Abstract
The Dmrt genes encode a large family of transcription factors whose function in sexual development has been well studied. However, their expression pattern is not restricted to the gonad, suggesting that Dmrt genes might regulate other developmental processes. Here, we report the expression and functional analysis of one member of this family: Xenopus Dmrt4 (XDmrt4). XDmrt4 is initially expressed in the anterior neural ridge and then becomes progressively restricted to part of the telencephalon and the olfactory placode/epithelium. XDmrt4 is induced at the anterior neural plate by a balance of neural inducers and caudalizing factors. Interference with XDmrt4 function by injection of a morpholino oligonucleotide or an inhibitory mutant resulted in a similar phenotype, the specific disruption of the olfactory placode expression of Xebf2 without affecting the expression of other placodal markers. Xebf2 belongs to a family of helix–loop–helix transcription factors implicated in neuronal differentiation, and later in embryogenesis XDmrt4-deficient embryos show impaired neurogenesis in the olfactory epithelium. Consistent with this finding, XDmrt4 is sufficient to activate neurogenin, Xebf2, and neural cell adhesion molecule expression in animal explants and is required for Noggin-mediated neuralization. Altogether, these results indicate that XDmrt4 is an important regulator of neurogenesis in the olfactory system upstream of neurogenin and Xebf2.
Keywords: placode, Xenopus, Xebf2, forebrain, neurogenin
Genes related to the Drosophila doublesex and Caenorhabditis elegans mab-3 genes encode transcription factors conserved during evolution (1). They constitute the Dmrt (doublesex and mab-3-related transcription factor) gene family, a class of molecules characterized by a signature zinc finger-like DNA-binding motif known as the DM domain (2).
Dmrt genes have been shown to regulate sexual development in arthropods, nematodes, and vertebrates and, as such, represent a rare example of genes whose function in sex regulation has been highly conserved during evolution (3). Dmrt1 was the first DM domain gene identified in vertebrates (4). In the mouse, Dmrt1 is expressed in the genital ridge, and upon sexual differentiation its expression decreases in the ovary and is maintained in the testis (4). Mice with a targeted deletion of Dmrt1 show normal development in XX individuals, whereas genetically male individuals have severely hypoplastic testes (5). In humans, DMRT1 maps to the short arm of chromosome 9, and hemizygous deletion of this region is associated with defective testicular development (6, 7). Dmrt1 has also been isolated in fish (8–13), amphibians (14), reptiles (15), and marsupials (16), and Dmrt1 expression pattern in these species is consistent with a role in testis differentiation.
There is accumulating evidence from different phyla that more than one Dmrt gene could be involved in sexual development. For example, Dmrt3, Dmrt5, and Dmrt7 also exhibit sexually dimorphic expression in the early embryonic gonad (17). Conversely, several Dmrt family members are expressed in multiple tissues in addition to the gonads, suggesting that Dmrt genes are not strictly involved in sexual development. For example, the zebrafish gene terra and its mouse homolog Dmrt2 both are expressed in the presomitic mesoderm and newly formed somites (18). Dmrt3 from chick and mouse (19) has similar expression domains in the forebrain, spinal cord, and nasal placode. However, the precise function of these genes in nongonadal tissues remains largely unknown.
Here, we analyze the expression and function of Xenopus Dmrt4 (XDmrt4). XDmrt4 is first detected in a domain within the preplacodal ectoderm and then becomes restricted to the forebrain and olfactory placode. Altered XDmrt4 function by injection of morpholino antisense oligonucleotide or an inhibitory mutant resulted in a specific inhibition of the olfactory placode expression of Xebf2, leading to impaired neurogenesis in the olfactory epithelium. We propose that XDmrt4 is a key regulator of neurogenesis in the olfactory system.
Materials and Methods
Isolation of XDmrt4 and Plasmid Constructions. Degenerate primers were designed within the DM domain (forward: TCSCCIMGSMYRCCSAAITGYGC; reverse: CYMARRGCIACYTGIGCIGCCAT) to amplify Dmrt genes from early neurula-stage Xenopus cDNA. PCR conditions were 30 s at 94°C, 40 s at 63°C, and 1 min at 72°C for 35 cycles. Several 160-bp products were isolated, cloned into pGEMTeasy (Promega), and sequenced. A second set of primers was generated (forward: CAACCATGGTGTCGTATCAGCC; reverse: TTTTGTCGCTGGAGGGACTGTTCA) to amplify the 3′ end by RACE (Clontech). This PCR product was used to screen a stage-17 lambda ZAPII cDNA library (a gift from Michael King, Indiana University, Terre Haute) to isolate a full-length clone. The sequence of Xenopus Dmrt4 has been deposited in the GenBank database (accession no. AY648303). The ORF of XDmrt4 cDNA was subcloned into pCS2+ expression plasmid. A mutated version of XDmrt4 (muXDmrt4) was generated by PCR. In this construct 6 bp were mutated 3′ of the ATG, (TATGGATTGTAAGTCCCCCAAACATG), within the recognition motif for the morpholino oligonucleotide (see below). These mutations did not affect the amino acid composition of XDmrt4 protein. An inhibitory mutant of XDmrt4 (XDmrt4ΔC), in which the C-terminal domain was deleted at amino acid 141, was generated by PCR and cloned into pCS2+. All constructs were sequenced and the corresponding proteins were monitored by using an in vitro transcription/translation-coupled rabbit reticulocyte lysate system (Promega) in the presence of [35S]methionine and resolved on a NuPAGE BIS-Tris gel (Invitrogen).
Xenopus Embryo Injections. Embryos were staged according to Nieuwkoop and Faber (20). Xwnt-3a (0.1 ng; ref. 21), Noggin (0.2 ng; ref. 22), fibroblast growth factor 8 (Fgf8) (0.1 ng; ref. 23), Xdkk1 (0.1 ng; ref. 2), β-gal (1 ng), XDmrt4 (1 ng), and XDmrt4ΔC(1ng) mRNAs were synthesized by using the Message Machine kit (Ambion, Austin, TX). For plasmid injections, a CS2+Xwnt-3a (0.1 ng) construct was used (25). XDmrt4 (CATGTTTGGGCTTTTGCAGTCCATG), β-catenin (26), and control morpholino oligonucleotides were obtained from Gene Tools, Philomath, OR. Synthetic mRNAs, plasmid DNA, or morpholinos were injected in one animal dorsal blastomere at the eight-cell stage to target the presumptive nasal placode (27). For animal explant experiments, both blastomeres of two-cell stage embryos were injected in the animal pole, and explants were dissected at the blastula stage, cultured in vitro, and analyzed by RT-PCR.
