with U0126 to prevent ERK activation. No effect was observed on the kinetics of lunatic fringe expression, suggesting that FGF/ERK activation is not required for cyclic genes oscillations (data not shown).
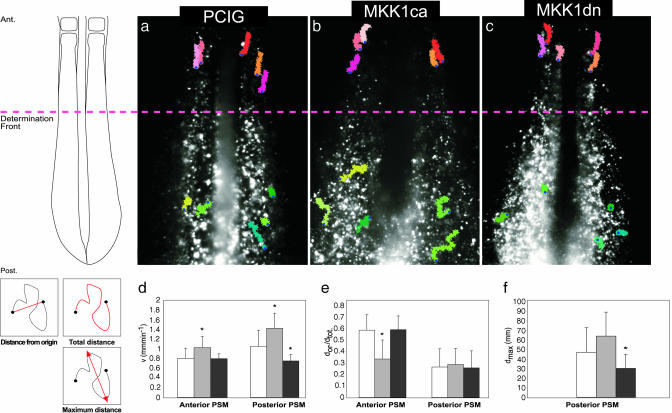
MAPK/ERK Pathway Regulates Morphogenetic Movements in the PSM. FGF signaling is known to regulate cell movements and migration during gastrulation in chick and mouse embryos (32, 33). To investigate the implication of the MAPK/ERK pathway in regulating cell movements within the PSM, we used time-lapse video microscopy to follow the migratory behavior of PSM cells after electroporation of GFP, MKK1ca, or MKK1dn constructs (see Movies 1–3, respectively, which are published as supporting information on the PNAS web site). In GFP control embryos, cells of the caudal PSM actively moved within the tissue (Fig. 4a and Movie 1), as previously reported (34). Their mean velocity in the caudal part of the PSM is 1.04 ± 0.33 μm·min–1, and these cells often change direction along any of the axes (Fig. 4 a, d, and e). As somitogenesis proceeds and as cells reside more and more anteriorly, their motile activity decreases progressively until these cells become static within the anterior, epithelialized PSM (Fig. 4 a and d). They nonetheless show a passive apparent anterior movement, due to the global elongation of the embryo (Fig. 4 a and e and Movie 1). Although difficult to quantify, this passive anterior movement can be deduced by the fact that, in this region of the PSM, two neighbor cells move at the same pace and keep their relative distance constant, whereas this is not the case in the posterior PSM. When cells were electroporated with MKK1ca, their motile activity was significantly increased. Their mean velocity rose to 1.41 ± 0.3 μm·min–1, with phases of dramatic accelerations and abrupt changes in direction (Fig. 4 b and d–f and Movie 2). This behavior was slightly attenuated as cells reached the anterior portion of the PSM, but, even in this region of the tissue, their motile activity remained higher than in controls, and they actively migrated out of the bulk of the paraxial mesoderm to become excluded at the edges of the tissue (data not shown). An opposite effect was observed when ERK signaling was inhibited by electroporating the dominant negative form of MKK1 in the PSM. Overexpressing cells displayed slower cell movements in the caudal part of the PSM (mean velocity of 0.74 ± 0.12 μm·min–1), and positive cells rapidly aggregated in the posterior PSM, forming patches that segregate from wild-type cells (Movie 3 and Movie 4, which is published as supporting information on the PNAS web site). These cells are, however, able to ultimately integrate the epithelial somites in clusters (Figs. 2 e and f and 4 c and d–f and Movies 3 and 4). These results indicate that the levels of ERK directly control the motile activity of PSM cells. Moreover, the observed aggregation of cells overexpressing MKK1dn and MKK1ca suggest that cells with an equal level of ERK activity tend to cluster together, whereas cells with a different ERK activity repulse each other. We propose that this behavior may trigger local rearrangements that ultimately distribute cells along the antero-posterior axis of the paraxial mesoderm according to their strict level of ERK activity. This process may favor a tidy progression of the wave of cell differentiation that sweeps the PSM.
Fig. 4.