Lineage Tracing and Whole-Mount in Situ Hybridization. In all experiments, embryos were coinjected with β-gal mRNA to identify the manipulated side. Antisense digoxigenin-labeled probes (Genius kit, Roche Diagnostics) were synthesized by using template cDNA encoding XDmrt4, XBF-1 (28), Xebf2 (29), XDlx5 (30), XEmx2 (31), Six1 (32), Xebf3 (33), Slug (34), and Pax6 (35). Embryos at the appropriate stage were successively processed for Red-Gal (Research Organics) staining and in situ hybridization (36).
Analysis of Gene Expression by RT-PCR. Total RNAs extracted from adult tissues or animal caps were reverse-transcribed and amplified by using primers for XDmrt4, XBF1, and elongation factor 1 α (EF-1α) (see Supporting Text, which is published as supporting information on the PNAS web site) and the products were analyzed on agarose gel. For real-time RT-PCR, total RNAs from animal caps were extracted by using a RNeasy micro kit (Qiagen, Valencia, CA), and real-time RT-PCR (LightCycler, Roche Diagnostics) was performed by using specific primer sets to quantify neurogenin, Xebf2, neural cell adhesion molecule (NCAM), and EF-1α transcript levels (see Supporting Text).
Results
Cloning of Xenopus Dmrt4. We used degenerate PCR to isolate Dmrt family members from neurula-stage Xenopus embryos. Among the several PCR products obtained, one was identified as Dmrt4, based on its homology to the human (37) and murine (17, 38) Dmrt4 DM domains. A 660-bp product was subsequently isolated by RACE and used to screen a stage-17 Xenopus cDNA library. All positive clones isolated contained a 1,344-bp ORF encoding a predicted 448-aa protein (see Fig. 7A, which is published as supporting information on the PNAS web site). The longest clone also contains 209 bp of 5′ UTR and 230 bp of 3′ UTR. At the amino acid level this clone shares highest identity with human Dmrt4 (46%; ref. 37), mouse Dmrt4 (43%; refs. 17 and 38), platyfish Dmrt4 (43%; ref. 39), and Medaka Dmrt4 (40%; ref. 39). Based on these observations we named this gene XDmrt4 (see Fig. 7B). RT-PCR analysis of adult organs (see Fig. 7C) reveals abundant XDmrt4 transcripts in the brain and the testis, whereas XDmrt4 is undetectable in the lung, liver, skeletal muscle, or heart. This finding is consistent with the adult tissue distribution reported for other Dmrt family members across species (17).
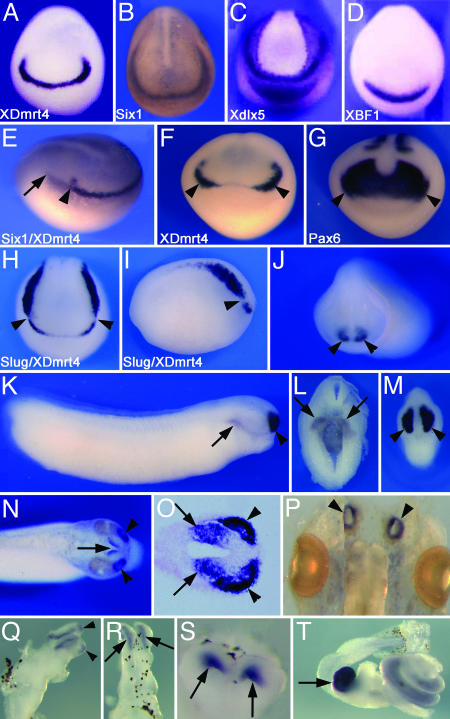
XDmrt4 Is Expressed in the Developing Olfactory System. In wholemount in situ hybridization XDmrt4 is first detected at the early neurula stage (stage 14) in the anterior-most region of the neural plate (Fig. 1A) in a subdomain of the preplacodal ectoderm known as the anterior neural ridge (40). Six1, a gene homologous to Drosophila sine oculis, is expressed in the entire preplacodal ectoderm (Fig. 1B and ref. 32). Double in situ hybridization using XDmrt4 and Six1 probes illustrates that XDmrt4 is expressed in a subdomain of the preplacodal ectoderm (Fig. 1E). Around stage 17, expression begins to fade at the midline but persists bilaterally in the prospective region of the olfactory placodes (Fig. 1F). The paired box gene Pax6 is expressed in the developing eye field and the prospective olfactory placode (35) and overlaps with XDmrt4 in the latter region (Fig. 1 F and G). Double in situ hybridization using the neural crest marker Slug (34) indicates that the posterior domain of expression of XDmrt4 abuts Slug anterior expression domain without any apparent overlap (Fig. 1 H and I). After neural tube closure XDmrt4 is strongly expressed in the olfactory placode and the forebrain (Fig. 1J). Around stage 29/30 XDmrt4 expression persists in the developing olfactory organ and forebrain and is also detected in the wall of the foregut (Fig. 1 K–M). At stage 35 the XDmrt4 expression domains in the olfactory organ and the telencephalon can be clearly identified (Fig. 1 N and O). At the swimming tadpole stage (stage 45), XDmrt4 is still detected in the olfactory epithelium (Fig. 1P), the olfactory bulb, and part of the telencephalon (Fig. 1 Q–S). XDmrt4 is also strongly expressed in the gall bladder and more diffusely throughout the intestine (Fig. 1T).
Fig. 1.
Developmental expression of XDmrt4 by whole-mount in situ hybridization. (A) XDmrt4 expression at the early neurula stage is detected at the anterior edge of the neural plate. (B–D) Six1 (B), Xdlx5 (C), and XBF1 (D) expression at a similar stage are shown for comparison. (E) Early neurula stage embryo stained with both XDmrt4 and Six1 probes show that XDmrt4 is expressed in a subdomain of the preplacodal ectoderm. The arrowhead indicates the posterior boundary of XDmrt4, and the arrow marks the posterior domain of Six1. (F and G) Comparison of XDmrt4 (F) and Pax6 (G) expression in sibling embryos illustrates that both genes are expressed in the presumptive olfactory placode (arrowheads). (H and I) Double in situ hybridization showing the relationship of the expression domain of the neural crest-specific gene Slug and XDmrt4 at stage 17. The arrowheads indicate the posterior and anterior boundary of XDmrt4 and Slug, respectively. Views are dorsal-anterior (A–D and F–H) and lateral (E and I, anterior to right). (J) At stage 23, XDmrt4 is detected bilaterally in the prospective nasal placode (arrowheads), anterior view. (K–M) By stages 29/30 XDmrt4 is strongly detected in the developing olfactory organ and the telencephalon (arrowheads). At this stage XDmrt4 is also detected in the foregut (arrows, K and L). Lateral (K) and anterior (M) views, dorsal to top. (L) Transverse section. (N and O) In stage-35 embryos XDmrt4 expression domains in the telencephalon (arrows) and the olfactory organ (arrowheads) are segregated. (N) Dorsal view, anterior to right. (O) Longitudinal section through the olfactory epithelium and the telencephalon. (P–S) At stage 45, XDmrt4 expression persists in the olfactory epithelium (arrowheads, P), the olfactory bulb (arrowheads, Q), and forebrain (arrows, R and S). Whole-brain lateral (Q) and dorsal (R) views. (S) Transverse section through the forebrain. (T) XDmrt4 is also strongly expressed in the gall bladder (arrow) and more diffusely throughout the intestine. (Magnifications: ×10 in A–D, H, and I; ×12 in E, F, and J–N; ×20 in O and S; and ×15 in P, Q, and T.)
Several genes are expressed in the developing olfactory placodes (41). Among these are the winged helix transcription factor XBF1 (28), the distal-less related homeobox gene XDlx5 (30), a gene related to Drosophila empty spiracles, XEmx2 (31), and the helix–loop–helix factor Xebf2 (29). At the neurula stage XDlx5 and XBF1 are also detected in the anterior neural ridge (Fig. 1 C and D). However, XBF1 expression appears broader than that of XDmrt4 at the anterior neural plate (Fig. 1 A and D), probably reflecting its expression in most of the prospective dorsal telencephalon (28). XDlx5 expression extends more posteriorly around the entire neural plate and shows expression in the prospective cement gland (Fig. 1C and ref. 30). The expression of Xebf2 and XEmx2 in the presumptive olfactory placode is weak at the neurula stage; however, expression becomes more prominent around stage 20 (data not shown and refs. 29 and 33).
We examined sections of stage-35 embryos stained by wholemount in situ hybridization to further analyze the forebrain and placodal expression domains of these marker genes. XDmrt4, XBF1, and XEmx2 are expressed in the olfactory placode and telencephalon. Whereas XBF1 expression in the forebrain includes the entire dorsal telencephalon, XEmx2 is excluded from the most anterior tip of the dorsal telencephalon and XDmrt4 localizes to the posterior half of the dorsal telencephalon (see Fig. 8, which is published as supporting information on the PNAS web site). Xebf2 is expressed in the olfactory placode and appears to be excluded from the telencephalon (see Fig. 8); however, it is also detected in the mesencephalon and the rhombencephalon (data not shown and ref. 29).
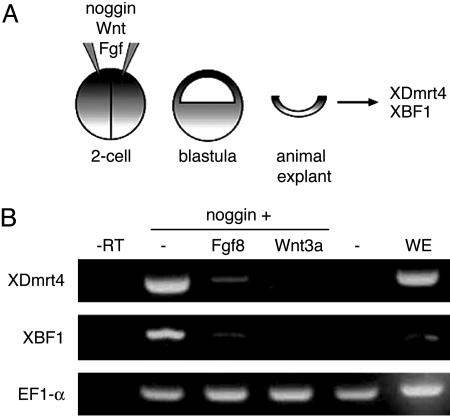
XDmrt4 Is Induced by Noggin and Repressed by Wnt and Fgf. XDmrt4 is expressed in the anterior region of the neural plate, suggesting that XDmrt4 is induced in response to neural-inducing signals. To test this possibility, blastula-stage animal explants were injected with mRNA encoding the neural inducer Noggin (22) and analyzed by RT-PCR at equivalent stage 17 (Fig. 2A). Noggin-injected explants showed strong activation of XDmrt4 and XBF1 (Fig. 2B) consistent with the view that induction of anterior fates requires inhibition of Bmp signaling in the ectoderm. Wnt and Fgf signaling have been shown to suppress anterior neural fate to generate posterior neural structures (42), and coninjection of Wnt-3a or Fgf8 mRNAs blocked Noggin-mediated induction of XDmrt4 and XBF1 in these explants (Fig. 2B). These results suggest that a balance of both neuralizing and caudalizing signals is likely to be involved in defining the XDmrt4 expression domain in vivo.
Fig. 2.
Signaling pathways involved in inducing XDmrt4. (A) Two-cell stage embryos are injected in both blastomeres with a combination of Noggin, Wnt3a, and Fgf8 mRNAs. Animal explants are dissected at the blastula stage, cultured until neurula stage, and analyzed by RT-PCR for expression of XDmrt4 and XBF1. (B) Noggin induces strong expression of XDmrt4 and XBF1. Coinjection of Fgf8 or Wnt3a severely reduces Noggin-mediated XDmrt4 and XBF1 induction. Controls include uninjected animal explant (–), whole embryo (WE) at stage 17, and reaction minus reverse transcriptase (–RT). Elongation factor 1α is used as a loading control.