The motility of PSM cells is controlled by ERK signaling. (a–c) Representative time-lapse analysis of embryos electroporated with pCIG (a), MKK1ca (b), or MKK1dn (c). In each case, the movements of five individual cells in the rostral PSM (in red) and five individual cells in the caudal PSM (in green) over 140 min is shown. The blue dots show the starting point of each cell path. The scheme on the Left illustrates the position of the cells in the PSM with respect to the determination front (red hatched line). Comparison of velocity (d), of the cell directionality (e) or of the cell dispersion (f) of the positive cells for pCIG (white bars), for MKK1ca (gray bars), or for MKK1dn (black bars), both in the anterior (d and e) and posterior (d, e, and f) PSM. dor., distance from origin; dtot., total distance; dmax, maximum distance ever reached between to points of the cell path. *, P < 0.05. These different values (in red) are illustrated on the schematic cell tracks shown on the Bottom Left. Ant., anterior; Post., posterior.
Based on its graded distribution in the posterior part of the mouse embryo, AKT has been proposed to act downstream of FGF signaling in the PSM (6). However, this role has not been examined at the functional level. No graded activity of ERK was reported in the mouse PSM (6, 10), but the failure to detect dpERK gradient in this species may likely be due to the very labile state of dpERK protein in mouse embryos (see Fig. 6, which is published as supporting information on the PNAS web site). Our data show that, in chicken embryo, regulation of PSM cell maturation is mediated by the activation level of MKK1/ERK signaling pathway. Together with observations in the fish embryo (4), it is therefore probable that ERK activity plays a conserved role in vertebrate somitogenesis. Further experiments will be required to analyze the function and regulation of other downstream effectors of FGF signaling, like PI3-K/AKT, in the PSM.
Our results strongly suggest that FGF/ERK signaling imposes a gradient of cell motility along the antero-posterior axis of the PSM. Posterior extension of the neural axis has been proposed to result from the growth of cells that localized in a posterior transition zone and that derived from a pool of tail bud precursor cells (21, 35). According to this model, the motor of posterior extension is not the deposition of cells at the tip of the axis but rather the growth of cells of the transition zone, which pushes the tail bud posteriorly. From cell-labeling experiments of the paraxial mesoderm precursors, it can be argued in favor of the existence in the posterior PSM of such a transition zone, populated by descendants of the streak/tail bud cells and contributing a progeny spanning several segments (34, 36). FGF signaling has been implicated in the control of this posterior extension process at two distinct levels: (i) in the maintenance of the immature state of tail bud cells, such that down-regulation of FGF signaling is required for cells to enter the transition zone and begin differentiation (21, 37, 38) and (ii) in the promotion of the epithelium to mesenchymal transition and in the control of the exit of cells from the precursor area (32, 33). Our data showing that ERK activation in the PSM promotes high cell motility is consistent with the idea that FGF signaling could provide a means for cells in the tail bud region to escape the precursor area and enter the transition zone to begin PSM differentiation. Cell motility progressively diminishes as cells read the FGF gradient along the PSM. Active movements stop when cells reach the determination front where they become allocated to their definitive somite, thus acquiring their positional identity along the AP axis. This arrest of cell movement also correlates with the beginning of the epithelialization of the PSM, which ultimately results in epithelial somite formation (39).
Supplementary Material
Acknowledgments
We thank Silvia Esteban for the artwork, Barbara Brede for her excellent technical help, and Paul Kulesa and the imaging facility for their help with movie analysis. We are grateful to members of the O.P. laboratory for helpful discussions and Christophe Marcelle for critical reading of the manuscript. We are very grateful to Natalie Ahn (Department of Chemistry and Biochemistry and Howard Hughes Medical Institute, University of Colorado, Boulder) for providing us the MKK1 constructs. We also thank Jean-Philippe Rey and the histology core facility of the Stowers Institute for Medical Research for their help in processing embryos for cryosection and GFP staining. M.C.D. was supported by the Association pour la Recherche contre le Cancer (ARC), by the Fondation Bettencourt Schueller, and by the Fondation Philippe. This work has been supported by National Institutes of Health Grant 1R01 HD043158-01 (to O.P.).