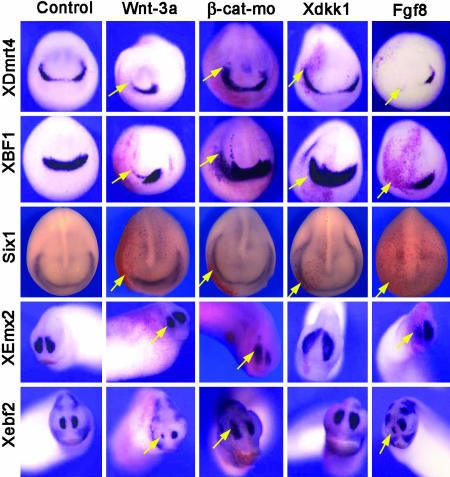
Anterior Expression of XDmrt4 Requires Inhibition of Caudalizing Factors. The exclusion of Wnt signaling from the anterior neural plate is required to establish anterior neural character (42). To investigate whether Wnt signaling functions in restricting XDmrt4 to the anterior neural ridge, we analyzed XDmrt4 expression in embryos in which Wnt signaling was experimentally manipulated. Embryos injected with Wnt-3a expression plasmid (used in lieu of RNA to avoid axis duplication) show a dramatic reduction of XDmrt4 (70% of the embryos; n = 49) on the injected side (Fig. 3). A reduction in the expression of XBF1 (71%, n = 63), Six1 (100%, n = 26), Xebf2 (58%, n = 36), and XEmx2 (85%, n = 39) was also observed in these embryos (Fig. 3). β-Catenin is a downstream component of the canonical Wnt pathway. Blockage of Wnt signaling by injection of a β-catenin morpholino in one animal dorsal blastomere at the eight-cell stage resulted in an enlargement and a posterior expansion of the XDmrt4 expression domain (Fig. 3) in 69% of the embryos (n = 77). A posterior expansion of Six1 (80%, n = 51) and XBF1 (92%, n = 12) was also observed in these embryos, which correlates with an enlargement of the olfactory placode expression domain of Xebf2 (47%, n = 114) and XEmx2 (68%, n = 41) at the tail bud stage (Fig. 3). Similarly, overexpression of Dickkopf1 (Xdkk1), a secreted Wnt inhibitor expressed in the head region (24), resulted in embryos with a dramatic expansion of all five marker genes, usually on both sides of the injected embryos (Fig. 3), presumably because of Xdkk1 protein diffusion.
Fig. 3.
Caudalizing factors restrict XDmrt4 expression domain. Activation of Wnt signaling pathway by injection of Wnt-3a blocks XDmrt4, XBF1, and Six1 expression at the neurula stage. Conversely, inhibition of Wnt signaling pathway by injection of β-catenin morpholino (β-cat-mo) or Xdkk1 mRNA expands posteriorly XDmrt4, XBF1, and Six1 placodal expression domains. At the tail bud stage, the same manipulations of the Wnt signaling pathway resulted in a reduction or an expansion of Xebf2 and XEmx2 expression in the nasal placode. In most cases, Xdkk1 injection leads to an expansion of these markers on both sides of the injected embryos presumably because of Xdkk1 protein diffusion. Overexpression of Fgf8 completely blocks XDmrt4, XBF1, and Six1 placodal expression at the neurula stage, as well as the nasal placode expression of Xebf2 and XEmx2 at the tail bud stage. Anterior views are shown, dorsal to the top. RNA encoding the lineage tracer β-gal was coinjected in all cases to identify the injected side (red staining, left, arrows). (Magnifications: ×8.)
Other factors involved in establishing anterior-posterior patterning in the neuroectoderm include Fgf and retinoic acid (RA) (42). Injection of Fgf8 mRNA resulted in a reduction of XDmrt4 (83%, n = 123), XBF1 (90%, n = 102), and Six1 (100%, n = 32) expression at stage 15 (Fig. 3), as well as a reduction of Xebf2 (82%, n = 28) and XEmx2 (98%, n = 41) at stage 25. Similarly, exposure to RA (see Supporting Text) led to a complete loss of XDmrt4 (see Fig. 9, which is published as supporting information on the PNAS web site). Altogether, these results indicate that XDmrt4 is induced in the anterior neural ridge by neural inducers (Bmp antagonists) and that a proper balance of caudalizing factors (Wnt, Fgf, and RA) is required for restricting the XDmrt4 expression domain anteriorly.
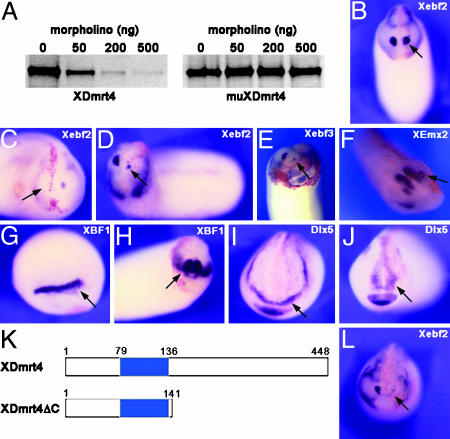
XDmrt4 Is Required for Xebf2 Expression in the Olfactory Placode. To determine whether XDmrt4 is required for olfactory placode development, we used a morpholino antisense oligonucleotide (XDmrt4-mo) designed to interfere with XDmrt4 translation. The specificity of XDmrt4-mo was tested in an in vitro transcription/translation reaction directed by WT XDmrt4 and muXDmrt4 (a construct that has 6-bp mutations within the morpholino recognition motif). Increasing amounts of morpholino can completely block translation of XDmrt4 mRNA in this assay, but failed to interfere with translation of muXDmrt4 mRNA (Fig. 4A).
Fig. 4.
XDmrt4-mo and XDmrt4ΔC specifically blocks Xebf2 expression. (A) In vitro-coupled transcription/translation reactions with plasmid encoding WT XDmrt4 and a mutated version of XDmrt4 (muXDmrt4) in the presence of increasing amounts of XDmrt4-mo. (B) Embryo injected in with a control morpholino shows normal expression of Xebf2 on the injected side. (C and D) Injection of 20 ng of XDmrt4-mo in one blastomere at the eight-cell stage results in a reduction or loss of Xebf2 on the injected side. (E) Another Olf/Ebf family member, Xebf3, is also reduced upon XDmrt4-mo injection. (F) However, the olfactory placode marker, XEmx2, remains unaffected under these conditions. (G–J) XBF1 (G and H) and Dlx5 (I and J) expression at stage 17 (G and I) or stage 25 (H and J) are unperturbed in XDmrt4-mo-injected embryos. (K) Schematic representation of WT and XDmrt4 deletion construct (XDmrt4ΔC). The blue box indicates the position of the DM domain. (L) Injection of 1 ng of XDmrt4ΔC mRNA results in a loss of the Xebf2 expression domain. (B–J and L) RNA encoding the lineage tracer β-gal was coinjected in all cases to identify the injected side (red staining, arrows). Anterior views are shown, dorsal to the top. (Magnifications: ×10.)
Upon injection of 20–40 ng of XDmrt4-mo in one animal dorsal blastomere at the eight-cell stage, 40% of the embryos (n = 163) showed a dramatic reduction or a complete loss of Xebf2 expression at stages 25–28 (Fig. 4 C and D and see Table 1, which is published as supporting information on the PNAS web site). Xebf3, another member of the Olf-1/Ebf family of helix–loop–helix transcription factors (33), was also reduced in 48% (n = 40) of XDmrt4-moinjected embryos (Fig. 4E). By contrast, XEmx2 expression was unperturbed in these embryos (Fig. 4F), as was the early (stage 14/17) and late (stage 25) expression of XBF1 and Dlx5 (Fig. 4 G–J). Similarly, Six1 expression was unaffected in these embryos (data not shown; 98%, n = 83). Importantly, injection of a control morpholino at the same concentration had no effect on the expression of Xebf2 (Fig. 4B and Table 1).
Attempts to rescue the Xebf2 phenotype by injection of an XDmrt4 expression plasmid or mRNA with a mutated morpholino recognition motif (muXDmrt4) were unsuccessful, presumably because sufficient levels of XDmrt4 proteins could not be obtained under these conditions without compromising the embryos survival. However, to further validate the phenotype of XDmrt4-depleted embryos we generated a truncated version of XDmrt4, lacking the C-terminal region (Fig. 4K, XDmrt4ΔC). This construct, which retains the DNA binding (DM) domain, is predicted to block XDmrt4 function, and possibly other Dmrt proteins, by acting as a dominant negative (see Fig. 6A). Embryos injected with XDmrt4ΔC mRNA displayed a strong reduction of Xebf2 expression at stages 25–28 (Fig. 4L and Table 1) without affecting the expression of other early and late placodal markers (data not shown), thereby mimicking the phenotype of morpholino-treated embryos. Because XDmrt4 morpholino and the XDmrt4ΔC inhibitory mutant both specifically interfere with Xebf2 expression, a well established regulator of neurogenesis (43), we predict that XDmrt4 is likely to function in the differentiation of the placode into an olfactory epithelium.
Fig. 6.
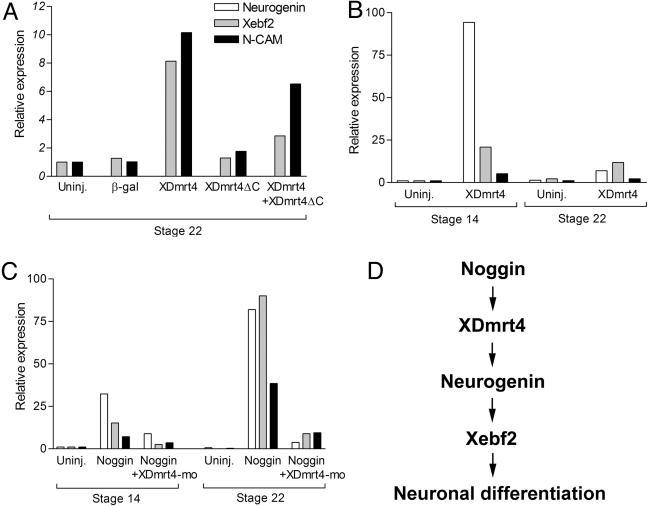
Regulation of neurogenin, Xebf2, and NCAM expression by XDmrt4. (A–C) Real-time RT-PCR of animal explants isolated from embryos injected with various combinations of mRNA and morpholinos as indicated and collected when sibling embryos reached stages 14 or 22. (A) Injection of XDmrt4 induces Xeb2 and NCAM expression. This activity is inhibited by coinjection of XDmrt4ΔC. (B) XDmrt4 induces robust expression of neurogenin at stage 14 and to a lesser extend at stage 22. (C) Noggin-mediated activation of neurogenin, Xebf2, and NCAM expression is blocked in the context of XDmrt4-depeleted embryos (+XDmrt4-mo). (D) Based on these findings and other studies that have positioned Xebf2 downstream of neurogenin (29) we propose that XDmrt4 is an upstream regulator of neurogenin and Xebf2 in the molecular cascade, leading to neuronal differentiation in the olfactory system.
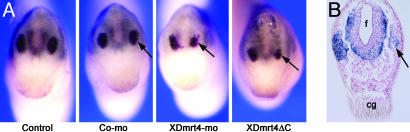
XDmrt4 Is Required for Neurogenesis in the Olfactory Epithelium. To further evaluate the consequences of compromised XDmrt4 function on neurogenesis in the olfactory system, embryos injected with either XDmrt4-mo or XDmrt4ΔC mRNA were analyzed for expression of the neuronal marker NCAM by immunocytochemistry at stage 35 (see Supporting Text). Both experimental manipulations of XDmrt4 resulted in embryos with reduced NCAM expression within the olfactory epithelium (Fig. 5), implying that the differentiation of olfactory neurons was impaired in these embryos. Injections of a control morpholino or a control mRNA (β-gal) had no effect on the level of NCAM (Fig. 5A and Table 1). Importantly, we found a strong correlation between the incidence of this phenotype at stage 35 (reduced NCAM expression) and the number of embryos that showed reduced Xebf2 expression at stages 25–28 (see Table 1), suggesting a link between XDmrt4, Xebf2, and active neurogenesis in the olfactory epithelium.
Fig. 5.
Impaired neurogenesis in the olfactory epithelium of embryos lacking XDmrt4 function. (A) XDmrt4-mo or XDmrt4ΔC mRNA-injected embryos show a marked reduction in NCAM expression in the olfactory epithelium (arrows). Injection of a control morpholino (Co-mo) has no effect on NCAM expression levels. Anterior views are shown, dorsal to the top. (B) Transverse section through the head of an XDmrt4-mo-injected embryo stained for NCAM shows a reduced number of neurons in the olfactory epithelium on the injected side (arrow). cg, cement gland; f, forebrain. (Magnifications: ×15.)