Author contributions: M.-C.D., J.D., and O.P. designed research; M.-C.D., J.D., P.M., and J.C. performed research; M.-C.D., J.D., P.M., J.C., and O.P. analyzed data; and M.-C.D., J.D., and O.P. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PSM, presomitic mesoderm; MAPK, mitogen-activated protein kinase; ERK, extracellular signal-regulated kinase; dpERK, activated diphosphorylated form of MAPK/ERK; MKK1, MAPK kinase 1; MKK1ca, constitutively active MKK1; MKK1dn, dominant-negative MKK1.
References
- 1.Saga, Y. & Takeda, H. (2001) Nat. Rev. Genet. 2, 835–845. [DOI] [PubMed] [Google Scholar]
- 2.Dubrulle, J. & Pourquié, O. (2004) Development (Cambridge, U.K.) 131, 5783–5793. [DOI] [PubMed] [Google Scholar]
- 3.Pourquié, O. (2003) Science 301, 328–330. [DOI] [PubMed] [Google Scholar]
- 4.Sawada, A., Shinya, M., Jiang, Y. J., Kawakami, A., Kuroiwa, A. & Takeda, H. (2001) Development (Cambridge, U.K.) 128, 4873–4880. [DOI] [PubMed] [Google Scholar]
- 5.Dubrulle, J., McGrew, M. J. & Pourquie, O. (2001) Cell 106, 219–232. [DOI] [PubMed] [Google Scholar]
- 6.Dubrulle, J. & Pourquié, O. (2004) Nature 427, 419–422. [DOI] [PubMed] [Google Scholar]
- 7.Deng, C. X., Wynshaw-Boris, A., Shen, M. M., Daugherty, C., Ornitz, D. M. & Leder, P. (1994) Genes Dev. 8, 3045–3057. [DOI] [PubMed] [Google Scholar]
- 8.Yamaguchi, T. P., Harpal, K., Henkemeyer, M. & Rossant, J. (1994) Genes Dev. 8, 3032–3044. [DOI] [PubMed] [Google Scholar]
- 9.Bottcher, R. T. & Niehrs, C. (2005) Endocr. Rev. 26, 63–77. [DOI] [PubMed] [Google Scholar]
- 10.Corson, L. B., Yamanaka, Y., Lai, K. M. & Rossant, J. (2003) Development (Cambridge, U.K.) 130, 4527–4537. [DOI] [PubMed] [Google Scholar]
- 11.Hamburger, V. (1992) Dev. Dyn. 195, 273–275. [DOI] [PubMed] [Google Scholar]
- 12.Pourquié, O. & Tam, P. P. (2001) Dev. Cell 1, 619–620. [DOI] [PubMed] [Google Scholar]
- 13.Palmeirim, I., Henrique, D., Ish-Horowicz, D. & Pourquié, O. (1997) Cell 91, 639–648. [DOI] [PubMed] [Google Scholar]
- 14.Niwa, H., Yamamura, K. & Miyazaki, J. (1991) Gene 108, 193–199. [DOI] [PubMed] [Google Scholar]
- 15.Megason, S. G. & McMahon, A. P. (2002) Development (Cambridge, U.K.) 129, 2087–2098. [DOI] [PubMed] [Google Scholar]
- 16.Henrique, D., Adam, J., Myat, A., Chitnis, A., Lewis, J. & Ish-Horowicz, D. (1995) Nature 375, 787–790. [DOI] [PubMed] [Google Scholar]
- 17.Knezevic, V., De Santo, R. & Mackem, S. (1997) Development (Cambridge, U.K.) 124, 411–419. [DOI] [PubMed] [Google Scholar]
- 18.Chambers, D. & Mason, I. (2000) Mech. Dev. 91, 361–364. [DOI] [PubMed] [Google Scholar]
- 19.Buchberger, A., Seidl, K., Klein, C., Eberhardt, H. & Arnold, H. H. (1998) Dev. Biol. 199, 201–215. [DOI] [PubMed] [Google Scholar]