XDmrt4 Is Sufficient to Promote Neurogenesis in Animal Explants. To determine whether XDmrt4 is sufficient to activate Xebf2 expression, embryos at the two-cell stage were injected in the animal pole region with XDmrt4, XDmrt4ΔC, or a combination of both XDmrt4+XDmrt4ΔC mRNAs. Animal explants were dissected at the blastula stage, cultured until stage 22, and analyzed by real-time RT-PCR for Xebf2 and NCAM expression. Strong induction of Xebf2 and NCAM was observed in XDmrt4-injected animal explants as compared with explants injected with XDmrt4ΔC or a control mRNA encoding β-gal (Fig. 6A). These results provide further evidence that Xebf2 and XDmrt4 are acting in the same pathway to promote neurogenesis. XDmrt4-mediated induction of Xebf2 and NCAM was inhibited by coinjection of XDmrt4ΔC, indicating that XDmrt4ΔC acts as a dominant interfering mutant for XDmrt4 function (Fig. 6A).
The basic helix–loop–helix factor neurogenin is an early neuronal determination gene in vertebrate (44). To further evaluate XDmrt4 neurogenic activity in animal explants we analyzed neurogenin expression in explants injected with XDmrt4 mRNA and collected at the early neurula stage (stage 14) or tailbud stage (stage 22). XDmrt4 induces strong expression of neurogenin at stage 14 and to a lesser extend at stage 22, thereby positioning XDmrt4 as a key regulator of neurogenesis in Xenopus (Fig. 6B). As Noggin is a strong inducer of XDmrt4 in animal explants (Fig. 2B), we tested whether Noggin-mediated neuralization depended on XDmrt4 function. Injection of Noggin mRNA induces strong expression of neurogenin, Xebf2, and NCAM in explants collected at stages 14 or 22; however, in the context of XDmrt4-depleted animal explants the induction of these genes by Noggin was strongly inhibited (Fig. 6C), suggesting that XDmrt4 function is required upstream of neurogenin and Xebf2 for Noggin-mediated neurogenesis.
Discussion
The results presented here strongly argue for a critical role of XDmrt4 in the development of the olfactory system in Xenopus. XDmrt4 is initially expressed in the anterior neural ridge and later becomes restricted to part of the developing telencephalon and the olfactory placode/epithelium where it promotes neurogenesis by regulating neurogenin and Xebf2 expression. Although Dmrt genes are well established regulators of sexual development, we demonstrate here a function for a Dmrt family member in nongonadal tissue.
The anterior neural ridge defines a subdomain of the preplacodal ectoderm at the rostral boundary of the neural plate, and the prospective olfactory placode and some forebrain tissues will develop from this region (40). At the early neurula stage XDmrt4 expression domain is restricted to the anterior neural ridge and abuts but does not overlap with the anterior domain of the cranial neural crest. As the neural tube closes, XDmrt4 expression is down-regulated at the midline and persists bilaterally in a region that eventually will segregate into olfactory placodes and part of the dorsal telencephalon. In the fish Medaka, Dmrt4 is also detected in the olfactory placode and the dorsal telencephalon (45). The expression pattern of Dmrt4 has not yet been described in other organisms to our knowledge. However, in both mouse and chick, Dmrt3 appears to be expressed in the telencephalon and the nasal pits (19). Interestingly, whereas Dmrt3 genes have additional expression domains in the neural tube of mouse, chick, and Medaka, the chicken Dmrt3 gene is also expressed in the presomitic mesoderm (19). This is not the case in Medaka in which the only Dmrt gene expressed in the presomitic mesoderm is Dmrt2, similarly to its zebrafish (18) and human (46) orthologs. These divergences in the expression of Dmrts across species illustrate a remarkable shift in expression pattern of this gene family during evolution.
Little is known about the signaling factors involved in olfactory placodes induction. However, given that these placodes derive from the anterior neural ridge, signals that induce forebrain are likely to be important for olfactory placodes induction. Conversely, factors repressing forebrain fate are expected to block olfactory placodes formation (47). Our results indicate that attenuation of Bmp signaling in the ectoderm and anterior activity of Wnt, Fgf, and RA antagonists (42) is likely to be involved in inducing and restricting XDmrt4 expression to the anterior neural ridge. The neural crest is found lateral to the neural plate but excluded from its most anterior region. Interestingly, the set of signals involved in restricting XDmrt4 anteriorly has also been implicated in the induction of the neural crest at the lateral neural plate border (48). By looking at the expression of the preplacodal gene Six1 and the neural crest marker FoxD3, a recent study indicates that these signaling pathways have opposite effects on placodal tissue and neural crest formation. Whereas Wnt and Fgf promote neural crest over placodal fate, inhibition of these signaling pathways favors placodal at the expense of neural crest fate (49). Therefore, the relative levels of these factors may influence cells at the neural plate border to become placode or neural crest cells.
Interference with XDmrt4 function by injection of a morpholino oligonucleotide or an inhibitory mutant resulted in a similar phenotype, the specific reduction/loss of the olfactory expression of Xebf2, suggesting a direct relationship between XDmrt4 and Xebf2 expression in the olfactory epithelium. Although Six1, XBF1, XDlx5, and XDmrt4 are coexpressed in the anterior neural ridge, loss of XDmrt4 did not affect the early or late expression of these genes, suggesting that XDmrt4 may act downstream of Six1, XBF1, and XDIx5 or that these factors are functioning in different pathways during olfactory placode development.
Although XDmrt4 is detected in the presumptive olfactory placode it appears to be dispensable for olfactory placode induction but required for the later development of the olfactory system. Compensatory activity of other Dmrt family members could account for this lack of an early phenotype in XDmrt4-deficient animals. Alternatively, this late phenotype may reflect the true function of this class of molecules. In that respect, there is an interesting parallel between our results and Dmrt1 function in the gonad. In mammals, Dmrt1 is expressed in the indifferent gonad; however, early events in testis differentiation occur normally in the knockout male mice (which have defects only in postnatal testis differentiation), indicating that Dmrt1 may not play a major early role in testis development in mammals (5).
Xebf2 belongs to the Olf/Ebf family of helix–loop–helix transcription factors (43). Both gain-of-function (29, 33) and loss-of-function (50–53) obtained in various model organisms have implicated Olf/Ebf genes in key stages of neural development (43). The loss of Xebf2 expression in Dmrt4-deficient embryos correlates with reduced NCAM expression in the olfactory epithelium, suggesting that Xebf2 controls neurogenesis in the olfactory system downstream of XDmrt4. Further analysis will be needed to determine whether these factors regulate the production, differentiation, and/or survival of olfactory neurons and whether other cell types within the olfactory epithelium (basal cells or support cells) are also affected.
The observation that XDmrt4 is sufficient to activate neurogenin, Xebf2, and NCAM expression in animal explants further supports the view that these factors are acting in the same pathway to promote neurogenesis in the olfactory epithelium. Moreover, Noggin-mediated induction of neurogenin, Xebf2, and NCAM is blocked in the absence of XDmrt4 function, further demonstrating that XDmrt4 is a key regulator of neurogenesis in the vertebrate embryo. Based on these findings and other studies that positioned Xebf2 downstream of neurogenin (29), we propose that XDmrt4 is an upstream regulator of neurogenin and Xebf2 in the molecular cascade leading to neuronal differentiation (Fig. 6D). Future studies should be directed at understanding the nature of the regulation of neurogenin and Xebf2 by XDmrt4 and defining the mechanism by which these factors control olfactory neuron development.
Supplementary Material
Acknowledgments
We thank Trish Labosky for comments on the manuscript; Christine Credidio for technical assistance; and Drs. Andrei Brandli (Swiss Federal Institute of Technology, Zurich), Giacomo Consalez (Scientific Institute San Raffaele, Milan), Xi He (Harvard Medical School, Boston), Michael King, Nancy Papalopulu (Wellcome Trust/Cancer Research UK Gurdon Institute, Cambridge), Thomas Sargent (National Institutes of Health, Bethesda), Monica Vetter (University of Utah, Salt Lake City), and Robert Vignali (University of Pisa, Pisa, Italy) for reagents. This work was supported by National Institutes of Health Grant DE14212.
Author contributions: X.H., C.-S.H., M.O., and J.-P.S.-J. designed research; X.H., C.-S.H., M.O., and J.-P.S.-J. performed research; X.H., C.-S.H., M.O., and J.-P.S.-J. analyzed data; and X.H., C.-S.H., and J.-P.S.-J. wrote the paper.
Abbreviations: Dmrt, doublesex and mab-3-related transcription factor; XDmrt4, Xenopus Dmrt4; Fgf, fibroblast growth factor; NCAM, neural cell adhesion molecule; RA, retinoic acid.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY648303).
References
- 1.Volff, J. N., Zarkower, D., Bardwell, V. J. & Schartl, M. (2003) J. Mol. Evol. 57, Suppl. 1, 241–249. [DOI] [PubMed] [Google Scholar]
- 2.Raymond, C. S., Shamu, C. E., Shen, M. M., Seifert, K. J., Hirsch, B., Hodgkin, J. & Zarkower, D. (1998) Nature 391, 691–695. [DOI] [PubMed] [Google Scholar]
- 3.Hodgkin, J. (2002) Genes Dev. 16, 2322–2326. [DOI] [PubMed] [Google Scholar]
- 4.Raymond, C. S., Kedttlewell, J. R., Hirsch, B., Bardwell, V. J. & Zarkower, D. (1999a) Dev. Biol. 215, 208–220. [DOI] [PubMed] [Google Scholar]
- 5.Raymond, C. S., Murphy, M. W., O'Sullivan, M. G., Bardwell, V. J. & Zarkower, D. (2000) Genes Dev. 14, 2587–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Veitia, R. A., Nunes, M., Quintana-Murci, L., Rappaport, R., Thibaud, E., Jaubert, F., Fellous, M., McElreavey, K., Goncalves, J., Silva, M., et al. (1998) Am. J. Hum. Genet. 63, 901–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raymond, C. S., Parker, E. D., Kedttlewell, J. R., Brown, L. G., Page, D. C., Kusz, K., Jaruzelska, J., Reinberg, Y., Flejter, W., Bardwell, V. J., et al. (1999) Hum. Mol. Genet. 8, 989–996. [DOI] [PubMed] [Google Scholar]
- 8.Marchand, O., Govoroun, M., D'Cotta, H., McMeel, O., Lareyre, J., Bernot, A., Laudet, V. & Guiguen, Y. (2000) Biochim. Biophys. Acta 1493, 180–187. [DOI] [PubMed] [Google Scholar]
- 9.Brunner, B., Hornung, U., Shan, Z., Nanda, I., Kondo, M., Zend-Ajusch, E., Haaf, T., Ropers, H. H., Shima, A., Schmid, M., et al. (2001) Genomics 77, 8–17. [DOI] [PubMed] [Google Scholar]
- 10.Guan, G., Kobayashi, T. & Nagahama, Y. (2000) Biochem. Biophys. Res. Commun. 272, 662–666. [DOI] [PubMed] [Google Scholar]
- 11.Nanda, I., Kondo, M., Hornung, U., Asakawa, S., Winkler, C., Shimizu, A., Shan, Z., Haaf, T., Shimizu, N., Shima, A., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 11778–11783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuda, M., Sato, T., Toyazaki, Y., Nagahama, Y., Hamaguchi, S. & Sakaizumi, M. (2003) Zool. Sci. 20, 159–161. [DOI] [PubMed] [Google Scholar]
- 13.Veith, A. M., Froschauer, A., Korting, C., Nanda, I., Hanel, R., Schmid, M., Schartl, M. & Volff, J. N. (2003) Gene 317, 59–66. [DOI] [PubMed] [Google Scholar]
- 14.Shibata, K., Takase, M. & Nakamura, M. (2002) Gen. Comp. Endocrinol. 127, 232–241. [DOI] [PubMed] [Google Scholar]
- 15.Kettlewell, J. R., Raymond, C. S. & Zarkower, D. (2000) Genesis 26, 174–178. [PubMed] [Google Scholar]
- 16.Pask, A. J., Behringer, R. R. & Renfree, M. B. (2003) Cytogenet. Genome Res. 101, 229–236. [DOI] [PubMed] [Google Scholar]
- 17.Kim, S., Kettlewell, J. R., Anderson, R. C., Bardwell, V. J. & Zarkower, D. (2003) Gene Expression Patterns 3, 77–82. [DOI] [PubMed] [Google Scholar]
- 18.Meng, A., Moore, B., Tang, H., Yuan, B. & Lin, S. (1999) Development (Cambridge, U.K.) 126, 1259–1268. [DOI] [PubMed] [Google Scholar]
- 19.Smith, C. A., Hurley, T. M., McClive, P. J. & Sinclair, A. H. (2002) Gene Expression Patterns 2, 69–72. [DOI] [PubMed] [Google Scholar]
- 20.Nieuwkoop, P. D. & Faber, J. (1967) Normal Table of Xenopus laevis (Daudin) (North–Holland, Amsterdam).