- 20.Sosic, D., Brand-Saberi, B., Schmidt, C., Christ, B. & Olson, E. N. (1997) Dev. Biol. 185, 229–243. [DOI] [PubMed] [Google Scholar]
- 21.Mathis, L., Kulesa, P. M. & Fraser, S. E. (2001) Nat. Cell Biol. 3, 559–566. [DOI] [PubMed] [Google Scholar]
- 22.Scheid, M. P. & Woodgett, J. R. (2001) Nat. Rev. Mol. Cell. Biol. 2, 760–768. [DOI] [PubMed] [Google Scholar]
- 23.Hanafusa, H., Torii, S., Yasunaga, T. & Nishida, E. (2002) Nat. Cell Biol. 4, 850–858. [DOI] [PubMed] [Google Scholar]
- 24.Mansour, S. J., Matten, W. T., Hermann, A. S., Candia, J. M., Rong, S., Fukasawa, K., Vande Woude, G. F. & Ahn, N. G. (1994) Science 265, 966–970. [DOI] [PubMed] [Google Scholar]
- 25.Johnson, G. L. & Lapadat, R. (2002) Science 298, 1911–1912. [DOI] [PubMed] [Google Scholar]
- 26.Minowada, G., Jarvis, L. A., Chi, C. L., Neubuser, A., Sun, X., Hacohen, N., Krasnow, M. A. & Martin, G. R. (1999) Development (Cambridge, U.K.) 126, 4465–4475. [DOI] [PubMed] [Google Scholar]
- 27.Burgess, R., Cserjesi, P., Ligon, K. L. & Olson, E. N. (1995) Dev. Biol. 168, 296–306. [DOI] [PubMed] [Google Scholar]
- 28.Diez del Corral, R., Olivera-Martinez, I., Goriely, A., Gale, E., Maden, M. & Storey, K. (2003) Neuron 40, 65–79. [DOI] [PubMed] [Google Scholar]
- 29.Moreno, T. A. & Kintner, C. (2004) Dev. Cell 6, 205–218. [DOI] [PubMed] [Google Scholar]
- 30.DeSilva, D. R., Jones, E. A., Favata, M. F., Jaffee, B. D., Magolda, R. L., Trzaskos, J. M. & Scherle, P. A. (1998) J. Immunol. 160, 4175–4181. [PubMed] [Google Scholar]
- 31.Pourquié, O. (2001) Annu. Rev. Cell Dev. Biol. 17, 311–350. [DOI] [PubMed] [Google Scholar]
- 32.Yang, X., Dormann, D., Munsterberg, A. E. & Weijer, C. J. (2002) Dev. Cell 3, 425–437. [DOI] [PubMed] [Google Scholar]
- 33.Ciruna, B. & Rossant, J. (2001) Dev. Cell 1, 37–49. [DOI] [PubMed] [Google Scholar]
- 34.Kulesa, P. M. & Fraser, S. E. (2002) Science 298, 991–995. [DOI] [PubMed] [Google Scholar]
- 35.Mathis, L. & Nicolas, J. F. (2000) Development (Cambridge, U.K.) 127, 1277–1290. [DOI] [PubMed] [Google Scholar]
- 36.Nicolas, J. F., Mathis, L., Bonnerot, C. & Saurin, W. (1996) Development (Cambridge, U.K.) 122, 2933–2946. [DOI] [PubMed] [Google Scholar]
- 37.Diez del Corral, R., Breitkreuz, D. N. & Storey, K. G. (2002) Development (Cambridge, U.K.) 129, 1681–1691. [DOI] [PubMed] [Google Scholar]
- 38.Bertrand, N., Medevielle, F. & Pituello, F. (2000) Development (Cambridge, U.K.) 127, 4837–4843. [DOI] [PubMed] [Google Scholar]
- 39.Nakaya, Y., Kuroda, S., Katagiri, Y. T., Kaibuchi, K. & Takahashi, Y. (2004) Dev. Cell 7, 425–438. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.