- 21.Wolda, S. L., Moody, C. J. & Moon, R. T. (1993) Dev. Biol. 155, 46–57. [DOI] [PubMed] [Google Scholar]
- 22.Smith, W. C. & Harland, R. M. (1992) Cell 70, 29–40. [DOI] [PubMed] [Google Scholar]
- 23.Christen, B. & Slack, J. M. (1997) Dev. Biol. 192, 455–466. [DOI] [PubMed] [Google Scholar]
- 24.Glinka, A., Wu, W., Delius, H., Monaghan, A. P., Blumenstock, C. & Niehrs, C. (1998) Nature 391, 357–362. [DOI] [PubMed] [Google Scholar]
- 25.Saint-Jeannet, J. P., He, X., Varmus, H. E. & Dawid, I. B. (1997) Proc. Natl. Acad. Sci. USA 94, 13713–13718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heasman, J., Kofron, M. & Wylie, C. C. (2000) Dev. Biol. 222, 124–134. [DOI] [PubMed] [Google Scholar]
- 27.Moody, S. A. (1987) Dev. Biol. 119, 560–578. [DOI] [PubMed] [Google Scholar]
- 28.Papalopulu, N. & Kintner, C. (1996) Development (Cambridge, U.K.) 122, 3409–3418. [DOI] [PubMed] [Google Scholar]
- 29.Dubois, L., Bally-Cuif, L., Crozatier, M., Moreau, J., Paquereau, L. & Vincent, A. (1998) Curr. Biol. 8, 199–209. [DOI] [PubMed] [Google Scholar]
- 30.Papalopulu, N. & Kintner, C. (1993) Development (Cambridge, U.K.) 117, 961–975. [DOI] [PubMed] [Google Scholar]
- 31.Pannese, M., Lupo, G., Kablar, B., Boncinelli, E., Barsacchi, G. & Vignali, R. (1998) Mech. Dev. 73, 73–83. [DOI] [PubMed] [Google Scholar]
- 32.Ghanbari, H., Seo, H.-C., Fjose, A. & Brandli, A. W. (2001) Mech. Dev. 101, 271–277. [DOI] [PubMed] [Google Scholar]
- 33.Pozzoli, O. A., Croci, L., Consalez, G. G. & Vetter, M. L. (2001) Dev. Biol. 233, 495–512. [DOI] [PubMed] [Google Scholar]
- 34.Mayor, R., Morgan, R. & Sargent, M. G. (1995) Development (Cambridge, U.K.) 121, 767–777. [DOI] [PubMed] [Google Scholar]
- 35.Hirsch, N. & Harris, W. A. (1997) J. Neurobiol. 32, 45–61. [PubMed] [Google Scholar]
- 36.Harland, R. M. (1991) Methods Cell Biol. 36, 685–695. [DOI] [PubMed] [Google Scholar]
- 37.Ottolenghi, C., Fellous, M., Barbieri, M. & McElreavey, K. (2002) Genomics 79, 333–343. [DOI] [PubMed] [Google Scholar]
- 38.Carninci, P., Shibata, Y., Hayatsu, N., Sugahara, Y., Shibata, K., Itoh, M., Konno, H., Okazaki, Y., Muramatsu, M. & Hayashizaki, Y. (2000) Genome Res. 10, 1617–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kondo, M., Froschauer, A., Kitano, A., Nanda, I., Hornung, U., Volff, J. N., Asakawa, S., Mitani, H., Naruse, K., Tanaka, M., et al. (2002) Gene 295, 213–222. [DOI] [PubMed] [Google Scholar]
- 40.Eagleson, G., Ferreiro, B. & Harris, W. A. (1995) J. Neurobiol. 28, 146–158. [DOI] [PubMed] [Google Scholar]
- 41.Schlosser, G. & Ahrens, K. (2004) Dev. Biol. 271, 439–466. [DOI] [PubMed] [Google Scholar]
- 42.Gamse, J. & Sive, H. (2000) BioEssays 22, 976–986. [DOI] [PubMed] [Google Scholar]
- 43.Dubois, L. & Vincent, A. (2001) Mech. Dev. 108, 3–12. [DOI] [PubMed] [Google Scholar]
- 44.Ma, Q., Kintner, C. & Anderson, D. J. (1996) Cell 87, 43–52. [DOI] [PubMed] [Google Scholar]
- 45.Winkler, C., Hornung, U., Kondo, M., Neuner, C., Duschl, J., Shima, A. & Schartl, M. (2004) Mech. Dev. 121, 997–1005. [DOI] [PubMed] [Google Scholar]
- 46.Ottolenghi, C., Veita, R., Barbieri, M., Fellous, M. & McElreavey, K. (2000) Genomics 64, 179–186. [DOI] [PubMed] [Google Scholar]
- 47.Baker, C. V. H. & Bronner-Fraser, M. (2001) Dev. Biol. 232, 1–61. [DOI] [PubMed] [Google Scholar]
- 48.Huang, X. & Saint-Jeannet, J.-P. (2004) Dev. Biol. 275, 1–11. [DOI] [PubMed] [Google Scholar]
- 49.Brugmann, S. A., Pandur, P. D., Kenyon, K. L., Pignoni, F. & Moody, S. A. (2004) Development (Cambridge, U.K.) 131, 5871–5881. [DOI] [PubMed] [Google Scholar]
- 50.Garel, S., Marin, F., Grosschedl, R. & Charnay, P. (1999) Development (Cambridge, U.K.) 126, 5285–5294. [DOI] [PubMed] [Google Scholar]
- 51.Prasad, B. C., Ye, B., Zackhary, R., Schrader, K., Seydoux, G. & Reed, R. R. (1998) Development (Cambridge, U.K.) 125, 1561–1568. [DOI] [PubMed] [Google Scholar]
- 52.Corradi, A., Croci, L., Broccoli, V., Zecchini, S., Previtali, S., Wurst, W., Amadio, S., Maggi, R., Quattrini, A. & Consalez, G. G. (2003) Development (Cambridge, U.K.) 130, 401–410. [DOI] [PubMed] [Google Scholar]
- 53.Wang, S. S., Lewcock, J. W., Feinstein, P., Mombaerts, P. & Reed, R. R. (2003) Development (Cambridge, U.K.) 131, 1377–1388. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.